Abstract
We show that an inducible rpoS antisense RNA complementary to the rpoS message can inhibit expression of RpoS in both exponential and stationary phases and can attenuate expression of the rpoS regulon in Escherichia coli. Plasmids containing rpoS antisense DNA expressed under the control of the T7lac promoter and T7 RNA polymerase were constructed, and expression of the rpoS antisense RNA was optimized in the pET expression system. rpoS antisense RNA levels could be manipulated to effectively control the expression of RpoS and RpoS-dependent genes. RpoS expression was inhibited by the expression of rpoS antisense RNA in both exponential and stationary phases in E. coli. RpoS-dependent catalase HPII was also downregulated, as determined by catalase activity assays and with native polyacrylamide gels stained for catalase. Induced RpoS antisense expression also reduced the level of RpoS-dependent glycogen synthesis. These results demonstrate that controlled expression of antisense RNA can be used to attenuate expression of a regulator required for the expression of host adaptation functions and may offer a basis for designing effective antimicrobial agents.
Naturally occurring antisense RNA can be an important regulator of gene expression in eukaryotic cells (53) and bacterial cells (35) by either blocking ribosome binding (13) or reducing mRNA stability (14, 16). Antisense RNA technology has successfully been used to manipulate gene expression in bacteria (17, 18, 38; for a review, see reference 56). For example, mar antisense RNA can inhibit expression of the multiple-antibiotic resistance (mar) operon in Escherichia coli (58), thus increasing the sensitivity of the cell to antibiotics. Expression of an hla antisense RNA inhibits alpha-toxin production in Staphylococcus aureus and thereby attenuates virulence (24). Antisense RNAs have also been used to manipulate metabolism in Clostridium acetobutylicum (14), Enterococcus faecalis (55), and Penicillium chrysogenum (60). Selectively expressed antisense RNA can effectively inhibit the growth of bacteriophage (51, 57).
RpoS (σs) is an alternative sigma subunit of bacterial RNA polymerase (23, 54). In response to environmental stress and nutrient starvation, RpoS mediates increases in the levels of expression of many genes (19). RpoS-dependent genes such as katE, which encodes catalase HPII, are induced to help the cell survive in stationary phase and during entry into stationary phase (33, 36, 44). Expression of the monocistronic gene glgS, required for glycogen synthesis, is also stimulated by RpoS (20, 32). rpoS mutants have a glycogen-negative phenotype (29), and overexpression of glgS stimulates excess glycogen synthesis in early stationary phase (20). Stationary-phase cells are resistant to multiple environmental stresses and undergo changes in cell morphology and physiology; thus, entry into stationary phase is accompanied by changes in gene expression and protein synthesis (28). Many factors regulate the expression of rpoS at the levels of transcription, translation, and protein stability (for a review, see reference 19). At the translational level, both positive and negative regulators have been identified. The small untranslated RNA OxyS (61) represses RpoS, while the small untranslated RNAs DsrA (35) and RprA (34) activate RpoS. The histone-like protein H-NS (4) and the LysR-like regulator LeuO (25) repress RpoS, but host factor HF-1 (61) and histone-like protein HU (3) activate RpoS. At the posttranslational level, the protease ClpPX (46), the response regulator RssB (5), and the chaperonin DnaK (40) negatively regulate RpoS stability. Posttranslational protein degradation has a major effect on RpoS levels during the course of growth (46, 47). The net consequence of these controls is low exponential-phase levels of RpoS, but during the transition to stationary phase, the levels increase and remain high (46). RpoS is an attractive target for new antimicrobial strategies because this regulator controls many genes that are likely important for adaptation to the host environment, including catalase HPII. Furthermore, RpoS is well suited as a target for antisense RNA because it controls a large regulon, ablation of its action can be easily assessed by several means, and finally, natural antisense RNA regulators are known to inhibit translation of RpoS.
The primary goal of this study was to block rpoS expression by using plasmid-encoded, inducible rpoS antisense RNA. RpoS was chosen as a target because it controls a large regulon, has well-established effects on the phenotype of the cell, and is a pathogenicity factor. Our hypothesis is that an rpoS antisense RNA complementary to rpoS mRNA could inhibit RpoS function and attenuate expression of the rpoS regulon in E. coli. The efficacy of antisense RNA was evaluated by measuring the expression of RpoS-dependent catalase HPII and glycogen. The results indicate that the expression of the RpoS regulon in E. coli can be effectively modulated by antisense RNA.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used and constructed in this study are listed in Table 1. The plasmids used and constructed in this study are listed in Table 2. Luria-Bertani (LB) liquid and solid media were prepared as described by Miller (37). For glycogen tests, E. coli strains were grown on Kornberg medium agar plates to allow maximal glycogen synthesis (20, 41). All strains were grown in LB broth containing the appropriate antibiotics in a shaker at 200 rpm and 37°C.
TABLE 1.
Bacterial strains used and constructed in this study
| E. coli strain | Genotype or description | Source or comment |
|---|---|---|
| DH5α | supE44 ΔlacU169(φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Stratagene |
| MC4100 | (argF-lac)205 araD139 flbB5301 relA1 rpsL150 thi ptsF25 | 45 |
| MC4100DE3 | Like MC4100, but expressing T7 RNA polymerase | This study |
| MC4100DE3Y | Like MC4100DE3, but osmY-lacZ | This study |
| HS1600 | Like MC4100, but rpoS13::Tn10 | Laboratory collection |
| HS1600DE3 | Like HS1600 but expressing T7 RNA polymerase | This study |
| BL21DE3 | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| MC4100DE3 (pET21) | Like MC4100DE3, but pET21 | This study |
| MC4100DE3Y(pET21) | Like MC4100DE3Y, but pET21 | This study |
| MC4100DE3(pSOPRL) | Like MC4100DE3 but carries rpoS antisense plasmid pSOPRL | This study |
| MC4100DE3Y(pSOPRL) | Like MC4100DE3Y, but pSOPRL | This study |
| MC4100DE3(pSOPR) | Like MC4100DE3 but carries rpoS antisense plasmid pSOPR | This study |
| MC4100DE3Y(pSOPR) | Like MC4100DE3Y, but pSOPR | This study |
| BL21DE3(pSOPR) | Like BL21DE3, but pSOPR | This study |
TABLE 2.
Plasmids used and constructed in this study
| Plasmid | Size (bp) | Description and resistance(s)a | Source |
|---|---|---|---|
| pGEM-T Easy | 3,001 | Ampr; blue or white color selection; single 3′ T overhangs to allow efficient ligation with PCR products | Promega |
| pET21 | 5,369 | Ampr; transcriptional plasmid; lacks the RBS and start codon | Novagen |
| pET22b | 5,493 | Ampr; translational plasmid; pe1B for protein export or folding plus His-Tag for binding to resin for purification | Novagen |
| pGC2 | 4,298 | pGEM-T Easy containing 1,278-bp rpoS fragment cloned in the antisense orientation with respect to the T7lac promoter | This study |
| pGC2a | 4,042 | pGEM-T Easy containing 1,022 bp of rpoS sequence in antisense orientation | This study |
| pSOPR | 6,411 | pET21 containing 1,042-bp EcoRI fragment from pGC2a in the antisense orientation with respect to the T7lac promoter | This study |
| pSOPRL | 6,667 | pET21 containing 1,278-bp rpoS EcoRI fragment from pGC2 in antisense orientation with respect to the T7lac promoter | This study |
| pG225a | 6,535 | pET22b with 1,042-bp EcoRI fragment from pGC2a in antisense orientation with respect to the T7lac promoter | This study |
| pGC226 | 6,791 | pET22b containing 1,298-bp EcoRI fragment from pGC2 in antisense orientation with respect to the T7lac promoter | This study |
Ampr, ampicillin resistant; RBS, ribosomal binding site.
Selection of λDE3 lysogens expressing the T7 RNA polymerase gene.
To make an expression host for the pET expression system, a phage lambda derivative (λDE3) carrying the T7 RNA polymerase gene under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter (15, 50) was integrated into the E. coli chromosome. A kit for lysogenization and verification of the presence of λDE3 (Invitrogen, Mississauga, Ontario, Canada) was used according to the instructions of the manufacturer. A T7 (RNA polymerase-negative) tester phage that can lyse cells only when it is supplied with T7 polymerase was used to confirm the successful integration of the T7 RNA polymerase-expressing phage. Both rpoS-positive and rpoS-negative lysogenic expression hosts were constructed.
Plasmid and genomic DNA isolation, manipulation, and transformation.
E. coli plasmid DNA was isolated by the alkaline lysis method (42) or with a Midi plasmid preparation kit (Qiagen Inc., Mississauga, Ontario, Canada). Genomic DNA isolation, endonuclease digestions, ligations, and transformations were performed by standard techniques (42).
PCR amplification and sequencing of rpoS antisense RNA.
Chromosomal DNA was isolated from E. coli MC4100 (42). PCR was used to amplify the rpoS fragments from E. coli DNA by using several rpoS-specific primers (the MOBIX lab, McMaster University, Hamilton, Ontario, Canada), as follows: a short rpoS fragment 5′ primer (5′-CTTGCATTTTGAAATTCGTTACA-3′), a short rpoS fragment 3′ primer (5′-GTGAGGCCAATTTCACGACCTA3′), a large rpoS antisense fragment 5′ primer (5′-CTTGCATTTTGAAATTCGTTACA-3′), and a large rpoS antisense fragment 3′ primer (5′-TTAACGACCATTCTCGGTTTTAC-3′).
Annealing temperatures were 5°C below the lowest melting temperature of each primer pair. PCR was performed with Taq DNA polymerase (Invitrogen) or Expand DNA polymerase (Roche Diagnostics, Laval, Quebec, Canada). The PCR products were purified with a QIAquick PCR purification kit (Qiagen Inc.) prior to further manipulation. All DNA used in cloning reactions and for probe preparation was extracted from the agarose gels with a QIAEX II DNA extraction kit (Qiagen Inc.). All amplified products were sequenced by the MOBIX lab, McMaster University. Sequences of short and large rpoS antisense fragments were aligned with the sequences from the complementary strand and analyzed by using the Gene Runner program (version 3.04; Hastings Software, Inc., Moraga, Calif.).
Construction of rpoS antisense plasmids for in vivo experiments.
Following PCR amplification and purification, a 1,278-bp fragment containing 3′ A residue overhangs was ligated with compatible overhangs into plasmid pGEM-T Easy plasmid (Promega Corporation, Madison, Wis.) overnight (4°C), and the ligation mixture was transformed into E. coli DH5α. Transformants were selected on ampicillin (100 μg/ml) plates. Plasmids were isolated from several ampicillin-resistant colonies and digested with EcoRI, HincII, and AccI to confirm insertion of the fragment. The orientations of the cloned fragments were confirmed by DNA sequencing. One plasmid containing the entire 1,278-bp sequence was designated pGC2. The EcoRI fragment of pGC2 was subcloned into the expression vectors pET21 and pET22b to yield pSOPRL (pET21 background) (Fig. 1) and pGC226 (pET22b background). The orientation of the rpoS gene was reversed with respect to that of the T7lac promoter on the plasmids, such that an antisense rpoS RNA is expressed in cells treated with IPTG.
FIG. 1.
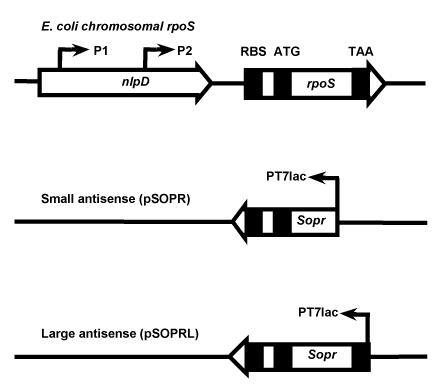
Schematic representation of the rpoS gene in the E. coli chromosome (top) and in antisense expression plasmids pSOPR and pSOPRL. The positions of promoters (P1, P2, and T7lac [PT7lac]) are indicated. The rpoS gene ribosomal binding site (RBS) and start (ATG) and stop (TAA) codons are also shown.
Similarly, a 1,022-bp PCR fragment of DNA was ligated with compatible overhangs of plasmid pGEM-T Easy to yield pGC2a. The 1,042-bp EcoRI fragment of pGC2a was subcloned into pET21 and pET22b, resulting in pSOPR (pET21 background) (Fig. 1 and 2) and pGC225a (pET22b background). The rpoS gene is in the reverse orientation with respect to that of the T7lac promoter.
FIG. 2.
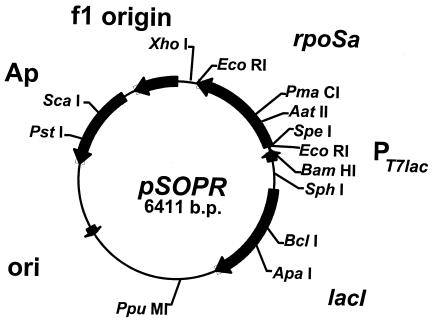
Antisense RNA plasmid pSOPR. The unique restriction endonuclease recognition sites are shown. Abbreviations: PT7lac, T7lac promoter; ori, plasmid origin of DNA replication; rpoSa, rpoS antisense RNA fragment; Ap, ampicillin resistance gene.
Bacterial growth and sampling conditions.
Single colonies were inoculated into 5 ml of LB medium with streptomycin (50 μg/ml) and ampicillin (100 μg/ml) where appropriate. Cultures were incubated overnight in a shaker at 200 rpm and 37°C. The overnight cultures were diluted 1/500 in fresh LB medium. After growth to an optical density at 600 nm (OD600) of 0.25, the cultures were again diluted 1/500 and grown to an OD600 of 0.01. The cultures were then divided into two flasks; one flask served as a control, and 0.3 mM IPTG (final concentration) was added to the other flask. The cultures were grown to an OD600 of 0.25, at which point 70 ml of the exponential-phase cells from each flask were collected. After further incubation to an OD600 of 1.5, 25 ml of the stationary-phase cells from each flask were collected.
Probe preparation, RNA extraction, and Northern blotting analyses.
To produce probes for Northern blotting, a 1,022-bp fragment of DNA corresponding to the short rpoS DNA sequence (above) was amplified from E. coli MC4100 chromosomal DNA by PCR. The PCR products were purified with a QIAquick PCR purification kit (Qiagen Inc.) and were labeled with [α-32P]dCTP by the random primer labeling method to generate probes with high specific activities (∼109 cpm/μg of DNA). The probes were purified with ProbeQuant G-50 micro columns (Amersham Biosciences, Inc., Piscataway, N.J.).
The cells were harvested and the RNA was extracted with an RNeasy Mini kit (Qiagen Inc.). The total RNA concentration was determined by measuring the absorbance at 260 nm (42). Following denaturing electrophoresis, the RNA was transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) by capillary action (42) and fixed by baking at 80°C for 2 h. Prehybridization and hybridization were performed at 60°C with gentle agitation. The membranes were probed with the 32P-labeled rpoS gene. The resulting blot was exposed to X-OMAT AR film (Eastman Kodak Company, Rochester, N.Y.) or to a Storage Phosphor Screen (Molecular Dynamics Inc., Sunnyvale, Calif.) for quantitation.
Protein extraction and determination.
Bacterial cell cultures were washed twice by centrifugation with 0.05 M phosphate buffer and were sonicated with a Heat Systems sonicator (Misonix Inc., Farmingdale, N.Y.) equipped with a cup horn (45). Cell debris was removed by centrifugation at 4°C for 15 min at 12,000 × g. The total protein concentration was determined by the method of Bradford (7) (Bio-Rad Laboratories, Hercules, Calif.).
Western blotting analysis.
Thirty micrograms of the proteins was separated on denaturing sodium dodecyl sulfate-10% polyacrylamide gels overnight with a Vertified Slab Gel unit (model SE400; Hoefer Scientific Instruments, San Francisco, Calif.) and then transferred to a Hybond-P membrane (Amersham Pharmacia Biotech Inc.). The membranes were stained with ponceau S to confirm efficient transfer. Following transfer, the blots were placed into blocking buffer (0.5% fraction V of bovine serum albumin and 5% skim milk in 0.1% Tween 20 in Tris-buffered saline [pH 7.6] [T-TBS]) overnight. The blots were then incubated with blocking buffer containing primary antibody (anti-σS antibody [polyclonal]; a gift from R. Hengge-Aronis) for 2 h at room temperature. After the blots were washed (three times with TBS-T or blocking buffer), the blots were placed in blocking buffer containing secondary antirabbit antibody (rabbit immunoglobulin and horseradish peroxidase-linked whole antibody from donkey; dilution, 1:1,000) and shaken for 1 h. The blots were again washed three times with TBS-T and incubated in 10 ml of enhanced chemiluminescence staining solution (detection reagent mixture; Amersham Pharmacia Biotech Inc.) and exposed to X-ray film (Kodak X-OMAT AR or BioMax MR film; Eastman Kodak Company) for 10 s to 10 min.
β-Galactosidase activity assays.
β-Galactosidase activity was assayed by using o-nitrophenyl-β-d-galactopyranoside as the substrate (37).
Catalase activity assays and detection.
Catalase activity on agar plates was qualitatively determined by adding a drop of 30% hydrogen peroxide to a colony and observing gas evolution. Catalase activity in cell extracts was assayed spectrophotometrically (45), as follows. One milliliter of hydrogen peroxide (0.5 ml of 30% H2O2 freshly diluted in 250 ml 50 mM potassium phosphate buffer [pH 7.0]) was added to 10 μl of the cell extract, and the decrease in the OD240 was monitored. The specific activity of catalase was calculated as [1,000 × (OD240/time of incubation)]/[43.6 × (milligrams of protein/milliliter of reaction mixture)] (6). To assess the catalase activity in the gels, protein samples (10 μg) were loaded into a 10% nondenaturing polyacrylamide gel for electrophoresis with a Mini-PROTEAN II system (Bio-Rad Laboratories). To detect the catalase activity, the gels were stained with horseradish peroxidase-diaminobenzidine (9). To confirm equal protein loading, parallel gels were stained for protein by using Coomassie blue (42).
Glycogen staining procedures.
The levels of glycogen accumulation in the bacterial cells were tested on Kornberg medium agar plates (20, 32) in the presence or absence of 0.3 mM IPTG. The plates were inverted and placed over a 500-ml beaker containing 3.3% iodine-6.6% potassium iodide solution. The colonies were stained with the iodine vapor by heating the solution for 3 to 5 min. Colonies in which glycogen had accumulated were dark brown in color, while those with little or no glycogen production stained yellow.
RESULTS
Construction of rpoS RNA expression system in pET21 and pET22b.
The DNA sequence of the rpoS gene (GenBank accession no. X16400) was used to design primers to amplify rpoS fragments. The objective was to produce an antisense RNA with properties similar to those of other recombinant antisense RNAs (35, 56). Thus, the sequence of the rpoS antisense RNA constructed is complementary to the sequence of a region that includes the translation initiation start codon, the Shine-Dalgarno sequence, other upstream untranslated sequences, and all or part of the coding sequences (Fig. 1). To test if rpoS antisense RNA can inhibit the expression of rpoS and RpoS-dependent genes, the rpoS PCR products were cloned into pET expression plasmids. The resulting plasmids in the antisense orientation with respect to that of the T7lac promoter were designated pSOPR (short antisense fragment in pET21 background) and pSOPRL (large antisense fragment in pET21 background) (Fig. 1 and 2). These plasmids were then transformed into various E. coli strains (Table 1) for in vivo expression of antisense RNAs.
Effect of IPTG on cell growth.
IPTG is normally used at a concentration of 1.0 mM for induction of protein synthesis in a pET plasmid system with the T7lac promoter, while 0.4 mM IPTG is used in a pET plasmid system carrying the plain T7 promoter (Novagen, Inc., Madison, Wis.). However, we found that concentrations of IPTG greater than 0.5 mM inhibited the growth of E. coli strains carrying pET plasmids with the T7lac promoter. To ensure that the results of the expression studies were not affected by growth inhibition, 0.3 mM IPTG was used in all experiments. At this concentration, IPTG had little effect on cell growth (data not shown).
Effect of IPTG on rpoS antisense RNA expression.
To measure the effect of the IPTG concentration on rpoS antisense RNA expression, rpoS antisense RNA expressed from cultures grown with various concentrations of IPTG was probed in Northern blots. In exponential phase (Fig. 3A), rpoS antisense mRNA was expressed from pSOPR in the presence of IPTG (lanes 14 to 18). Expression was not detectable in the absence of IPTG (lane 13) or in strains carrying control plasmid pET21 (lanes 7 to 12) or no plasmid (lanes 1 to 6). rpoS antisense RNA was also expressed in BL21DE3(pSOPR) in the presence of IPTG (lanes 19), which is a positive control. In MC4100DE3(pSOPR) cells in stationary phase (Fig. 3B), rpoS antisense RNA was expressed only in the presence of IPTG (lanes 14 to 18). Expression was not detectable in the absence of IPTG (lane 13). Expression in MC4100DE3 and its transformants containing control plasmid pET21 was not detectable in the absence or the presence of IPTG (lanes 1 to 12). The levels of rpoS mRNA, which is similar in size to antisense rpoS RNA encoded on pSOPR, are relatively low compared to those of the highly expressed, plasmid-derived antisense RNA, and therefore, rpoS mRNA was not detected in Northern blots under the conditions used. The optimal concentration of IPTG for induction of rpoS antisense RNA in exponential phase and stationary phase was determined to be 0.3 mM, which is below the level that inhibits cell growth.
FIG. 3.
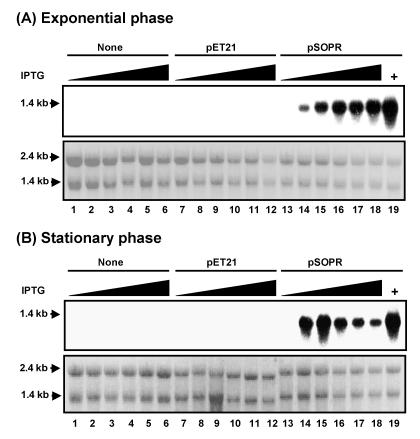
Northern blot analysis of rpoS antisense RNA induced with IPTG in the MC4100DE3 series of strains in exponential (A) and stationary (B) phases showing the effect of IPTG on rpoS antisense RNA expression. Total RNA was isolated from rpoS wild-type strain MC4100DE3 and its transformants containing control plasmid pET21 and plasmid pSOPR with antisense rpoS RNA. RNA was isolated from cultures grown in the absence (lanes 1, 7, and 13) or in the presence of 0.2, 0.3, 0.5, 0.75, and 1 mM IPTG (lanes 2 to 6, 8 to 12, and 14 to 18, respectively). As a positive control, strain BL21DE3(pSOPR) was induced with 0.75 mM IPTG (lane 19). The lower panels in panels A and B show that equal amounts of RNA were applied to formaldehyde-agarose gels and then transferred to Hybond-N+ membranes. The expression of antisense RNA was demonstrated with an rpoS-specific [α-32P]dCTP-labeled double-stranded DNA probe.
Induction of short and large fragments of rpoS antisense RNA expression in MC4100DE3 and BL21DE3.
To increase the ratio of antisense RNA to rpoS mRNA, the cultures were diluted (see above for experimental procedures) to reduce the level of background rpoS mRNA and were induced with IPTG at a very early stage of cell growth. Antisense rpoS RNA should thus be available to sequester rpoS mRNA as it is generated during cell growth. The level of antisense products expressed by the cells is high due to the high-copy-number nature of plasmid pET21 and the strong T7lac promoter.
In exponential and stationary phases (Fig. 4A and B, respectively), expression of rpoS antisense RNA was induced by IPTG in strains carrying the antisense transcriptional plasmids. In the exponential-phase samples, the short and large fragments of rpoS antisense RNA were highly induced from antisense expression plasmids pSOPR and pSOPRL, respectively, in the presence of IPTG, as indicated on Northern blots probed with rpoS DNA. The level of short-fragment mRNA was higher than that of the large-fragment RNA, especially in MC4100DE3. In stationary phase, antisense mRNA was also transcribed (data for large-fragment mRNA are not shown for MC4100DE3). In all cases, antisense RNA levels were not detectable in the absence of IPTG. Antisense RNA was also not detectable in rpoS-positive and rpoS-negative control strains without a plasmid or strains that harbor control plasmid pET21 in the presence or absence of IPTG. The ratio of the levels of antisense RNA expressed by the cells to the levels of rpoS mRNA expressed by the cells was high. Short-fragment antisense construct pSOPR was selected for further study.
FIG. 4.
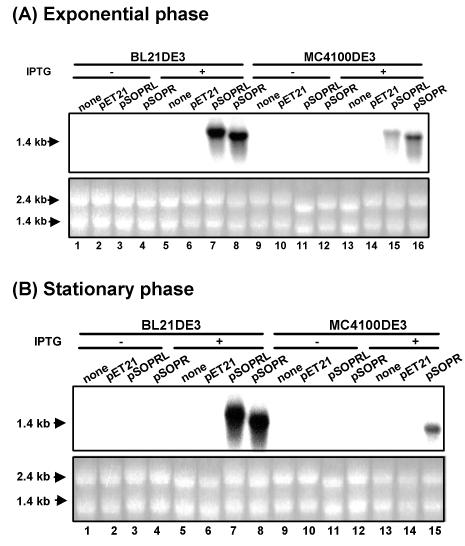
Northern blot analysis of short and large fragments of rpoS antisense RNAs induced by IPTG in BL21DE3 and MC4100DE3 in exponential (A) and stationary (B) phases. Total RNA was isolated from rpoS wild-type strains BL21DE3 and MC4100DE3 and their transformants containing control plasmid pET21 and plasmids pSOPRL and pSOPR with antisense rpoS RNA. RNA was isolated from a culture grown in the absence of IPTG (−) (lanes 1 to 4 and 9 to 12) or in the presence of IPTG (+) (lanes 5 to 8 and 13 to 16). The lower panels in panels A and B show that equal amounts of RNA were applied to formaldehyde-agarose gels and then transferred to Hybond-N+ membranes. The expression of antisense RNA was demonstrated with an rpoS-specific [α-32P]dCTP-labeled double-stranded DNA probe.
For maximal inhibition of RpoS expression, we found that antisense RNA must be induced in early exponential phase (OD600 = 0.01). In this way, antisense RNA is present in the cell before rpoS is transcriptionally activated and can thus effectively sequester rpoS mRNA before it is translated. However, the antisense rpoS RNA expressed from pET translational plasmid pET22b in strains BL21DE3 and MC4100DE3 (carrying pGC226 or pGC225a) did not inhibit highly RpoS-dependent katE and osmY expression (data not shown). In plasmid pET22b, the short and large fragments of rpoS antisense RNA were probably translated into a chimeric protein. pET22b contains its own ribosomal binding site located downstream of the T7lac promoter.
Antisense RNA-repressed RpoS protein expression.
In stationary phase, RpoS protein was produced in rpoS-positive control cells (Fig. 5, lanes 9 and 10), cells carrying control plasmid pET21 (lanes 13 and 14), and uninduced rpoS-positive cells carrying the antisense construct (lane 15). However, when antisense RNA was expressed (following induction with IPTG), the level of RpoS protein was substantially reduced (lane 16). RpoS protein was not produced in the rpoS-negative control strain in the presence or absence of IPTG (lanes 11 and 12). The very faint bands present below the RpoS band (lanes 9 to 15) may be due to a cross-reaction with the polyclonal anti-RpoS antibody. The levels of RpoS protein were higher in strain MC4100DE3 than in MC4100DE3(pET21). Proteins expressed from control plasmid pET21 (for example, β-lactamase and LacI) may interfere with RpoS protein synthesis.
FIG. 5.
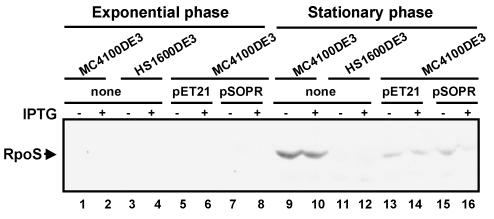
Western blot analysis of RpoS levels in E. coli MC4100 derivatives with anti-σS antiserum. Protein was extracted from cultures grown to exponential phase (OD600 = 0.25) and stationary phase (OD600 = 1.5). The polyvinylidene difluoride membrane was stained with enhanced chemiluminescence staining solution and exposed to X-OMAT film (Eastman Kodak Company).
The ClpXP protease rapidly degrades RpoS in exponential-phase cells, resulting in extremely small amounts of this sigma factor (46). Consistent with this, we found that in exponential-phase cultures of all strains tested (Fig. 5, lanes 1 to 8), RpoS protein levels were too low to be detected by Western blotting.
Inhibition of RpoS-dependent osmY expression by rpoS antisense RNA.
To measure the effect of rpoS antisense RNA on the expression of the highly RpoS-dependent osmY gene (27), an osmY-lacZ operon fusion was transduced into MC4100DE3. Expression of osmY during the growth of various strains (Table 1) was quantified by measuring the levels of β-galactosidase expression. The level of OsmY expression was reduced by approximately 50% by the addition of IPTG to cultures containing rpoS antisense RNA-expressing constructs, and OsmY was normally expressed in control cultures incubated in the absence of IPTG or in strains containing only the pET21 plasmid vector (data not shown).
Inhibition of RpoS-dependent catalase HPII expression by rpoS antisense RNA expression.
E. coli catalase HPII, encoded by katE, can be used as a target protein to evaluate highly RpoS-dependent gene expression (44). Catalase activity was examined on LB and M9 agar plates supplemented with IPTG by adding 30% hydrogen peroxide to 24-h-old colonies (45). The lack of significant gas evolution indicated that catalase activity was reduced in antisense RNA-expressing strains in the presence of IPTG. In addition to qualitative catalase assays, catalase enzyme activity was measured to confirm the effect of rpoS antisense RNA expression on catalase expression. As shown in Fig. 6A and B, catalase activity was greatly inhibited in rpoS antisense RNA-expressing strains [compare the open bars to the filled bars for strain MC4100DE3(pSOPR)] in both exponential and stationary phases, respectively. Control strain MC4100DE3 and transformants containing the control plasmid exhibited similar levels of activity, and these were much higher than those in the antisense RNA-producing strain [compare the filled bars for strains MC4100DE3(pSOPR), MC4100DE3, and MC4100DE3(pET21)]. Expression of the HPII catalase is known to be much more dependent on RpoS than that of the HPI catalase (45). To test whether downregulation of catalase activity in exponential and stationary phases by induced rpoS antisense RNA is due to reduced levels of expression of the RpoS-dependent catalase HPII, the levels of HPII and HPI were assessed by nondenaturing polyacrylamide gel electrophoresis, followed by catalase staining. In strains expressing rpoS [MC4100DE3, MC4100DE3(pET21), and MC4100DE3(pSOPR)], the level of HPII expression was normal: the levels were low in exponential-phase cultures and high in stationary-phase cultures (Fig. 7A and B, respectively). As expected, induction of rpoS antisense RNA inhibited RpoS-dependent HPII expression in stationary phase (Fig. 7B, lane 8), although HPII was not present in exponential phase in any sample in native catalase gels (Fig. 7A).
FIG.6.
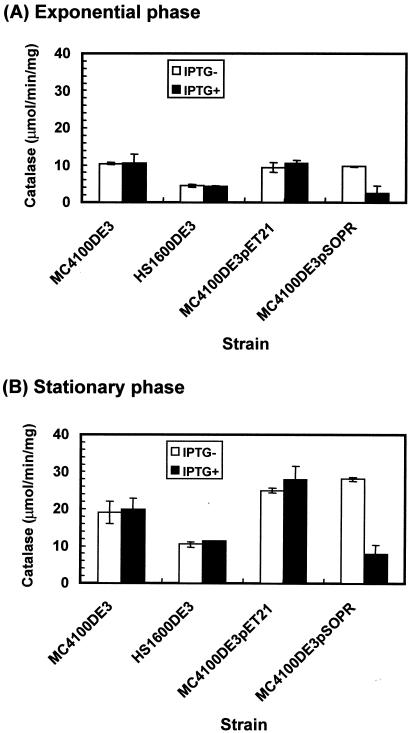
Catalase-specific activity of rpoS wild-type strain MC4100DE3 containing control plasmid pET21 and antisense construct pSOPR grown in the absence (−) and the presence (+) of 0.3 mM IPTG in LB medium. Induced rpoS antisense RNA downregulated expression of RpoS-dependent gene katE.
FIG. 7.
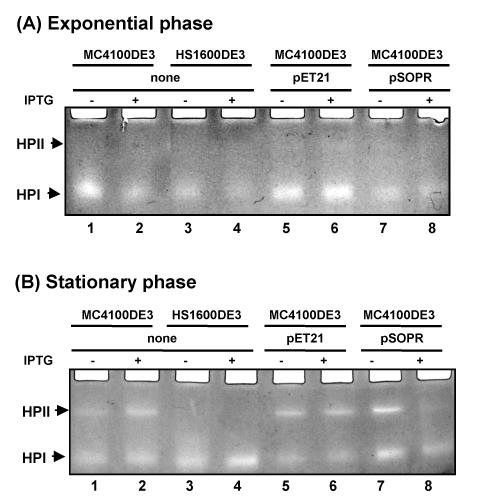
Catalase activity in polyacrylamide gels containing protein extracted from rpoS wild-type strain MC4100DE3 carrying control plasmid pET21 and antisense construct pSOPR grown in the absence (−) and the presence (+) of 0.3 mM IPTG in LB medium. Induced rpoS antisense RNA downregulated RpoS-dependent katE expression (compare lanes 7 and 8 in panel B).
Inhibition of glycogen accumulation by rpoS antisense RNA.
Glycogen accumulation is stimulated by RpoS during entry into stationary phase (20, 32), as expression of glgS is entirely dependent on RpoS in vivo in E. coli. RpoS-deficient mutants do not produce GlgS, and overexpression of glgS stimulates higher levels of glycogen synthesis in early stationary phase (20). Glycogen accumulation in bacterial colonies can be visualized on Kornberg medium, a nitrogen-limited, glucose-rich medium (20, 41), by a dark brown stain in the presence of iodine; when little or no glycogen is present the colonies are yellow in color (32). As expected, MC4100DE3 cells (rpoS positive) accumulated glycogen, while RpoS-deficient mutants (HS1600DE3) did not (Fig. 8A). However, induced RpoS antisense RNA expression reduced the level of glycogen synthesis, resulting in yellow colonies following iodine staining (Fig. 8B).
FIG. 8.
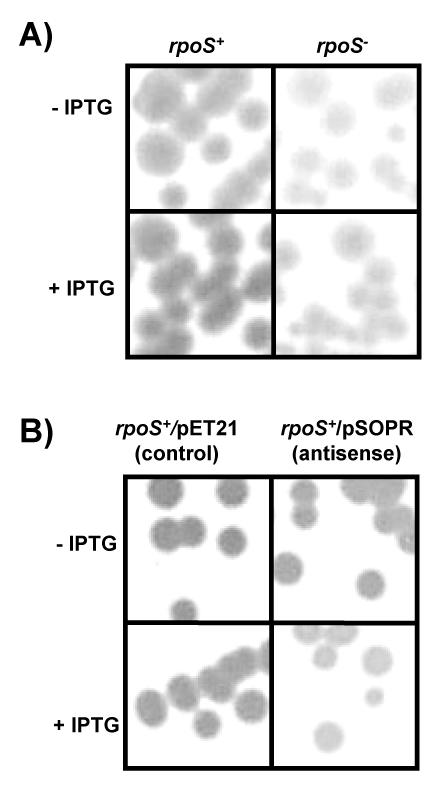
Glycogen accumulation of strains during growth on Kornberg medium plates in the presence (+) or absence (−) of IPTG. The strains were grown for 1 to 2 days at 37°C, and intracellular glycogen was stained with iodine vapors. rpoS−, rpoS-negative strain; rpoS+, rpoS-positive strain.
DISCUSSION
During the last decade, antisense RNA research has focused on increasing the efficiency of antisense RNA, manipulating cell metabolic pathways, and investigating unknown gene functions when gene disruption is not possible under certain conditions (8). The use of antisense RNA to attenuate gene expression has several advantages over gene inactivation: lethal mutations in essential genes can be avoided, genes can be inactivated when homologous recombination is weak (8), and antisense RNA expression is controllable (59). Therefore, proteins can be transiently downregulated. Regulation of RpoS through plasmid-encoded rpoS antisense RNA can help us understand the bacterial pathogenesis and environmental adaptation responses. RpoS plays an important role in pathogenicity in enterobacteria and is induced under stress conditions. For example, RpoS is required for the virulence that is dependent on spvABCD by Salmonella enterica serovar Typhimurium in a mouse model (10, 11, 26). RpoS is also required for the maintenance of chronic lung infection with Pseudomonas aeruginosa in a rat model (52). In addition, RpoS controls potential antimicrobial agent and drug efflux operon yhiUV in E. coli (39, 43).
By reducing the level of expression of transcriptional regulator RpoS with controllable antisense RNA during bacterial infection, expression of a number of RpoS-dependent genes would be prevented. Control of expression of RpoS-dependent genes by antisense RNA may offer a good model for the regulation of RpoS translation and could provide a new tool with which bacterial infection processes can be studied. One limitation of this study is that high-copy-number plasmid pET21 may have an effect on E. coli cell growth and the expression of rpoS. Future work will include insertion of the rpoS antisense RNA construct in a low-copy-number plasmid or as a single copy in the host chromosome.
Expression of antisense RNA from a multicopy plasmid resulting in reduced levels of mRNA translation has been successfully used in previous studies. Introduction of a marA antisense RNA-expressing plasmid into E. coli cells carrying a marORA-lacZ fusion reduces the level of lacZ expression and subsequently increases the level of multiple-antibiotic susceptibility. Furthermore, the antisense RNA that most efficiently represses expression of marORA is complementary to a region encompassing 20 bases of untranslated sequence upstream of marR, the AUG initiation codon, and 92 bases of marA-coding sequences (58), indicating that sequestration of mRNA control sequences can ablate translation. In S. aureus, expression of an antisense hla RNA from a plasmid reduces the level of alpha-toxin virulence up to 17-fold relative to that of the wild-type strain carrying a control plasmid and, as a consequence, eliminates lethality in a mouse model (22, 24). In this study, the levels of RpoS-dependent catalase HPII expression and glycogen accumulation were dramatically decreased in antisense RNA-expressing cells compared to those in cells that did not express rpoS antisense RNA. This suggests that antisense technology can be used successfully to reduce the levels of a transcriptional regulator and, consequently, attenuate expression of a downstream target. An rpoS antisense RNA is complementary to a region that includes the 5′ untranslated region (93 bases) and part of the coding region of rpoS (907 bases), while another one is complementary to a region that includes the 5′ untranslated region (93 bases) and all of the coding region of rpoS with the 156-bp transcription termination sequence. Expression of antisense RNA was sufficient to inhibit rpoS gene expression. The construct carrying the antisense sequence spanning the 5′ untranslated region and a part of the coding region of rpoS mRNA was found to be the most effective in attenuating the expression of the rpoS gene (data not shown).
Several natural antisense RNA regulators of rpoS gene expression and the resulting phenotypes have been identified in E. coli. In many case their modes of action have been described. OxyS RNA represses rpoS translation by preventing RNA-binding protein Hfq from activating rpoS translation (61). OxyS RNA is proposed to bind to Hfq protein through an A-rich linker region between two stem-loops in OxyS RNA (61), thereby preventing formation of a translationally active complex with rpoS mRNA (19). OxyS represses osmotic induction of RpoS in strains carrying rpoS-lacZ translational fusions treated with high salt concentrations (61). OxyS also represses transcriptional activator FhlA by binding to the Shine-Dalgarno sequence or the coding region of fhlA, resulting in stable sense-antisense complex (1, 2). DsrA stimulates rpoS translation at a low temperature (20°C) (30) by stabilizing rpoS mRNA (48). A stem-loop of DsrA binds to the 5′ untranslated leader sequence of rpoS mRNA just before the translation initiation site, resulting in increased RpoS protein levels (31, 35). DsrA RNA acts in trans by RNA-RNA interactions with rpoS mRNA (30). Hfq is important for DsrA-activated regulation of rpoS (49).
We propose the following model to explain rpoS antisense RNA repression of rpoS mRNA translation. The ratio of the amount of rpoS antisense RNA expressed from the high-copy-number plasmid to the amount of rpoS mRNA generated from the chromosome is very high. Thus, antisense RNA is sufficient to bind to the sense rpoS RNA strand and produce a double-stranded RNA molecule. The double-stranded RNA molecule is degraded by RNases (21). The 5′ untranslated region of the rpoS mRNA is self-complementary, forming a secondary structure (12, 34, 35) which is inaccessible to ribosomes and which therefore blocks translation of the rpoS mRNA into the RpoS protein. Induced expression of rpoS antisense RNA inhibits the expression of rpoS and RpoS-dependent genes such as katE and glgS by binding to rpoS mRNA and inhibiting translation, thereby ablating synthesis of the RNA polymerase sigma factor required for RpoS-dependent gene transcription.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research (to H.E.S.).
We thank R. Hengge-Aronis for the RpoS antibody used in these studies.
REFERENCES
- 1.Altuvia, S., A. Zhang, L. Argaman, A. Tiwari, and G. Storz. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17:6069-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argaman, L., and S. Altuvia. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol. Biol. 300:1101-1112. [DOI] [PubMed] [Google Scholar]
- 3.Balandina, A., L. Claret, R. Hengge-Aronis, and J. Rouviere-Yaniv. 2001. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 39:1069-1079. [DOI] [PubMed] [Google Scholar]
- 4.Barth, M., C. Marschall, A. Muffler, D. Fischer, and R. Hengge-Aronis. 1995. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of sigma S and many sigma S-dependent genes in Escherichia coli. J. Bacteriol. 177:3455-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 96:6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beers, R. F., Jr., and I. W. Sizer. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195:133. [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Casqueiro, J., S. Gutierrez, O. Banuelos, M. J. Hijarrubia, and J. F. Martin. 1999. Gene targeting in Penicillium chrysogenum: disruption of the lys2 gene leads to penicillin overproduction. J. Bacteriol. 181:1181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clare, D. A., M. N. Duong, D. Darr, F. Archibald, and I. Fridovich. 1984. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 140:532-537. [DOI] [PubMed] [Google Scholar]
- 10.Coynault, C., V. Robbe-Saule, and F. Norel. 1996. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (sigma S) regulon. Mol. Microbiol. 22:149-160. [DOI] [PubMed] [Google Scholar]
- 11.Coynault, C., V. Robbe-Saule, M. Y. Popoff, and F. Norel. 1992. Growth phase and SpvR regulation of transcription of Salmonella typhimurium spvABC virulence genes. Microb. Pathog. 13:133-143. [DOI] [PubMed] [Google Scholar]
- 12.Cunning, C., L. Brown, and T. Elliott. 1998. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 180:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daugherty, B. L., K. Hotta, C. Kumar, Y. H. Ahn, J. D. Zhu, and S. Pestka. 1989. Antisense RNA: effect of ribosome binding sites, target location, size, and concentration on the translation of specific mRNA molecules. Gene Anal. Tech. 6:1-16. [DOI] [PubMed] [Google Scholar]
- 14.Desai, R. P., and E. T. Papoutsakis. 1999. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum. Appl. Environ. Microbiol. 65:936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubendorff, J. W., and F. W. Studier. 1991. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 219:45-59. [DOI] [PubMed] [Google Scholar]
- 16.Gill, R. T., J. J. Valdes, and W. E. Bentley. 1999. Reverse transcription-PCR differential display analysis of Escherichia coli global gene regulation in response to heat shock. Appl. Environ. Microbiol. 65:5386-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good, L., and P. E. Nielsen. 1998. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16:355-358. [DOI] [PubMed] [Google Scholar]
- 18.Guerrier-Takada, C., R. Salavati, and S. Altman. 1997. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc. Natl. Acad. Sci. USA 94:8468-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 1996. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887-893. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R., and D. Fischer. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 6:1877-1886. [DOI] [PubMed] [Google Scholar]
- 21.Inouye, M. 1988. Antisense RNA: its functions and applications in gene regulation—a review. Gene 72:25-34. [DOI] [PubMed] [Google Scholar]
- 22.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kernodle, D. S., R. K. Voladri, B. E. Menzies, C. C. Hager, and K. M. Edwards. 1997. Expression of an antisense hla fragment in Staphylococcus aureus reduces alpha-toxin production in vitro and attenuates lethal activity in a murine model. Infect. Immun. 65:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klauck, E., J. Bohringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 26.Kowarz, L., C. Coynault, V. Robbe-Saule, and F. Norel. 1994. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J. Bacteriol. 176:6852-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange, R., M. Barth, and R. Hengge-Aronis. 1993. Complex transcriptional control of the sigma S-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J. Bacteriol. 175:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange, R., and R. Hengge-Aronis. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 173:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 30.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. USA 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, M. Y., H. Yang, and T. Romeo. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 177:2663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loewen, P. C., J. Switala, and B. L. Triggs-Raine. 1985. Catalases HPI and HPII in Escherichia coli are induced independently. Arch. Biochem. Biophys. 243:144-149. [DOI] [PubMed] [Google Scholar]
- 34.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 35.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 173:4188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Nellen, W., and C. Lichtenstein. 1993. What makes an mRNA anti-sense-itive? Trends Biochem. Sci. 18:419-423. [DOI] [PubMed] [Google Scholar]
- 39.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romeo, T., and J. Preiss. 1989. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5′-diphosphate 3′-diphosphate and analysis of in vivo transcripts. J. Bacteriol. 171:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schellhorn, H. E., J. P. Audia, L. I. Wei, and L. Chang. 1998. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J. Bacteriol. 180:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schellhorn, H. E., and H. M. Hassan. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 170:4286-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schellhorn, H. E., and V. L. Stones. 1992. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J. Bacteriol. 174:4769-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (sigma S) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweder, T., H. Y. Lin, B. Jurgen, A. Breitenstein, S. Riemschneider, V. Khalameyzer, A. Gupta, K. Buttner, and P. Neubauer. 2002. Role of the general stress response during strong overexpression of a heterologous gene in Escherichia coli. Appl. Microbiol. Biotechnol. 58:330-337. [DOI] [PubMed] [Google Scholar]
- 48.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 49.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 51.Sturino, J. M., and T. R. Klaenhammer. 2002. Expression of antisense RNA targeted against Streptococcus thermophilus bacteriophages. Appl. Environ. Microbiol. 68:588-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamm, I., B. Dorken, and G. Hartmann. 2001. Antisense therapy in oncology: new hope for an old idea? Lancet 358:489-497. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA 90:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres, V. C., S. Tsiodras, H. S. Gold, E. P. Coakley, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., and R. T. Inouye. 2001. Restoration of vancomycin susceptibility in Enterococcus faecalis by antiresistance determinant gene transfer. Antimicrob. Agents Chemother. 45:973-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner, E. G., and R. W. Simons. 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 48:713-742. [DOI] [PubMed] [Google Scholar]
- 57.Walker, S. A., and T. R. Klaenhammer. 2000. An explosive antisense RNA strategy for inhibition of a lactococcal bacteriophage. Appl. Environ. Microbiol. 66:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, D. G., K. Maneewannakul, E. von Hofe, M. Zillman, W. Eisenberg, A. K. Field, and S. B. Levy. 1997. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yatzkan, E., B. Szoor, Z. Feher, V. Dombradi, and O. Yarden. 1998. Protein phosphatase 2A is involved in hyphal growth of Neurospora crassa. Mol. Gen. Genet. 259:523-531. [DOI] [PubMed] [Google Scholar]
- 60.Zadra, I., B. Abt, W. Parson, and H. Haas. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 66:4810-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17:6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]


