Abstract
In 1999, 39 of 2,599 isolates of the family Enterobacteriaceae (1.5%) collected by eight private laboratories in the Aquitaine region in France produced an extended-spectrum β-lactamase (ESBL). Among these were 19 Enterobacter aerogenes isolates; 8 Klebsiella pneumoniae isolates; 6 Escherichia coli isolates; 3 Proteus mirabilis isolates; and 1 isolate each of Serratia marcescens, Morganella morganii, and Providencia stuartii. ESBL producers were isolated from 38 patients, including 33 residents of 11 clinics or nursing homes and 5 ambulatory patients. Seven different ESBLs were characterized. These mainly consisted of TEM-24 (25 isolates) and TEM-21 (9 isolates), but TEM-15 (2 isolates) and TEM-3, TEM-19, SHV-4, and CTX-M-1 (1 isolate each) were also characterized. Seven strains showed the coexistence of different TEM- and/or SHV-encoding genes, including a new SHV-1 variant, SHV-44, defined by the substitution R205L previously reported for SHV-3 in association with S238G. The epidemiology of the ESBL producers was investigated by random amplification of polymorphic DNA, typing by enterobacterial repetitive intergenic consensus PCR, analysis of resistance cotransferred with the ESBL, and analysis of the restriction profiles of the ESBL-encoding plasmids. Of the TEM-24-expressing strains, 18 were E. aerogenes isolates, including 9 from the same clinic, that were representatives of the epidemic clone disseminating in France. Of the TEM-21-producing strains that belonged to different species of the family Enterobacteriaceae (E. coli, K. pneumoniae, and P. mirabilis), 8 were isolated in the same nursing home. Outbreaks due to strain and/or plasmid dissemination in these clinic and nursing home were demonstrated. The presence of ESBL producers in five ambulatory patients probably resulted from nosocomial acquisition. Our data highlight the serious need to monitor patients for ESBL-producing Enterobacteriaceae in general practice.
Extended-spectrum β-lactamases (ESBLs) have been observed in virtually all species of the family Enterobacteriaceae. These enzymes are predominantly plasmid mediated and are derived from broad-spectrum β-lactamase TEM-1, TEM-2, or SHV-1 by a limited number of mutations (13). Other newly emerging class A enzymes, such as members of the CTX-M family, can also be encountered in these organisms (23). ESBL-producing Enterobacteriaceae are typically nosocomial pathogens and are often responsible for outbreaks, particularly in intensive care units (ICUs). Thus, their presence in the hospital is regularly monitored, and due to control efforts, the prevalence of ESBLs producers has been drastically reduced in some centers (29, 38). However, many studies have demonstrated the great potential for the spread of ESBL-producing strains and also ESBL-encoding plasmids to different hospitals (16) and even different countries (19). Most patients hospitalized in ICUs are discharged to a general acute-care unit and then go to a rest, nursing, or retirement home. Some of them continue to carry ESBL-producing Enterobacteriaceae over prolonged periods, and continued carriage of such strains may contribute to their extrahospital propagation (12). In addition, community-acquired strains possessing ESBLs might be selected from the existing gastrointestinal flora when it is exposed to broad-spectrum antimicrobial agents (27). Thus, ESBL producers are expected to be present in general practice, but their occurrence has rarely been reported, probably because screening for ESBLs requires special tests and operator expertise.
In France, besides public hospitals, the private health care sector includes community and private health care centers, i.e., clinics and nursing homes (NHs). Clinics are essentially inpatient facilities which tend to be smaller than hospitals; they similarly encompass various acute-, intermediate-, and long-term-care facilities (LTCFs) or occasionally may be more specialized (e.g., functional reeducation or psychiatric facilities). A variety of NHs, including rest, nursing, and retirement homes, accommodate different populations with a wide spectrum of clinical disabilities. ESBL producers have been detected and have even been demonstrated to cause authentic outbreaks in LTCFs and NHs in the United States but rarely elsewhere. On the other hand, few studies have reported on the presence of ESBL producers in the community, particularly among uropathogens (14, 20, 25, 28, 39). However, ESBL-producing strains found in the community have not been characterized at the molecular level. Molecular characterization would offer important information that, together with strain typing and genetic support analysis, might help provide an understanding of the route of dissemination of ESBL-producing organisms in extrahospital practice via strain, plasmid, or gene propagation.
During a previous survey conducted over a 5-month period in 1999 in the Aquitaine region of France by eight private laboratories (C. Quentin et al., submitted for publication), 39 of 2,599 consecutive, nonredundant strains of the family Enterobacteriaceae deemed to cause infections produced an ESBL. The aim of the present study was to analyze the origins of these strains, to identify the ESBLs that they produce, and to investigate their epidemiological relationships by molecular and nonmolecular methods.
MATERIALS AND METHODS
Bacterial strains.
Thirty-nine ESBL-producing strains of the family Enterobacteriaceae isolated in a previous survey (Quentin et al., submitted) were analyzed (Table 1). Identification to the species level was confirmed with the API 20E system (bioMérieux, Marcy l'Etoile, France). Eight clinically unrelated strains (two Enterobacter aerogenes isolates, two Escherichia coli isolates, two Klebsiella pneumoniae isolates, and two Proteus mirabilis isolates) expressing or not expressing a β-lactamase were used as controls for molecular typing analysis. In addition, two TEM-24b-producing E. aerogenes strains belonging to the clone responsible for the present epidemic in France (12) were also included for comparative purposes. E. coli strains encoding the blaTEM-1 (45), blaSHV-1 (7), and blaCTX-M-3 (V. Dubois, personal communication) β-lactamase genes and Pseudomonas aeruginosa strains carrying the acc(6′)-I and aac(3)-II genes (21, 22) were used as positive controls for the PCR amplifications.
TABLE 1.
Distribution of isolates of the family Enterobacteriaceae according to setting
| Clinical setting | Total no. of isolates
|
|
|---|---|---|
| Enterobacteriaceae | ESBL-producing Enterobacteriaceae (species)a | |
| Community | 1,584 | 5 (2 Ea, 2 Ec, 1 Kp) |
| Clinic | ||
| CLI-1 | 127 | 11 (9 Ea, 1 Kp, 1 Mm) |
| CLI-1 | 100 | 3 (2 Ea, 1 Kp) |
| CLI-3 | 78 | 2 (2 Ea) |
| CLI-4 | 77 | 2 (1 Ec, 1 Sm) |
| CLI-5 | 68 | 1 (1 Kp) |
| CLI-6 | 56 | 2 (2 Ea) |
| CLI-7 | 44 | 1 (1 Ps) |
| CLI-8 | 6 | 1 (1 Ea) |
| Others (n = 11) | 331 | 0 |
| NH | ||
| NH-1 | 19 | 8 (3 Ec, 3 Pm, 2 Kp) |
| NH-2 | 3 | 2 (2 Kp) |
| NH-3 | 1 | 1 (1 Ea) |
| Others (n = 15) | 105 | 0 |
The number of isolates of each species in given in parentheses. the species are abbreviated as follows: Ea, E. aerogenes; Ec, E. coli; Kp, K. pneumoniae; Mm, M. morganii; Pm, P. mirabilis; Ps, P. stuartii; Sm, S. marcescens.
Antimicrobial susceptibility testing.
The antibiotic susceptibility patterns of the ESBL-producing strains and their transconjugants or transformants were determined by the disk diffusion method in Mueller-Hinton agar with 27 disks (32). The production of ESBL was controlled by the double-disk synergy test between clavulanic acid and ceftazidime, cefotaxime, or cefepime. The MICs for the recombinant clone producing the SHV-1 variant were determined by an agar dilution method (32). The β-lactam agents were kindly supplied by their manufacturers, as follows: ceftazidime, GlaxoSmithKline; cefotaxime, Aventis Pharma; and aztreonam and cefepime, Bristol-Myers Squibb.
Isoelectric focusing.
Isoelectric focusing was performed in polyacrylamide gels containing ampholines (Amersham Biosciences, Orsay, France) with a pH range of 3.5 to 10.0, as described previously (3). β-Lactamase activities were detected by an iodine-starch procedure in an agar gel with benzylpenicillin (75 μg/ml), which is hydrolyzed by all β-lactamases, and ceftriaxone (0.25 μg/ml), which is an elective substrate for the ESBLs and other broad-spectrum cephalosporin-hydrolyzing enzymes. The isoelectric points (pIs) of the β-lactamases studied were determined by comparison with those for reference β-lactamases whose pIs were known.
Transfer experiments and plasmid analysis.
Conjugation assays were carried out by a broth mating procedure in brain heart infusion medium with a nalidixic acid-resistant (Nalr) and rifampin-resistant (Rifr) mutant of E. coli K-12 as the recipient (15). Transconjugants were selected on Mueller-Hinton agar containing nalidixic acid and/or rifampin (100 μg/ml) plus ceftazidime (2 μg/ml) or cefotaxime (4 μg/ml). For each conjugation experiment, three transconjugants were tested for their antibiotic susceptibilities. When the initial results were negative, mating procedures were repeated by using the more efficient filter method (15). Transformation assays were performed by electroporation (Gene Pulser; Bio-Rad, Marnes la Coquette, France) with E. coli strain DH5α as the recipient, and transformants were cultivated on medium containing ampicillin (100 μg/ml). Plasmid DNA was extracted by an alkaline lysis method (10) and analyzed by electrophoresis on 0.8% (wt/vol) agarose gels after digestion with the EcoRI endonuclease (Promega, Charbonnière-les-Bains, France).
PCR amplifications.
PCR experiments were performed with crude lysates obtained after boiling according to the instructions of the supplier (Applied Biosystems Division, Perkin-Elmer, Courtaboeuf Cedex, France) under standard conditions (94°C for 5 min and 35 subsequent cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, with a final step at 72°C for 10 min). The pairs of primers specific for blaTEM, blaSHV, blaCTX-M-1, aac(6′)-I, and aac(3)-II that were used are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Primera | Sequence (5′→3′)b | Nucleotide positionc | Reference or source |
|---|---|---|---|
| blaTEM | |||
| Amplification and sequencing | |||
| TEM-A2(F) | GTATCCGCTCATGAGACAATA | 148 | 22 |
| TEM-ext(R) | TCTAAAGTATATATGAGTAAC | 1103 | 22 |
| Amplification and cloning | |||
| HIII-TEM(F) | CACACAAAGCTTGAAGACGAAAGGGCCTCGTG | 6 | This study |
| EI-TEM(R) | CACACAGAATTCTCTAAAGTATATATGAGTAAAC | 1103 | This study |
| Sequencing | |||
| TEM-C(F) | GGGCAAGAGCAACTCGG | 461 | 5 |
| TEM-D(F) | CAGCAATGGCAACAACGTTG | 753 | 5 |
| TEM-F(R) | CAACGTTGTTGCCATTGCTGCAG | 772 | 5 |
| TEM-G(R) | ACCGAGTTGCTCTTGCCC | 478 | 5 |
| blaSHV | |||
| Amplification and sequencing | |||
| 0S0(F) | CTCGCCTTTATCGGCCCTCAC | 85 | 4 |
| 0S5(R) | CGGCCACGCGGGTTAGCG | 1000 | 4 |
| Amplification and cloning | |||
| HIII-0S0(F) | CACACAAAGCTTGATGAAAAATGATGAAGGAAAAAAGAG | 8 | 4 |
| EI-0S5(R) | CACACAGAATTCGGTGGCCACGTTTATGGCGTTACCTTTGA | 1127 | 4 |
| Sequencing | |||
| 0S1(F) | GGACTACTCGCCGGTCAGC | 421 | 4 |
| 0S2(R) | GCTGACCGGCGAGTAGTCC | 439 | 4 |
| 0S3(F) | GATTGTCGCCCTGCTTTGG | 846 | 4 |
| 0S4(R) | CCAAGCAGGGCGACAATC | 863 | 4 |
| blaCTX-M-1 | |||
| Amplification and sequencing | |||
| CTX-MA | CGCTTTGCGATGTGCAG | 264 | 11 |
| CTX-MB | ACCGCGATATCGTTGGT | 814 | 11 |
| aac(6′)-I | |||
| Amplification | |||
| AAC6-I(F) | GTGACCAACAGCAACGATTCCG | 431 | This study |
| AAC6(R) | CCTCGATGGAAGGGTTAGGC | 999 | 22 |
| aac(3)-II | |||
| Amplification | |||
| AAC3-II(F) | ATATCGCGATGCATACGCGG | −8 | This study |
| AAC3-II(R) | GACGGCCTCTAACCGGAAGG | 869 | This study |
F, forward primers; R, reverse primers.
The HindIII and EcoRI restriction sites are underlined. The numbering of primers HIII-TEM(F), EI-TEM(R), HIII-0S0(F), and EI-0S5(R) begins at the base indicated by a double underline.
The primer positions are given as the first 5′ base according to the numbering of Sutcliffe (45) for blaTEM, Mercier and Lévêsque (33) for blaSHV (33), Bonnet et al. (11) for blaCTX-M-1, GenBank accession number M21682 for aac(6′)-I, and Vliegenthart et al. (47) for aac(3)-II.
Sequence analysis.
The PCR products were purified by using microcolumns of the Microspin Sephacryl S-400 purification system (Amersham Biosciences); and both strands were sequenced by using sets of custom-made specific primers (Eurogentec, Angers, France), an automated fluorescent method based on dye terminator chemistry (AmpliTaq DNA polymerase FS Dye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems Division, Perkin-Elmer), and the ABI Prism 310 sequencer (Applied Biosystems Division, Perkin-Elmer). For each cloned PCR product, the sequences of three independent clones were determined to avoid Taq polymerase errors during the amplification step.
Cloning experiments.
The entire nucleotide sequences coding for TEM or SHV β-lactamases were cloned after PCR amplification by using oligonucleotides carrying restriction sites at their ends, that is, the HindIII site in the forward primers and the EcoRI site in the reverse primers (Table 2). The digested amplification products were then ligated into the pBK-CMV cloning vector (Stratagene-Europe/BIOCREST, Amsterdam, The Netherlands) cut with the same enzymes. The ligation mixture was used to electrotransform E. coli DH5α cells, which were subsequently plated on medium containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml).
RAPD and ERIC-PCR typing.
The epidemiological relationships between multiple strains belonging to the same species were analyzed by random amplified polymorphic DNA (RAPD) analysis with primer AP12h (3, 16) and enterobacterial repetitive intergenic consensus (ERIC) PCR (ERIC-PCR) with primers ERIC2 and ERIC1R (16). The amplification conditions were as follows: 94°C for 5 min and 45 subsequent cycles of 1 min at 94°C, 1 min at 37°C, and 1 min at 72°C, with a final step at 72°C for 10 min.
Nucleotide sequence accession number.
The nucleotide sequence of blaSHV-44 is available in the GenBank database under accession number AY259119.
RESULTS
Distribution of ESBL producers.
The 39 ESBL producers detected among the 2,599 isolates of the family Enterobacteriaceae (1.5%) consisted of 34 of 1,015 (3.3%) strains from institutions, including 23 of 887 (2.6%) strains from clinics and 11 of 128 strains (8.6%) from NHs, and 5 of 1,584 (0.3%) strains from the community, with all of the strains from the community being from urine samples (5 of 1,432 [0.3%] urine samples) (Table 1). Most strains from clinics were provided by a single center (clinic 1 [CLI-1]; 11 of 23 [47.8%] strains), as was the case for the majority of strains from NHs (NH-1; 8 of 11 [72.7%] strains) (Table 1). These strains were collected from 38 patients, including 33 residents of 11 institutions (8 of 19 [42.1%] clinics and 3 of 18 [16.7%] NHs) and 5 ambulatory individuals. The isolates were mainly recovered from urine (28 samples), but they were also recovered from the respiratory tract (8 samples) and pus (3 samples, including 2 bedsores) (Table 3). ESBL producers mainly comprised 19 of 39 (49%) E. aerogenes isolates, 8 of 39 (21%) K. pneumoniae isolates, 6 of 39 (15%) E. coli isolates, and 6 of 39 (15%) isolates of miscellaneous species (3 P. mirabilis isolates and 1 isolate each of Serratia marcescens, Morganella morganii, and Providencia stuartii) (Table 3).
TABLE 3.
Characteristics of ESBL-producing enterobacteria isolated in general practice
| Species (total no. of isolates) | Total no. (%) of ESBL-producing isolates | Molecular typea | Locationb,c | Specimenc | Plasmid profile | β-Lactamase contentd | Antibiotypee |
|---|---|---|---|---|---|---|---|
| E. coli (1,920) | 6 (0.3) | Ec1 | C-1 | Urine | A-1 | TEM-24b | KTNtA CHL SSS TMP |
| Ec2 | NH-1 | Urine | B | TEM-21, TEM-1 | KGTNt(A) CHL SSS TMP TET NAL | ||
| Ec3 | NH-1 | Urine | B | TEM-21 TEM-1 | KGTNt(A) CHL SSS TMP TET | ||
| Ec4 | NH-1 | Urine | B | TEM-21 | KGTNt(A) CHL SSS NAL | ||
| Ec5 | C-2 | Urine | B | TEM-21, TEM-1 | KGTNt(A) CHL SSS TET OFX | ||
| Ec6 | CLI-4 | Urine | NDf | CTX-M-1, TEM-1 | CHL TET SSS | ||
| P. mirabilis (208) | 3 (1.4) | Pm1 (2) | NH-1 (2) | Pus (2) | B | TEM-21 | KGTNt(A) CHL SSS TET |
| Pm2 | NH-1 | Urine | B | TEM-21, TEM-1 | KGTNt(A) CHL SSS TET FOF OFX | ||
| K. pneumoniae (104) | 8 (7.7) | Kp1 | CLI-1 | Urine | A-2 | TEM-24b, SHV-1 | KTNt(A) CHL SSS TMP (TET) |
| Kp2 | NH-2 | Urine | A-3 | TEM-24b, SHV-1 | KTNt(A) SSS TMP NAL | ||
| Kp2 | NH-2 | Urine | A-3 | TEM-24b, SHV-1 | KTNt(A) CHL SSS TMP (TET) OFX | ||
| Kp3 | NH-1 | Urine | B | TEM-21, SHV-1 | KGTNt(A) CHL SSS | ||
| Kp4 | NH-1 | Urine | B | TEM-21, SHV-1 | KGTNt(A) CHL SSS | ||
| Kp5 | CLI-2 | Urine | ND | TEM-19, SHV-1 | KGTNt(A) SSS (TET) NAL | ||
| Kp6 | CLI-5 | Respiratory tract | ND | TEM-15, SHV-44, SHV-1, TEM-1 | GTNt SSS TMP NAL | ||
| Kp7 | C-3 | Urine | ND | TEM-15, SHV-4, SHV-1 | KTNtA CHL SSS TMP TET OFX | ||
| E. aerogenes (48) | 19 (39.6) | Ea1 (11) | CLI-1 (6), CLI-3 (2), CLI-6, NH-3, C-4 | Urine (6), respiratory tract (4), pus (1) | A-1 | TEM-24b, AmpC | KTNt(A) CHL SSS TMP (TET) OFX |
| Ea1 | CLI-2 | Urine | A-1 | TEM-24b, AmpC | KTNt(A) CHL SSS TMP (TET) OFX MOX | ||
| Ea1 (3) | CLI-1, CLI-2, CLI-6 | Respiratory tract (2), urine | A-2 | TEM-24b, AmpC | KTNt(A) CHL SSS TMP (TET) OFX | ||
| Ea1 | C-5 | Urine | A-2 | TEM-24b, AmpC | KTNt(A) CHL SSS TMP (TET) OFX MOX IPM | ||
| Ea1 | CLI-1 | Urine | A-4 | TEM-24b, AmpC | KTNt(A) CHL SSS (TET) OFX MOX IPM | ||
| Ea1 | CLI-1 | Respiratory tract | A-5 | TEM-24b, AmpC | KTNt(A) SSS (TET) OFX | ||
| Ea2 | CLI-8 | Urine | ND | TEM-3, AmpC | KTNt(A) CHL SSS TMP (TET) OFX | ||
| S. marcescens (41) | 1 (2.4) | Sm | CLI-4 | Urine | A-4 | TEM-24b, AmpC | KTNt(A) CHL SSS TMP TET OFX |
| M. morganii (36) | 1 (2.8) | Mm | CLI-1 | Urine | A-2 | TEM-24b, AmpC | KTNt(A) CHL SSS TMP TET OFX |
| P. stuartii (24) | 1 (4.2) | Ps | CLI-7 | Urine | A-1 | TEM-24b | KGTNt(A) CHL SSS TMP TET |
A single number was assigned according to the concordant results obtained by three molecular typing methods (RAPD with primer AP12h and ERIC-PCR with primers ERIC2 and ERIC1R).
The different locations are NHs, clinics (CLI), and the community (C).
The number of isolates is given in parentheses if more than one isolate was recovered.
AmpC and SHV-1, species-specific cephalosporinase and SHV-1-like chromosomal penicillinase, respectively.
The resistance cotransferred with the ESBL(s) is indicated in boldface. G, K, T, Nt, and A, gentamicin, kanamycin, tobramycin, netilmicin, and amikacin, respectively. CHL, chloramphenicol; SSS, sulfamethoxazole; TMP, trimethoprim; TET, tetracycline; NAL, nalidixic acid; OFX, ofloxacin; FOF, fosfomycin, MOX, moxalactam; IPM, imipenem. Parentheses indicate a low level of resistance.
ND, not determined.
Epidemiological typing.
The strains were first compared with regard to their antibiotic resistance phenotypes (Table 3). Then, in order to type the isolates by a molecular method, the 36 strains of E. coli, P. mirabilis, K. pneumoniae, and E. aerogenes were analyzed by RAPD analysis (with primer AP12h) and ERIC-PCR (with primers ERIC2 and ERIC1R). The results obtained with these three primers were strictly concordant, allowing assignment of the three molecular types obtained for each strain to a single number in Table 3. Furthermore, these profiles were different from those of two clinically unrelated control strains belonging to the same species, validating the results of our molecular typing method. The results showed that the E. coli isolates were distinct by antibiotyping as well as by molecular typing (molecular types Ec1 to Ec6). Among the three P. mirabilis isolates collected from different patients living in the same NH (NH-1), two strains with the same antibiotype gave identical RAPD and ERIC-PCR profiles (type Pm1), in contrast to the remaining strain (type Pm2), which additionally expressed a TEM-1 β-lactamase and resistance to fosfomycin and fluoroquinolone. The patterns of all except two K. pneumoniae strains differed; two isolates from the same patient exhibited profile Kp2, and the isolates were considered nonduplicates because of differences in chloramphenicol, tetracycline, and quinolone resistance. Conversely, two strains with different fingerprints (types Kp3 and Kp4) showed the same antibiotype. Fifteen multiresistant E. aerogenes isolates had identical antibiotypes, while four strains exhibited different patterns, with differences in susceptibility to chloramphenicol, trimethoprim, moxalactam, and imipenem (Table 3). They had two fingerprints: either the Ea1 profile (18 isolates, including the 4 strains with distinct antibiotypes) or the Ea2 profile (1 strain producing a TEM-3 enzyme) (Fig. 1). It is noteworthy that the Ea1 profile was identical to the profiles of two isolates belonging to the prevalent clone described in France (12).
FIG. 1.
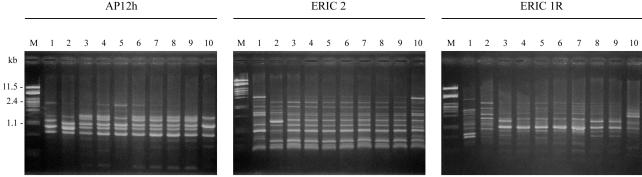
RAPD and ERIC-PCR fingerprints of E. aerogenes strains. The profiles obtained with primers AP12h, ERIC2, and ERIC1R are shown. Lanes 1 and 2, unrelated strains of E. aerogenes used as controls; lanes 3 and 4, the TEM-24b-producing E. aerogenes strain belonging to the clone responsible for the present epidemic in France; lanes 5 to 9, representative TEM-24-producing strains (profile Ea-1); lane 10, TEM-3-producing strain (profile Ea-2); lanes M, molecular weight marker (lambda phage DNA digested with PstI).
β-Lactamase characterization.
The β-lactamase contents of the strains were first analyzed by isoelectric focusing. According to the pIs of the β-lactamases suspected to be ESBLs, PCR experiments were performed with primers specific for different classes of β-lactamases (TEM, SHV, or CTX-M), and the amplification products were directly sequenced. For strains containing several enzymes belonging to the same type (TEM or SHV), mating assays with E. coli K-12 Nalr Rifr as the recipient were carried out. In two cases (i.e., for strains with molecular types Ec2 and Kp7), the coexisting enzymes could not be separated by conjugation experiments, and PCR amplification followed by cloning and sequencing of the amplification products was undertaken.
Seven varieties of ESBLs were characterized by this procedure. The TEM derivatives were widely predominant since they were present in almost all strains (38 of 39). They were distributed into five different TEM types (Table 3): TEM-24 encoded by the blaTEM-24b subtype gene (26), which differs from the blaTEM-24a subtype by a silent mutation (T682C); TEM-21; TEM-15; TEM-19; and TEM-3. The TEM-24 enzyme was present in 18 strains of E. aerogenes and 7 other TEM-24-producing isolates belonging to other species (1 E. coli isolate, 3 K. pneumoniae isolates, and 1 isolate each of S. marcescens, M. morganii, and P. stuartii) (Table 3). The TEM-21 enzyme was expressed by nine strains (four E. coli strains, three P. mirabilis strains, and two K. pneumoniae strains). Two strains of K. pneumoniae contained TEM-15, another one synthesized TEM-19, and the remaining strain of E. aerogenes produced the TEM-3 ESBL (Table 3). The single SHV-type ESBL was an SHV-4 enzyme produced by a K. pneumoniae strain (40). Likewise, one strain of E. coli that appeared to be highly resistant to cefotaxime but susceptible to ceftazidime produced a CTX-M-1 β-lactamase (8). In addition, the derepressed chromosomal cephalosporinase (AmpC) was visible by isoelectric focusing in E. aerogenes, S. marcescens (pI > 8.0), and M. morganii (pI 7.1) strains; and the SHV-1-like chromosomal penicillinase (pI 7.6) was visible by isoelectric focusing in K. pneumoniae strains. Moreover, seven strains simultaneously elaborated several β-lactamases: one K. pneumoniae strain (molecular type Kp7) exhibited two ESBLs (TEM-15 and SHV-4), and another strain of K. pneumoniae (molecular type Kp6) contained a new plasmid-encoded SHV-1 variant named SHV-44 (http://www.lahey.org/studies/webt.htm) which is associated with a TEM-15 ESBL. Four of the six E. coli strains and one of the three P. mirabilis strains possessed an ESBL in combination with the TEM-1 β-lactamase (pI 5.4).
The nucleotide sequence of the gene for the SHV-44 enzyme (pI 7.0) differed from that of the blaSHV-1 gene (http://www.lahey.org/studies/webt.htm) by four mutations, at positions 729 (G→T), 832 (G→A), 889 (T→C), and 913 (C→G), according to the numbering of Mercier and Lévêsque (33). The first nucleotide change led to an amino acid variation, i.e., R205L (according to the ABL numbering of Ambler et al. [1]). To assess the effect of this substitution on resistance to broad-spectrum cephalosporins, the blaSHV-1 and blaSHV-44 genes were cloned under the same conditions described above into the pBK-CMV vector, and the recombinant plasmids were transformed into E. coli DH5α. All amino acid sequences and promoter regions except the mutation concerned (i.e., R205L) were verified to be isogenic. The MICs for SHV-44- and SHV-1-producing transformants were not significantly different (ceftazidime, 0.047 and 0.025 μg/ml, respectively; cefotaxime, 0.375 and 0.187 μg/ml, respectively; aztreonam, 0.1875 and 0.1875 μg/ml, respectively; and cefepime, 0.094 and 0.047 μg/ml, respectively), indicating that the leucine at position 205 did not significantly expand the enzyme's spectrum of activity.
ESBL cotransfer of antibiotic resistance and plasmid restriction analysis.
Mating assays allowed the transfer of ESBLs from all strains except K. pneumoniae strains of molecular type Kp7. Conjugation frequencies varied between 10−5 and 10−8 transconjugants per donor cell, with the lowest rates of transfer obtained for TEM-24 and the highest ones obtained for TEM-21. In all TEM-24- and TEM-3-producing transconjugants, the phenotype of resistance to kanamycin, tobramycin, netilmicin, and amikacin was associated with the presence of the aac(6′)-I gene, as demonstrated by PCR amplification (data not shown); sulfamethoxazole resistance also was always transferred, whereas chloramphenicol resistance and trimethoprim resistance were lacking in four and three transconjugants, respectively (Table 3). Plasmids from TEM-24-producing transconjugants, which all carried the genes for the cotransfer of resistance mentioned above, could be divided into two profiles: the most frequent profile (profile A-1) was found in 12 E. aerogenes isolates and 1 E. coli isolate; and the other one (profile A-2) was observed in 4 E. aerogenes isolates, 1 K. pneumoniae isolate, and 1 M. morganii isolate. Both profiles were similar since they consisted of at least five common EcoRI fragments (Fig. 2). Furthermore, the two strains belonging to the prevalent clone harbored either the A-1 or the A-2 profile. The transconjugants lacking chloramphenicol and/or trimethoprim resistance (two E. aerogenes isolates, two K. pneumoniae isolates, one S. marcescens isolate, and one P. stuartii isolate) also possessed similar plasmids with profiles A-3 to A-5. The transconjugant of the P. stuartii strain, however, which lacked chloramphenicol resistance, had the predominant profile, profile A-1. All TEM-21-expressing transconjugants contained both the aac(6′)-I and the aac(3)-II genes (according to PCR amplifications; data not shown), which together conferred the phenotype of resistance to gentamicin, kanamycin, tobramycin, netilmicin, and amikacin; chloramphenicol resistance and sulfamethoxazole resistance were also cotransferred. Plasmid analysis revealed an identical profile, profile B (Fig. 2). However, transformation experiments were necessary for two transconjugant plasmids (from molecular types Ec4 and Ec5) due to the simultaneous transfer of two plasmids. Noteworthy was the fact that the transconjugant plasmid from molecular type Ec2 gave profile B (Fig. 2, lane 14), although it contained an additional blaTEM-1 gene. The resistance phenotype cotransferred with β-lactamase TEM-19 was the same as that associated with the TEM-21 enzyme, except for chloramphenicol resistance. The blaTEM-15 gene found in two strains of K. pneumoniae was located either on a nonconjugative plasmid (molecular type Kp7) or on two transferable plasmids (molecular type Kp6). Indeed, in the latter strain, blaTEM-15 was found either alone or in association with the blaSHV-44 gene. Finally, chloramphenicol, tetracycline, and sulfonamide resistance was cotransferred with the CTX-M-1 enzyme with but not with the TEM-1 β-lactamase.
FIG. 2.
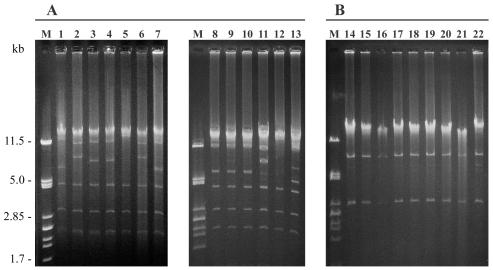
Plasmid profile analysis of TEM-24b-producing (A) and TEM-21-producing (B) members of the family Enterobacteriaceae. EcoRI restriction plasmid patterns were obtained from transconjugants or transformants. Lane 1, E. coli strain of molecular type Ec1 (profile A-1); lane 2, K. pneumoniae strain of molecular type Kp1 (profile A-2); lanes 3 and 4, K. pneumoniae strain of molecular type Kp2 (profiles A-3); lane 5, S. marcescens (profile A-4); lane 6, M. morganii (profile A-2); lane 7, P. stuartii (profile A-1); lanes 8 to 13, TEM-24-producing E. aerogenes strain of molecular type Ea1 divided into profile A-1 (lanes 8 to 10), profile A-2 (lane 11), profile A-4 (lane 12), and profile A-5 (lane 13); lanes 14 to 22, TEM-21-producing strains with the B profile. Lanes M, molecular weight marker (lambda phage DNA digested with PstI).
DISCUSSION
In a previous survey performed in 1999 in the Aquitaine region of France, ESBL-producing members of the family Enterobacteriaceae represented 1.5% of all strains collected in general practice; but 3.3% of the strains were from institutions, whereas 0.3% of all strains or strains from urine were from the community. The prevalences in private health care centers were similar to those found at the same time in French hospitals (2.2 to 3.3%) (18, 39, 44), and the prevalences in the community were in agreement with those in the literature (0.2 and 0.6% of all isolates and urine isolates from the community, respectively) (25, 39). However, there was a great disparity between the frequencies of ESBL-producing strains in clinics (2.6%) and NHs (8.6%). While ESBL producers were encountered in 42.1% of the clinics and 16.7% of the NHs, most of the strains were recovered from one large clinic with several surgical wards, a geriatric department, and an ICU (CLI-1) and one NH which serves primarily as a long-term residence for patients, the majority of whom are highly dependent elderly patients (NH-1), suggesting the existence of outbreaks in these establishments.
ESBL producers were mainly E. aerogenes (39.6%), as is observed at present in France (18, 39, 44), in contrast to other countries, where K. pneumoniae generally prevails (24). Of the 19 E. aerogenes isolates, 18 contained the same TEM-24b enzyme, and 16 of these came from six institutions (five clinics, including 9 strains from CLI-1, and 1 from an NH) and 2 came from the community. Such observations suggest the sporadic or epidemic occurrence of a strain which has disseminated in France since the early 1990s (12, 16) and more recently in a neighboring country, Belgium (19). Actually, the RAPD and ERIC-PCR typing methods, which have already been successfully used in previous studies (3, 17, 31), have confirmed that all of these 18 E. aerogenes isolates were identical to the clone prevalent throughout France and abroad. The extrahospital diffusion of this strain had previously been suspected, since patients were demonstrated to harbor it on admission to hospital (12), but the present study provides evidence for such an assumption. This clone probably possesses selective factors responsible for a strong capacity for adaptation and geographic dissemination (12). Most TEM-24b enzymes were encoded by a comparably large conjugative plasmid associated with the cotransfer of resistance to aminoglycosides (kanamycin, tobramycin, netilmicin, and amikacin), chloramphenicol, sulfamethoxazole, and trimethoprim, similar to the prevalent clone. The lack of transfer of chloramphenicol and/or trimethoprim resistance in two clonal E. aerogenes strains might result from plasmid DNA rearrangements or the loss of resistance-encoding transposons (42); such events may or may not lead to a modification of the restriction plasmid profile. Furthermore, seven strains belonging to different species of the family Enterobacteriaceae also produced the TEM-24b enzyme, encoded by identical or similar plasmids (profiles A-1 to A-4). These strains came from four clinics (including two strains in CLI-1), one NH, and the community. They probably resulted from the interspecies horizontal transfer of the plasmid carrying the TEM-24b-encoding gene. Indeed, plasmids with TEM-24b-encoding genes have previously been demonstrated to spread in vivo very easily among the Enterobacteriaceae (34). Thus, an outbreak occurred in CLI-1, mainly due to strain dissemination and occasionally to plasmid transfer. This is the first outbreak reported in a French clinic.
Nine strains belonging to different species of the Enterobacteriaceae produced a TEM-21 ESBL. Of these strains, eight were collected from patients living in the same NH (NH-1). The gene responsible for the ESBL production phenotype conjugated at high frequencies together with genes for resistance to aminoglycosides (gentamicin, kanamycin, tobramycin, netilmicin, and amikacin), chloramphenicol, and sulfamethoxazole. Furthermore, all TEM-21-encoding plasmids gave identical profiles, although one of them probably acquired an additional TEM-1-encoding transposon (42). The results of both conjugation and plasmid analysis strongly suggested epidemics cause by plasmid transfer in this NH. Very few outbreaks of ESBL-producing Enterobacteriaceae have been described in the NH setting (48), probably because they are not looked for as systematically in NHs as they are in the hospital setting. However, NHs have been identified as an important reservoir of ESBL-producing Enterobacteriaceae (36). The use of broad-spectrum oral antibiotics, often associated with poor infection control practices, may facilitate this dissemination (36). Furthermore, this environment is considered a microbiologic extension of the hospital environment, and patients are likely to be readmitted to the hospital, where they can serve as a source of these resistant microorganisms for other patients (27, 36). It is noteworthy that the TEM-21 enzyme has previously been described in an M. morganii strain in the University of Bordeaux Hospital (46) and in a P. aeruginosa strain in NH-1 (21).
Five strains produced the much less frequently occurring ESBLs TEM-3, TEM-15, TEM-19, SHV-4, and CTX-M-1 (three K. pneumoniae strains and one strain each of E. coli and E. aerogenes). These isolates were scattered in three clinics, one NH, and the community. In the early 1990s, SHV-4-producing K. pneumoniae strains were highly prevalent in France (2) and in the Aquitaine University Hospital (6, 9). A single SHV-4-producing K. pneumoniae strain was found in the present study, reflecting the decline of this enzyme in our region. On the other hand, a single TEM-3 enzyme synthesized by an E. aerogenes strain was also found, although this ESBL was recently observed in 36.7% of ESBL producers in France (18), showing the great geographic diversity of the distribution of ESBLs. Until now, the TEM-15 and TEM-19 ESBLs have rarely been encountered in France (30, 43), but they have recently been reported in a neighboring country, Italy, principally in P. mirabilis and Klebsiella oxytoca (41). Likewise, only one CTX-M-type ESBL was isolated in our study, but a recent report suggests the establishment and diffusion of CTX-M-encoding plasmids in Europe (23). It is difficult to speculate on the origins of these ESBL-producing strains, but it is noteworthy that TEM-19 derives from TEM-1A by a single mutation and therefore may have arisen under antibiotic selection pressure.
Our experimental approach for the detection of multiple β-lactamase-encoding genes in the same isolate allowed precise assessment of the contents of the strains. By this procedure, one strain of K. pneumoniae was found to produce two ESBLs (TEM-15 and SHV-4) and another one produced an ESBL (TEM-15) together with the SHV-44 enzyme. Furthermore, several of the strains investigated in this study produced both an ESBL and a TEM-1 β-lactamase. The production of such a combination was found in two-thirds of the E. coli strains tested and one-third of the P. mirabilis strains tested. Thus, despite the presence of a huge TEM reservoir, only a low proportion of E. coli strains producing TEM-type ESBLs (0.3%) were generated, as noted previously (18). The SHV-1 variant SHV-44 was defined by a single substitution (R205L), which modified the pI (7.0 instead of 7.6). The effect of this mutation on the enzyme's spectrum of activity has previously been analyzed only indirectly by comparing SHV-2 (G238S) and SHV-3 (R205L, S238G) (35), and it was concluded that the mutation results in a small increase in the level of ceftazidime resistance (37). We report here for the first time the isolation of a clinical strain harboring the R205L variant of SHV-1, and we have shown that this mutation alone does not enhance the resistance to broad-spectrum cephalosporins enough to consider SHV-44 an ESBL.
Of the five patients in the community harboring ESBL-producing strains of the Enterobacteriaceae, four were infected with a TEM-24b- or a TEM-21-producing strain, and the fifth one was infected with an SHV-4- and TEM-15-producing isolate, i.e., ESBL-producing strains that were simultaneously or previously responsible for epidemics in health care centers in our region. Among the isolates tested, one TEM-24b-producing E. aerogenes strain was imipenem resistant, as was another strain from CLI-1. The TEM-24b- and TEM-21-producing E. coli strains carried the same plasmids as other strains producing the same enzymes in the institutions. Moreover, the nucleotide sequences of the ESBL-encoding genes were identical to those of the isolates collected in clinics or NHs. Finally, all five patients in the community had been hospitalized in regional health care centers during the year preceding the isolation of the ESBL producer. Thus, even if the domestic emergence of ESBLs is possible (27), these data together strongly suggest that in our study the patients in the community acquired the ESBL-producing strains from a nosocomial (hospital, clinic, or NH) source.
In conclusion, this study shows that a variety of ESBLs and ESBL producers are present in the extrahospital setting. Most of the ESBL-producing members of the family Enterobacteriaceae were TEM-24b-producing E. aerogenes strains, representative of the clone responsible for the present epidemic in France, and TEM-21-producing Enterobacteriaceae strains were essentially found in only one NH. Outbreaks due to these ESBL-producing organisms by strain and/or plasmid dissemination were demonstrated in a clinic and an NH. The spread of ESBL-producing organisms to the community seems to be related to previous nosocomial acquisition. These data emphasize the need for private laboratories to adequately screen for ESBL-producing strains of the family Enterobacteriaceae that may appear to be falsely susceptible to broad-spectrum cephalosporins while the infections caused by these organisms are not efficiently treated with these antibiotics.
Acknowledgments
We thank Davin-Regli, who provided us with the two TEM-24-producing strains of E. aerogenes belonging to the prevalent French clone.
This work was supported by grants from the French Network on β-Lactamase Study and from the Ministère de l'Education Nationale et de la Recherche (grant EA-525), Université de Bordeaux 2, Bordeaux, France.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlet, G., M. Rouveau, I. Casin, P. J. Bouvet, P. H. Lagrange, and A. Philippon. 1994. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 β-lactamase and which were isolated in 14 French hospitals. J. Clin. Microbiol. 32:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arpin, C., C. Coze, A. M. Rogues, J. P. Gachie, C. Bebear, and C. Quentin. 1996. Epidemiological study of an outbreak due to multidrug-resistant Enterobacter aerogenes in a medical intensive care unit J. Clin. Microbiol. 34:2163-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arpin, C., R. Labia, C. Andre, C. Frigo, Z. El Harrif, and C. Quentin. 2001. SHV-16, a β-lactamase with a pentapeptide duplication in the omega loop. Antimicrob. Agents Chemother. 45:2480-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arpin, C., R. Labia, V. Dubois, P. Noury, M. Souquet, and C. Quentin. 2002. TEM-80, a novel inhibitor-resistant β-lactamase in a clinical isolate of Enterobacter cloacae. Antimicrob. Agents Chemother. 46:1183-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arpin, C., A. M. Rogues, S. Kabouche, G. Boulard, C. Quesnel, J. P. Gachie, and C. Quentin. 2000. Prospective survey of colonization and infection caused by SHV-4 producing Klebsiella pneumoniae in a neurosurgical intensive care unit. Epidemiol. Infect. 124:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barthélémy, M., J. Peduzzi, and R. Labia. 1988. Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1). Biochem. J. 251:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bermudes, H., C. Arpin, F. Jude, Z. El-Harrif, C. Bebear, and C. Quentin. 1997. Molecular epidemiology of an outbreak due to extended-spectrum β-lactamase-producing enterobacteria in a French hospital. Eur. J. Clin. Microbiol. Infect. Dis. 16:523-529. [DOI] [PubMed] [Google Scholar]
- 10.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnet, R., C. Dutour, J. L. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosi, C., A. Davin-Regli, C. Bornet, M. Mallea, J. M. Pages, and C. Bollet. 1999. Most Enterobacter aerogenes strains in France belong to a prevalent clone. J. Clin. Microbiol. 37:2165-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormican, M., D. Morris, G. Corbett-Feeeney, and J. Flynn. 1998. Extended spectrum β-lactamase production and fluoroquinolone resistance in pathogens associated with community acquired urinary tract infection. Diagn. Microbiol. Infect. Dis. 32:317-319. [DOI] [PubMed] [Google Scholar]
- 15.Courvalin, P., F. Goldstein, A. Philippon, and J. Sirot. (ed.). 1985. In L'antibiogramme. MPC-Vidéom ed. Paris, France.
- 16.Davin-Regli, A., D. Monnet, P. Saux, C. Bosi, R. Charrel, A. Barthelemy, and C. Bollet. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J. Clin. Microbiol. 34:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davin-Regli, A., P. Saux, C. Bollet, F. Gouin, and P. De Micco. 1996. Investigation of outbreaks of Enterobacter aerogenes colonisation and infection in intensive care units by random amplification of polymorphic DNA. J. Med. Microbiol. 44:89-98. [DOI] [PubMed] [Google Scholar]
- 18.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and The French Study Group. 2000. A 1998 survey of extended-spectrum β-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Gheldre, Y., M. J. Struelens, Y. Glupczynski, P. De Mol, N. Maes, C. Nonhoff, H. Chetoui, C. Sion, O. Ronveaux, and M. Vaneechoutte. 2001. National epidemiologic surveys of Enterobacter aerogenes in Belgian hospitals from 1996 to 1998. J. Clin. Microbiol. 39:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Moüy, D., J. D. Cavallo, R. Fabre, E. Garrabe, F. Grobost, M. Armengaud, R. Labia, and les Membres de l'AFORCOPIBIO. 1997. Les entérobactéries isolées d'infections urinaires en pratique de ville: étude AFORCOPIBIO 1995. Med. Mal. Infect. 27:642-645. [Google Scholar]
- 21.Dubois, V., C. Arpin, P. Noury, and C. Quentin. 2002. Clinical strain of Pseudomonas aeruginosa carrying a blaTEM-21 gene located on a chromosomal interrupted TnA type transposon. Antimicrob. Agents Chemother. 46:3624-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutour, C., R. Bonnet, H. Marchandin, M. Boyer, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gniadkowski, M. 2001. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin. Microbiol. Infect. 7:597-608. [DOI] [PubMed] [Google Scholar]
- 25.Goldstein, F. W., and the Multicentre Study Group. 2000. Antibiotic susceptibility of bacterial strains isolated from patients with community-acquired urinary tract infections in France. Eur. J. Clin. Microbiol. Infect. Dis. 19:112-117. [DOI] [PubMed] [Google Scholar]
- 26.Goussard, S., and P. Courvalin. 1999. Updated sequence information for TEM β-lactamase genes. Antimicrob. Agents Chemother. 43:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heseltine, P. 2000. Has resistance spread to the community? Clin. Microbiol. Infect. 6:11-16. [DOI] [PubMed] [Google Scholar]
- 28.Hryniewicz, K., K. Szczypa, A. Sulikowska, K. Jankowski, K. Betlejewska, and W. Hryniewicz. 2001. Antibiotic susceptibility of bacterial strains isolated from urinary tract infections in Poland. J. Antimicrob. Chemother. 47:773-780. [DOI] [PubMed] [Google Scholar]
- 29.Lucet, J. C., D. Decré, A. Fichelle, M. L. Joly-Guillou, M. Pernet, C. Deblangy, M. J. Kosmann, and B. Régnier. 1999. Control of a prolonged outbreak of extended-spectrum β-lactamase-producing Enterobacteriaceae in a university hospital. Clin. Infect. Dis. 29:1411-1418. [DOI] [PubMed] [Google Scholar]
- 30.Mabilat, C., and P. Courvalin. 1990. Development of “oligotyping” for characterization and molecular epidemiology of TEM β-lactamases in members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 34:2210-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mammeri, H., G. Laurans, M. Eveillard, S. Castelain, and F. Eb. 2001. Coexistence of SHV-4- and TEM-24-producing Enterobacter aerogenes strains before a large outbreak of TEM-24-producing strains in a French hospital. J. Clin. Microbiol. 39:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Members of the SFM Antibiogram Committee. 2003. Comité de l'Antibiogramme de la Société Francaise de Microbiologie report 2003. Int. J. Antimicrob. Agents 21:364-391. [DOI] [PubMed] [Google Scholar]
- 33.Mercier, J., and R. C. Lévêsque. 1990. Cloning of SHV-2, OHIO-1, and OXA-6 β-lactamases and cloning and sequencing of SHV-1 β-lactamase. Antimicrob. Agents Chemother. 34:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuwirth, C., E. Siebor, A. Pechinot, J. M. Duez, M. Pruneaux, F. Garel, A. Kazmierczak, and R. Labia. 2001. Evidence of in vivo transfer of a plasmid encoding the extended-spectrum β-lactamase TEM-24 and other resistance factors among different members of the family Enterobacteriaceae. J. Clin. Microbiol. 39:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolas, M. H., V. Jarlier, N. Honore, A. Philippon, and S. T. Cole. 1989. Molecular characterization of the gene encoding SHV-3 β-lactamase responsible for transferable cefotaxime resistance in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 33:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolle, L. E., L. J. Strausbaugh, and R. A. Garibaldi. 1996. Infections and antibiotic resistance in nursing homes. Clin. Microbiol. Rev. 9:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nüesch-Inderbinen, M. T., H. Hächler, and F. H. Kayser. 1995. New system based on site-directed mutagenesis for highly accurate comparison of resistance levels conferred by SHV β-lactamases. Antimicrob. Agents Chemother. 39:1726-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paterson., D. L. 2001. Extended-spectrum β-lactamases: the European experience. Curr. Opin. Infect. Dis. 14:697-701. [DOI] [PubMed] [Google Scholar]
- 39.Péan, Y., F. W. Goldstein, M. L. Guerrier, F. De Bels, and les Membres de VIGIL'ROC. 1999. Sensibilité aux β-lactamines des bactéries isolées en ville et à l'hôpital au cours d'une enquête multicentrique française. Antibiotiques 1:165-170. [Google Scholar]
- 40.Péduzzi, J., M. Barthélémy, K. Tiwari, D. Mattioni, and R. Labia. 1989. Structural features related to hydrolytic activity against ceftazidime of plasmid-mediated SHV-type CAZ-5 β-lactamase. Antimicrob. Agents Chemother. 33:2160-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perilli, M., E. Dell'Amico, B. Segatore, M. R. de Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sirot, D., C. De Champs, C. Chanal, R. Labia, A. Darfeuille-Michaud, R. Perroux, and J. Sirot. 1991. Translocation of antibiotic resistance determinants including an extended-spectrum β-lactamase between conjugative plasmids of Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 35:1576-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sirot, D., C. Recule, E. B. Chaibi, L. Bret, J. Croize, C. Chanal-Claris, R. Labia, and J. Sirot. 1997. A complex mutant of TEM-1 β-lactamase with mutations encountered in both IRT-4 and extended-spectrum TEM-15, produced by an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 41:1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirot, J., M. H. Nicolas-Chanoine, H. Chardon, J. L. Avril, C. Cattoen, J. C. Croix, H. Dabernat, T. Fosse, J. C. Ghnassia, E. Lecaillon, A. Marmonier, M. Roussel-Delvallez, C. J. Soussy, A. Trevoux, F. Vandenesch, C. Dib, N. Moniot-Ville, and Y. Rezvani. 2002. Susceptibility of Enterobacteriaceae to β-lactam agents and fluoroquinolones: a 3-year survey in France. Clin. Microbiol. Infect. 8:207-213. [DOI] [PubMed] [Google Scholar]
- 45.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322 Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tessier, F., C. Arpin, A. Allery, and C. Quentin. 1998. Molecular characterization of a TEM-21 β-lactamase in a clinical isolate of Morganella morganii. Antimicrob. Agents Chemother. 42:2125-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vliegenthart, J. S., P. A. Ketelaar-van Gaalen, and J. A. van de Klundert. 1989. Nucleotide sequence of the aacC2 gene, a gentamicin resistance determinant involved in a hospital epidemic of multiply resistant members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 33:1153-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiener, J., J. P. Quinn, P. A. Bradford, R. V. Goering, C. Nathan, K. Bush, and R. A. Weinstein. 1999. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 281:517-523. [DOI] [PubMed] [Google Scholar]


