Abstract
A total of 88 salmonella isolates (72 clinical isolates for which the ciprofloxacin MIC was >0.06 μg/ml, 15 isolates for which the ciprofloxacin MIC was ≤0.06 μg/ml, and Salmonella enterica serotype Typhimurium ATCC 13311) were studied for the presence of genetic alterations in four quinolone resistance genes, gyrA, gyrB, parC, and parE, by multiplex PCR amplimer conformation analysis. The genetic alterations were confirmed by direct nucleotide sequencing. A considerable number of strains had a mutation in parC, the first to be reported in salmonellae. Seven of the isolates sensitive to 0.06 μg of ciprofloxacin per ml had a novel mutation at codon 57 of parC (Tyr57→Ser) which was also found in 29 isolates for which ciprofloxacin MICs were >0.06 μg/ml. Thirty-two isolates had a single gyrA mutation (Ser83→Phe, Ser83→Tyr, Asp87→Asn, Asp87→Tyr, or Asp87→Gly), 34 had both a gyrA mutation and a parC mutation (29 isolates with a parC mutation of Tyr57→Ser and 5 isolates with a parC mutation of Ser80→Arg). Six isolates which were isolated recently (from 1998 to 2001) were resistant to 4 μg of ciprofloxacin per ml. Two of these isolates had double gyrA mutations (Ser83→Phe and Asp87→Asn) and a parC mutation (Ser80→Arg) (MICs, 8 to 32 μg/ml), and four of these isolates had double gyrA mutations (Ser83→Phe and Asp87→Gly), one parC mutation (Ser80→Arg), and one parE mutation (Ser458→Pro) (MICs, 16 to 64 μg/ml). All six of these isolates and those with a Ser80→Arg parC mutation were S. enterica serotype Typhimurium. One S. enterica serotype Typhi isolate harbored a single gyrA mutation (Ser83→Phe), and an S. enterica serotype Paratyphi A isolate harbored a gyrA mutation (Ser83→Tyr) and a parC mutation (Tyr57→Ser); both of these isolates had decreased susceptibilities to the fluoroquinolones. The MICs of ciprofloxacin, levofloxacin, and sparfloxacin were in general the lowest of those of the six fluoroquinolones tested. Isolates with a single gyrA mutation were less resistant to fluoroquinolones than those with an additional parC mutation (Tyr57→Ser or Ser80→Arg), while those with double gyrA mutations were more resistant.
Salmonellosis remains a major public health problem worldwide. In contrast to gastroenteritis caused by salmonellae, invasive salmonellosis requires prompt antibiotic therapy. The antibiotics used for the treatment of salmonellosis have traditionally been chloramphenicol, sulfamethoxazole-trimethoprim, and ampicillin. However, with the development of resistance to these drugs, fluoroquinolones have been used and have been successful in treating many clinical cases. Unfortunately, fluoroquinolone-resistant strains have rapidly developed in recent years, and treatment failures have been reported (2, 8, 13, 20, 37, 44, 47).
Resistance to the fluoroquinolones in salmonellae has mainly been attributed to mutations in the gyrA gene. Mutations have rarely been reported in the gyrB gene, and none have been reported in the parC gene (7, 12, 36, 48, 51). Other resistance mechanisms such as efflux and reduced uptake of drugs have also been demonstrated (9, 10, 22).
All salmonellae in Hong Kong had remained susceptible to the fluoroquinolones (26-29). However, we saw a trend of increasing MICs (from 0.03 to 2 μg/ml) of fluoroquinolones for salmonellae isolated from 1993 to 1998 (unpublished observation). In 1998, we isolated the first fluoroquinolone-resistant salmonella strain in the Prince of Wales Hospital in Hong Kong. Clinical isolates of salmonellae with significant fluoroquinolone resistance (MICs, ≥8 μg/ml) remain extremely rare worldwide (36). It is important to understand the underlying mechanisms that cause high-level resistance in salmonellae and how they differ from those of the low-level resistance phenotypes in order to elucidate factors associated with increases in resistance to fluoroquinolones among these organisms. We therefore studied mutations in the gyrase and topoisomerase IV genes that lead to fluoroquinolone resistance in clinical salmonella isolates and the association of such mutations with resistance phenotypes and serotypes.
MATERIALS AND METHODS
Bacterial strains.
A total of 2,348 salmonella isolates (71% of all salmonella isolates recovered) recovered from patients in the Prince of Wales Hospital from 1990 to 2001 were tested for their susceptibilities to the fluoroquinolones. Of these, 72 (3%) that were resistant to 0.06 μg of ciprofloxacin per ml were studied. Fifteen isolates for which ciprofloxacin MICs were ≤0.06 μg/ml and which were isolated throughout the study period and randomly selected and a standard strain of Salmonella enterica serotype Typhimurium (ATCC 13311) were included as sensitive controls.
Antimicrobial susceptibility testing.
Screening for resistance to 0.06 μg of ciprofloxacin per ml was performed by an agar dilution method (32) by using a breakpoint concentration of 0.06 μg/ml. The MICs of six fluoroquinolones, ciprofloxacin, ofloxacin, sparfloxacin, levofloxacin, moxifloxacin, and gemifloxacin, for isolates resistant to 0.06 μg of ciprofloxacin per ml and the 15 isolates for which the ciprofloxacin MICs were ≤0.06 μg/ml were then determined by the agar dilution method (32). The geometric mean MICs (GMMs) were calculated by the formula
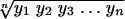 |
where y represents the individual MIC and n is the number of MICs used in the calculation.
MPAC analysis.
Total DNA was extracted by suspending a few overnight colonies in 0.5 ml of double-distilled water and heating the mixture at 100°C for 10 min. The extracted DNA was subjected to amplification by PCR with primers specific for the quinolone resistance-determining regions of gyrA, gyrB, parC, and parE (Table 1). Amplification was performed in a thermal cycler (PTC-100; MJ Research, Watertown, Mass.) by using the following protocol: an initial denaturation step at 95°C for 5 min, followed by 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 52°C, and 1 min of extension at 72°C, with a final extension of 5 min at 72°C.
TABLE 1.
Primers used for PCR and their nucleotide positions and sizes of amplified products
| Gene | Primer (nucleotide position)
|
Product size (bp) | |
|---|---|---|---|
| Forward | Backward | ||
| gyrAa | 5′-TGTCCGAGATGGCCTGAAGC-3′ (108-127) | 5′-TACCGTCATAGTTATCCACG-3′ (454-435) | 347 |
| gyrAa,b | 5′-TACACCGTCGCGTACTTTACGCCA-3′ (131-154) | 5′-CCATCAGTTCGTGGGCGATTTTCG-3′ (406-383) | 275 |
| gyrBc | 5′-CAAACTGGCGGACTGTCAGG-3′ (1215-1234) | 5′-TTCCGGCATCTGACGATAGA-3′ (1560-1541) | 345 |
| parCd | 5′-ATGAGCGATATGGCAGAGCG-3′ (14-33) | 5′-TGACCGAGTTCGCTTAACAG-3′ (426-407) | 412 |
| parEe | 5′-GACCGAGCTGTTCCTTGTGG-3′ (1285-1304) | 5′-AGCAGAGTAGCGATATGCAA-3′ (1557-1538) | 272 |
The PCR products were subjected to multiplex PCR amplimer conformation (MPAC) analysis by the method of McIlhatton et al. (31), with slight modifications. Briefly, the method used standard single-stranded conformational polymorphism analysis procedures, except that a reference PCR product, such as that of the ATCC 13311 control strain, was mixed with the test sample in equal quantities prior to analysis. Alternatively, two PCR products with an overlapping region amplified from the same test sample product were mixed prior to heat denaturation. MPAC analysis was found to greatly enhance the sensitivity of mutation detection. A denaturing solution containing 94% formamide, 0.05% xylene cyanol, and 0.04% bromophenol blue was added to the mixture, which was then heated at 95°C for 5 min, immediately placed on ice, and loaded onto 12.5% ExcelGel (Amersham Biosciences, Uppsala, Sweden). The samples were electrophoresed in a Multiphor II electrophoresis system (Amersham) at 15°C and 600 V for 80 min. The MPAC patterns were detected by silver staining (PlusOne silver staining kit; Amersham).
Direct nucleotide sequencing.
The amplified products were purified by using GFX columns (Amersham) and were then sequenced by using the Silver Sequence DNA sequencing system (Promega, Madison, Wis.). Approximately 120 fmol of purified PCR products and 4.5 pmol of sequencing primer were included in each reaction mixture. The sequencing primer was either the forward or the reverse primer used in the PCR. The sequencing reaction conditions were the same as those used for the PCR, except that a total of 50 amplification cycles were used. The sequencing reaction products were analyzed on a 6% polyacrylamide gel (acrylamide-bisacrylamide [19:1], 7 M urea) at 1,700 V with an S2 sequencing apparatus (Life Technologies, Rockville, Md.). DNA bands were detected by silver staining (Promega).
Pulsed-field gel electrophoresis.
Isolates of the same serotypes were typed by pulsed-field gel electrophoresis, as described previously (28).
RESULTS
Of the 72 isolates that were resistant to 0.06 μg of ciprofloxacin per ml, 14 were S. enterica serotype Typhimurium, 12 were S. enterica serotype Enteritidis, 12 were S. enterica serotype Blockley, 10 were S. enterica serotype Virchow, and 1 to 3 isolates each were of 15 other serotypes (Table 2). Six isolates were shown to have similar or identical pulsed-field gel electrophoresis patterns. These comprised three S. enterica serotype Blockley isolates, two S. enterica serotype Enteritidis isolates, and one S. enterica serotype Typhimurium isolate. Thus, there were a total of 66 nonidentical isolates. Since the numbers were small, the total numbers of isolates are provided and the numbers of nonidentical isolates are indicated in parentheses in Table 2. The MIC ranges and GMMs of the six fluoroquinolones tested for these isolates are shown in Table 3. Only six isolates showed high-level resistance to the drugs (MICs, ≥8 μg/ml). Fifteen isolates for which ciprofloxacin MICs were 0.015 to 0.06 μg/ml and S. enterica serotype Typhimurium ATCC 13311 were included as sensitive controls.
TABLE 2.
Distribution of mutations in gyrA, parC, and parE genes in salmonellae in Hong Kong
| Gene, mutationa | Yr | No. of isolates
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group B
|
Group C
|
Group D
|
Othersd | Total | ||||||||
| Typhimurium | Otherb | Blockley | Braenderup | Haardt | Virchow | Otherc | Enteritidis | Typhi | ||||
| parC, Tyr57→Ser (ACC→AGC) | 2000 | 2 | 7 | |||||||||
| 2001 | 5 | |||||||||||
| gyrA, Ser83→Phe (TCC→TTC) | 1992 | 1 | 8 | |||||||||
| 1996 | 1 | |||||||||||
| 1998 | 1 | |||||||||||
| 2000 | 1 | |||||||||||
| 2001 | 1 | 2 | 1 | |||||||||
| gyrA, Ser83→Phe | 2001 | 1 (Agona) | 2 (Hadar) | 3 | ||||||||
| parC, Tyr57→Ser (ACC→AGC) | ||||||||||||
| gyrA, Ser83→Tyr (TCC→TAC) | 1998 | 1 | 1 | |||||||||
| gyrA, Ser83→Tyr | 2000 | 1 (Paratyphi A) | 1 | |||||||||
| parC, Tyr57→Ser | ||||||||||||
| gyrA, Asp87→Asn (GAC→AAC) | 1998 | 3 (2)e | 1 | 8 (7) | ||||||||
| 2000 | 1 | |||||||||||
| 2001 | 3 | |||||||||||
| gyrA, Asp87→Asn | 1997 | 4 (3) | 1 | 19 (16) | ||||||||
| parC, Tyr57→Ser | 1998 | 1 | 1 | |||||||||
| 1999 | 6 (4) | 1 (Rissen) | ||||||||||
| 2000 | 1 | 1 | ||||||||||
| 2001 | 1 (Derby) | 1 | 1 (Newport) | |||||||||
| gyrA, Asp87→Tyr (GAC→TAC) | 1994 | 1 | 11 | |||||||||
| 1995 | 1 | |||||||||||
| 1996 | 1 | |||||||||||
| 1998 | 2 | 1 | ||||||||||
| 1999 | 1 | 1 | ||||||||||
| 2000 | 1 | |||||||||||
| 2001 | 1 | 1 | ||||||||||
| gyrA, Asp87→Tyr | 2000 | 1 | 6 | |||||||||
| parC, Tyr57→Ser | 2001 | 2 (Indiana) | 2 | 1 | ||||||||
| gyrA, Asp87→Gly (GAC→GGC) | 2001 | 4 (2) | 4 (2) | |||||||||
| gyrA, Ser83→Phe | 1990 | 1 | 5 | |||||||||
| parC, Ser80→Arg (AGC→CGC) | 1992 | 1 | ||||||||||
| 1993 | 2 | |||||||||||
| 1995 | 1 | |||||||||||
| gyrA, Ser83→Phe, Asp87→Asn | 1998 | 1 | 2 | |||||||||
| parC, Ser80→Arg | 2001 | 1 | ||||||||||
| gyrA, Ser83→Phe, Asp87→Gly | 1998 | 1 | 4 | |||||||||
| parC, Ser80→Arg | 1999 | 1 | ||||||||||
| parE, Ser458→Pro (TCG→CCG) | 2000 | 2 | ||||||||||
| Total | 14 (13) | 16 | 12 (9) | 3 | 3 | 10 | 6 | 12 (10) | 1 | 2 | 79 (73) | |
Underscored nucleotides represent the mutated nucleotide.
Includes serotypes Agona, Derby, Indiana, Panama, Reading, and Stanley.
Includes serotypes Emek, Hadar, Newport, Rissen, and Thompson.
Includes serotypes Paratyphi A and Weltevreden.
The numbers of strains with different pulsed-field gel electrophoresis types are given in parentheses.
TABLE 3.
Correlation of gene mutations and antimicrobial susceptibilities
| Mutation | No. of strains | MIC (μg/ml)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin
|
Ofloxacin
|
Sparfloxacin
|
Levofloxacin
|
Moxifloxacin
|
Gemifloxacin
|
GMM for all isolates | ||||||||
| Range | GMM | Range | GMM | Range | GMM | Range | GMM | Range | GMM | Range | GMM | |||
| None | 9 | 0.015-0.12 | 0.03 | 0.06-0.25 | 0.10 | 0.0075-0.12 | 0.03 | 0.03-0.25 | 0.06 | 0.06-0.25 | 0.11 | 0.03-0.25 | 0.07 | 0.06 |
| Thr57→Ser (parC) | 7 | 0.06 | 0.06 | 0.25-0.5 | 0.28 | 0.06-0.25 | 0.13 | 0.06-0.25 | 0.12 | 0.12-0.25 | 0.23 | 0.03-0.25 | 0.10 | 0.13 |
| Asp87→Gly (gyrA) | 4 | 0.12-0.25 | 0.14 | 1 | 1.00 | 0.12-0.25 | 0.21 | 0.12-0.25 | 0.21 | 0.5-1 | 0.84 | 0.12 | 0.12 | 0.29 |
| Asp87→Asn (gyrA) | 8 | 0.12-0.5 | 0.21 | 0.5-1 | 0.65 | 0.0075-0.5 | 0.13 | 0.25-0.5 | 0.30 | 0.5-1 | 0.71 | 0.12-1 | 0.23 | 0.31 |
| Asp87→Tyr (gyrA) | 11 | 0.12-1 | 0.22 | 0.25-2 | 0.53 | 0.12-2 | 0.26 | 0.12-2 | 0.32 | 0.5-2 | 0.57 | 0.12-1 | 0.53 | 0.38 |
| Ser83→Tyr (gyrA) | 1 | 0.25 | 1 | 0.25 | 0.5 | 0.5 | 1 | 0.50 | ||||||
| Asp87→Asn (gyrA), Tyr57→Ser (parC), Ser83→Phe (gyrA) | 19 | 0.12-0.5 | 0.36 | 0.25-2 | 0.96 | 0.0075-2 | 0.36 | 0.12-1 | 0.52 | 0.5-2 | 1.08 | 0.12-2 | 0.67 | 0.60 |
| 8 | 0.25-1 | 0.35 | 0.5-2 | 0.92 | 0.25-1 | 0.55 | 0.25-1 | 0.55 | 0.25-2 | 0.65 | 0.12-4 | 0.99 | 0.63 | |
| Ser83→Phe (gyrA) Ser80→Arg(parC) | 5 | 0.5 | 0.50 | 1 | 1.00 | 0.5-1 | 0.66 | 0.5 | 0.50 | 1 | 1.00 | 0.5-1 | 0.76 | 0.71 |
| Ser83→Phe (gyrA) Tyr57→Ser (parC) | 3 | 0.25-2 | 0.50 | 1-2 | 1.26 | 0.5-1 | 0.79 | 0.5-1 | 0.63 | 1 | 1.00 | 0.25-4 | 0.79 | 0.79 |
| Asp87→Tyr (gyrA) Tyr57→Ser (parC) | 6 | 0.25-1 | 0.50 | 0.5-2 | 1.26 | 0.5-1 | 0.63 | 0.25-2 | 0.63 | 0.5-2 | 1.12 | 0.25-4 | 0.89 | 0.79 |
| Ser83→Tyr (gyrA) Tyr57→Ser (parC) | 1 | 0.5 | 2 | 1 | 1 | 2 | 0.5 | 1.00 | ||||||
| Ser83→Phe and Asp87→Asn (gyrA), Ser80→Arg (parC) | 2 | 8-16 | 11.30 | 16 | 16.00 | 8-16 | 11.31 | 8 | 8.00 | 16 | 16.00 | 16-32 | 22.63 | 13.45 |
| Ser83→Phe and Asp87→Gly (gyrA), Ser80→Arg (parC), Ser458→Pro (parE) | 4 | 16-32 | 19.03 | 32-64 | 38.05 | 32-64 | 45.25 | 16 | 16.00 | 32-64 | 45.25 | 32-64 | 45.25 | 32.55 |
A total of 122 mutations in gyrA, parC, and parE were found in these 72 isolates. No gyrB mutations were detected. Seven of the 16 fluoroquinolone-sensitive isolates also harbored a single mutation each. The most common mutation found in 36 isolates (28% of mutations) was at codon 57 of parC, where a C→G transversion led to substitution of serine for tyrosine (Tyr57→Ser) (Table 2). Twenty-nine of these had an additional gyrA mutation. The next most common mutation was at codon 87 of gyrA, in which a G→A transversion led to substitution of asparagine for aspartate (Asp87→Asn) and which was found in 29 isolates (22% of mutations), with 19 isolates having an additional parC mutation (Tyr57→Ser) and 2 isolates having two additional mutations, one at gyrA (Ser83→Phe) and another at parC (Ser80→Arg). A mutation at the same codon involving a G→T transversion leading to a tyrosine substitution was found in 17 isolates. The mutation at codon 83 (C→T transversion) resulting in Ser83→Phe was found in 22 isolates, with 11 isolates having one to three additional mutations. Asp87→Gly and Ser83→Tyr changes in gyrA were found in eight and two isolates, respectively. Thirty-four isolates had double mutations, 32 isolates had one mutation, and 2 isolates had three mutations. Four isolates had four mutations, two in gyrA (Ser83→Phe, Asp87→Gly) and one each in parC (Ser80→Arg) and parE (Ser458→Pro).
The ciprofloxacin MICs for all isolates without any mutations and those with a single parC mutation (Tyr57→Ser) were ≤0.06 μg/ml, although the GMMs of the six fluoroquinolones tested and the overall GMMs were two- to more than fourfold higher for isolates with the parC mutation (Table 3). The MICs for isolates with a single mutation at codon 87 of gyrA were about twofold higher than those for isolates with a single parC mutation (Tyr57→Ser). The MICs for the isolate with Ser83→Tyr ranged from 0.25 to 1 μg/ml, and the GMM was 0.5 μg/ml, which was similar to that for isolates with Asp87→Asn plus Tyr57→Ser or Ser83→Phe mutations. A parC mutation in addition to a gyrA mutation caused isolates to be more resistant, with the effect of Tyr57→Ser being slightly more pronounced than that of Ser80→Arg (Table 3). An additional Tyr57→Ser mutation caused the MICs for the isolate with the Ser83→Tyr mutation to be twofold higher (GMMs, 0.5 versus 1.0 μg/ml).
Other than the ciprofloxacin GMMs for isolates with a single parC mutation (Tyr57→Ser), the ciprofloxacin GMMs for isolates with a single gyrA mutation at either codon 83 or codon 87 were the lowest (0.14 to 0.35 μg/ml), with the MIC range being 0.12 to 1 μg/ml (Table 3). The GMMs of sparfloxacin and levofloxacin for these isolates were 0.13 to 0.55 μg/ml, with the GMMs of ofloxacin being the highest (0.53 to 1 μg/ml). For isolates with more than two mutations (n = 6), the MICs of ciprofloxacin were 8 to 32 μg/ml, the levofloxacin MICs were up to 16 μg/ml, and the MICs of the other four fluoroquinolones tested were up to 64 μg/ml.
Mutations appeared as early as 1990. A double mutation (Ser83→Phe in gyrA and Ser80→Arg in parC) was found only in strains isolated from 1990 to 1995. Instead, the gyrA mutation (Ser83→Phe) was found in strains isolated from 1992 to 2001, whereas Asp87→Tyr was first detected in 1994 and Asp87→Asn was first detected in 1997. Triple mutations (Ser83→Phe and Asp87→Asn of gyrA, Ser80→Arg of parC) and quadruple mutations (Ser83→Phe and Asp87→Gly of gyrA, Ser80→Arg of parC, Ser458→Pro of parE) appeared only in recent years (1998 to 2001). Similarly, the parC mutation (Tyr57→Ser) was detected only after 1997.
S. enterica serotypes Typhimurium, Enteritidis, Blockley, and Virchow were the serotypes in which mutations were most commonly found (14 to 19%). The parC mutation Tyr57→Ser was not detected in S. enterica serotype Typhimurium but was detected in the rarer serotypes (Table 2). In contrast, all except one of the isolates resistant to 4 μg of the six fluoroquinolones tested per ml were S. enterica serotype Typhimurium; the exception was an S. enterica serotype Agona strain. S. enterica serotypes Typhi and Paratyphi A had decreased susceptibilities to the fluoroquinolones, with MICs ranging from 0.12 μg/ml for gemifloxacin to 0.5 μg/ml for ofloxacin and from 0.5 μg/ml for ciprofloxacin and gemifloxacin to 2 μg/ml for ofloxacin and moxifloxacin, respectively.
DISCUSSION
Mutations in gyrA.
An increase incidence of clinical salmonella isolates with reduced susceptibilities to the fluoroquinolones (i.e., ciprofloxacin MICs ranging from 0.12 to 2 μg/ml) has been seen in recent years (16, 30, 42). Although they are susceptible to the NCCLS breakpoint for susceptible strains (32), treatment failures have been encountered when fluoroquinolones are used to treat infections caused by these strains (5, 15, 46). A number of mechanisms of fluoroquinolone resistance in different bacterial species have been suggested, and the most common ones are mutations in the gyrA gene, usually at either codon 83 or 87, and/or the parC gene (6, 12, 34, 36, 39, 49). The mutations that we found in the gyrA region of our isolates (Ser83→Phe; Ser83→Tyr; Asp87→Gly, Asn, or Tyr) have been reported previously (12, 14, 36). Ser83 is suggested to be an important site for fluoroquinolone resistance (12, 23, 33, 39, 49). This is supported by our results, in that of the five single gyrA mutations, the presence of Ser83→Phe was associated with the highest level of resistance (GMMs, 0.63 versus ≤0.5 μg/ml) (Table 3). Other mutations in gyrA that have been reported include Ser83→Leu (6, 14); Gly81→Asp, Gly81→Cys, and Asp82→Gly (43, 49); Asp87→His (6); Asp87→Tyr (6, 12, 14); and Ala119→Glu (12). All of these mutations conferred low-level resistance to fluoroquinolones. It appears that a gyrA mutation alone, regardless of the mutation type, may not contribute to a resistance level greater than 2 μg of ciprofloxacin per ml. Instead, strains with double amino acid changes in GyrA were more resistant than those with single amino acid changes in GyrA (4, 49).
Mutations in parC.
ParC and ParE are two subunits of topoisomerase IV, with the former being homologous with GyrA and the latter being homologous with GyrB (21, 35, 40). High-level fluoroquinolone resistance was found to be due to both parC and gyrA mutations (13). Other workers (4, 14, 24) have reported on the parC mutation causing the substitution Ser80→Arg found in this study in organisms other than salmonellae. This mutation caused only slight decreases in the susceptibilities of the isolates when it was present together with one other gyrA mutation. Recently, it has been suggested that Glu84 of parC plays an important role in topoisomerase IV-DNA interactions and that a mutation at this site appears to have more deleterious effects than one at Ser80 (17). That the resistance levels of the isolates were elevated to >4 μg of ciprofloxacin per ml in the presence of two gyrA mutations (Ser83→Phe, Asp87→Asn) was probably due to the gyrA mutations rather than the parC mutation (4). Thus, Ser80→Arg did not appear to play an important role in conferring resistance.
The Thr57→Ser mutation in parC detected in many of our isolates in this study has not been reported previously. Besides being the first report of a parC mutation in salmonellae, it is also the first report of a single parC mutation in gram-negative bacteria without a gyrA mutation and in isolates for which ciprofloxacin MICs are <0.12 μg/ml. In general, mutations in parC are rarer in gram-negative bacteria and usually arise later than gyrA mutations, probably because in these organisms, gyrase rather than topoisomerase IV is the primary target of fluoroquinolones (38, 41). Hence, changes to gyrA mostly precede those to parC (6), as there is probably less selective advantage to single mutations in parC. Other mutations in parC that have been reported in other organisms include Gly78→Asp (14); Asp79→Ala (3); Ser80→Ile (4, 6, 24, 45); and Glu84→Gly and Glu84→Lys (3, 4, 6, 14, 24). Further studies on other resistance mechanisms such as alterations in membrane permeability, changes in influx or efflux, etc., are required to evaluate the contribution of parC mutations to fluoroquinolone resistance in salmonellae.
Mutations in gyrB.
We did not detect gyrB mutations in our salmonella isolates. This is not surprising, as gyrB mutations remain extremely rare in most bacterial species, even among highly resistant strains, although they have been reported in salmonellae (7, 13). It may be useful to investigate whether the GyrB protein, along with essential regions of the GyrA, ParC, and ParE proteins where amino acid changes are not tolerated, represents a good target for future drug designs. Alternatively, the importance of amino acids 83 and 87 of the GyrA protein in fluoroquinolone resistance may infer that drugs targeting the altered GyrA proteins can be helpful in eliminating fluoroquinolone-resistant strains.
Mutations in parE.
While most workers detected mutations in gyrA in highly fluoroquinolone-resistant strains of Escherichia coli, salmonellae, and Shigella spp., mutations in gyrB and parE are rare (3, 6, 9, 12, 25). We detected a mutation in parE (Ser458→Pro) that has not been reported in salmonellae. It is likely that this mutation might not be directly responsible for high-level resistance but rather increased the resistance levels in isolates already harboring two gyrA mutations and one parC mutation. Gensberg and colleagues (7) detected a gyrB mutation in S. enterica serotype Typhimurium (Ser463→Tyr). Breines et al. (1) reported a parE mutation (Leu445→His) in E. coli, and González et al. (11) reported a Pro424→Gln mutation in viridans group streptococci. It could be postulated that amino acids in the region from positions 463 to 468 in gyrB and those in the region from positions 424 to 460 of parE played a role in conferring fluoroquinolone resistance.
Sequential development of mutations in topoisomerase genes.
Our results are in agreement with the observations of other workers that the gyrA mutations Ser83→Phe and Asp87→Tyr were the first to develop in salmonellae (12, 39), since they were first detected in our isolates in 1990 and 1994, respectively. Subsequently, other mutations accumulated, including the parE mutation (Ser458→Pro), so that triple and quadruple mutations were observed after 1998. It is interesting that the parC mutation (Tyr57→Ser) was also a recent development.
Gene mutations and Salmonella serotypes.
S. enterica serotype Typhimurium was the serotype in which mutations were first detected. It was also the serotype that had more than two mutations and that did not have the parC mutation (Tyr57→Ser). In contrast, other than the Tyr57→Ser (parC) mutation, mutations in both parC and parE were not found in salmonellae other than S. enterica serotype Typhimurium. This was probably due to the minimal effect that this mutation has on fluoroquinolone resistance, which was rarely seen in salmonellae other than S. enterica serotype Typhimurium. However, it would be difficult to explain the absence of this mutation in S. enterica serotype Typhimurium. The development of decreased fluoroquinolone susceptibilities in S. enterica serotypes Typhi and Paratyphi A is a cause for concern. Hirose and colleagues (18) also recently detected gyrA mutations in S. enterica serotype Typhi and Paratyphi strains that were less susceptible to the fluoroquinolones. Continuous surveillance must be carried out to monitor the development of fluoroquinolone resistance in these organisms.
Effects of mutations on fluoroquinolone susceptibility.
All mutations had the least, although an important, effect on susceptibility to ciprofloxacin, as the GMMs of ciprofloxacin were usually the lowest of those of all drugs tested, followed by those of levofloxacin, while the effect on susceptibility to ofloxacin was most prominent, as the ofloxacin GMMs were frequently the highest of those among the six fluoroquinolones tested, closely followed by the GMMs of moxifloxacin. Since fluoroquinolones preferentially bind to different target regions of gyrase or topoisomerase IV (19, 41, 50, 52), it can be postulated that fluoroquinolones with binding properties similar to those of ciprofloxacin would be most effective even against organisms with mutated genes.
We have also tested the susceptibilities of the isolates to nalidixic acid. The nalidixic acid GMM for isolates without any mutation was 6 μg/ml and that for isolates with a single parC mutation (Tyr57→Ser) was 11 μg/ml, while the nalidixic acid MICs for all other isolates were >128 μg/ml (results not shown). Isolates with a novel parC mutation (Tyr57→Ser) were also less susceptible to the other fluoroquinolones than those without any mutations. The susceptibility to nalidixic acid and the other fluoroquinolones therefore might be a marker of low-level resistance to fluoroquinolones.
Conclusion.
This study has provided evidence that isolates with reduced susceptibilities to fluoroquinolones might be important in the clinical development of resistance, as they could become highly resistant upon sequential accumulation of target gene mutations. Further studies involving analysis of mutations in clonally related organisms or in mutants developed in vitro would be required to confirm this observation. Mutations in gyrA conferred low-level fluoroquinolone resistance, while addition of another gyrA mutation together with a parC and/or a parE mutation increased the resistance to a high level. The presence of gyrA, parC, and parE mutations in S. enterica serotype Typhimurium, the most common salmonella serotype resistant to high concentrations of fluoroquinolones, and the presence of gyrA mutations in S. enterica serotypes Typhi and Paratyphi are a serious concern and call for continuous monitoring of fluoroquinolone-resistant salmonellae.
Acknowledgments
This work was supported by a research grant (grant CUHK4049/00 M) from the Research Grants Council.
We thank GlaxoSmithKline (United Kingdom) for the gift of gemifloxacin, Hong Kong Medical Supplies Ltd. (Hong Kong) for levofloxacin, and Bayer AG (Wuppertal, Germany) for moxifloxacin.
REFERENCES
- 1.Breines, D. M., S. Ouabdesselam, E. Y. Ng, J. Tankovic, S. Shah, C. J. Soussy, and D. C. Hooper. 1997. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob. Agents Chemother. 41:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, N. M., M. R. Millar, J. A. Frost, and B. Rowe. 1994. Ciprofloxacin resistance in Salmonella paratyphi A. J. Antimicrob. Chemother. 33:1258-1259. [DOI] [PubMed] [Google Scholar]
- 3.Chu, Y.-W., E. T. S. Houang, and A. F. B. Cheng. 1998. Novel combination of mutations in the DNA gyrase and topoisomerase IV genes in laboratory-grown fluoroquinolone-resistant Shigella flexneri mutants. Antimicrob. Agents Chemother. 42:3051-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deguchi, T., A. Fukuoka, M. Yasuda, M. Nakano, S. Ozeki, E. Kanematsu, Y. Nishino, S. Ishihara, Y. Ban, and Y. Kawada. 1997. Alterations in the GyrA subunit of DNA gyrase and ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 41:699-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutta, P., U. Mitra, S. Dutta, A. De, M. K. Chatterjee, and S. K. Bhatta-charya. 2001. Ceftriaxone therapy in ciprofloxacin treatment failure in typhoid fever in children. Indian J. Med. Res. 113:210-213. [PubMed] [Google Scholar]
- 6.Everett, M. J., Y. F. Jin, V. Ricci, and L. J. V. Piddock. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gensberg, K., Y. F. Jin, and L. J. V. Piddock. 1995. A novel gyrB mutation in a fluoroquinolone-resistant clinical isolate of Salmonella typhimurium. FEMS Microbiol. Lett. 132:57-60. [DOI] [PubMed] [Google Scholar]
- 8.Gibb, A. P., C. S. Lewin, and O. J. Garden. 1991. Development of quinolone resistance in and multiple antibiotic resistance in Salmonella bovismorbificans in a pancreatic abscess. J. Antimicrob. Chemother. 28:318-321. [DOI] [PubMed] [Google Scholar]
- 9.Giraud, E., A. Brisabois, J. L. Martel, and E. Chaslus-Dancla. 1999. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43:2131-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González, I., M. Georgiou, F. Alcaide, D. Balas, J. Linares, and A. G. de la Campa. 1998. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents Chemother. 42:2792-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griggs, D. J., K. Gensberg, and L. J. V. Piddock. 1996. Mutations in gyrA of quinolone-resistant salmonella serotypes isolated from humans and animals. Antimicrob. Agents Chemother. 40:1009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heisig, P. 1993. High-level fluoroquinolone resistance in a Salmonella typhimurium isolate due to alterations in both gyrA and gyrB genes. J. Antimicrob. Chemother. 32:367-378. [DOI] [PubMed] [Google Scholar]
- 14.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40:879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helms, M., P. Vastrup, P. Gerner-Smidt, and K. Malbak. 2002. Excess mortality associated with antimicrobial drug-resistant Salmonella typhimurium. Emerg. Infect. Dis. 8:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heurtin-Le Corre, C., P.-Y. Donnio, M. Perrin, M.-F. Travert, and J.-L. Avril. 1999. Increasing incidence and comparison of nalidixic acid-resistant Salmonella enterica subsp. enterica serotype Typhimurium isolates from humans and animals. J. Clin. Microbiol. 37:266-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiasa, H. 2002. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry 41:11779-11785. [DOI] [PubMed] [Google Scholar]
- 18.Hirose, K., A. Hashimoto, K. Tamura, Y. Kawamura, T. Ezaki, H. Sagara, and H. Watanabe. 2002. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob. Agents Chemother. 46:3249-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houssaye, S., L. Gutmann, and E. Varon. 2002. Topoisomerase mutations associated with in vitro selection of resistance to moxifloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2712-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard, A. J., T. D. Joseph, L. L. Bloodworth, J. A. Frost, H. Chart, and B. Rowe. 1990. The emergence of ciprofloxacin resistance in Salmonella typhimurium. J. Antimicrob. Chemother. 26:296-298. [DOI] [PubMed] [Google Scholar]
- 21.Kato, J.-I., H. Suzuki, and H. Ikeda. 1992. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 267:25676-25684. [PubMed] [Google Scholar]
- 22.Kern, W. V., M. Oethinger, A. S. Jellen-Ritter, and S. B. Levy. 2000. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 44:814-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. H., E. H. Cho, K. S. Kim, H. Y. Kim, and Y. M. Kim. 1998. Cloning and nucleotide sequence of the DNA gyrase gyrA gene from Serratia marcescens and characterization of mutations in gyrA of quinolone-resistant clinical isolates. Antimicrob. Agents Chemother. 42:190-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumagai, Y., J.-I. Kato, K. Hoshino, T. Akasaka, K. Sato, and H. Ikeda. 1996. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob. Agents Chemother. 40:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linde, H.-J., F. Notka, M. Metz, B. Kochanowski, P. Heisig, and N. Lehn. 2000. In vivo increase in resistance to ciprofloxacin in Escherichia coli associated with deletion of the C-terminal part of MarR. Antimicrob. Agents Chemother. 44:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling, J. M., N. W. S. Lo, Y. M. Ho, K. M. Kam, C. H. Ma, S. C. Wong, and A. F. Cheng. 1996. Emerging resistance in Salmonella enterica serotype Typhi in Hong Kong. Int. J. Antimicrob. Agents 7:161-166. [DOI] [PubMed] [Google Scholar]
- 27.Ling, J. M., I. C. Koo, K. M. Kam, and A. F. Cheng. 1998. Antimicrobial susceptibilities and molecular epidemiology of Salmonella enterica serotype Enteritidis in Hong Kong from 1986 to 1996. J. Clin. Microbiol. 36:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling, J. M., I. C. Koo, and A. F. Cheng. 2001. Epidemiological analysis of Salmonella enterica serotype Typhimurium from Hong Kong by ribotyping and PFGE. Scand. J. Infect. Dis. 33:272-278. [DOI] [PubMed] [Google Scholar]
- 29.Ling, J. M., E. W. C. Chan, and A. F. Cheng. 2001. Molecular epidemiologic analysis of Salmonella enterica serotype Derby infections in Hong Kong. J. Infect. 42:145-153. [DOI] [PubMed] [Google Scholar]
- 30.Malbak, K., P. Gerner-Smidt, and H. C. Wegener. 2002. Increasing quinolone resistance in Salmonella enterica serotype Enteritidis. Emerg. Infect. Dis. 8:514-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIlhatton, B. P., C. Keating, M. D. Curran, M. F. McMullin, J. G. Barr, J. A. Madrigal, and D. Middleton. 2002. Identification of medically important pathogenic fungi by reference strand-mediated conformational analysis (RSCA). J. Med. Microbiol. 51:468-478. [DOI] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.Ouabdesselam, S., D. C. Hooper, J. Tankovic, and C. J. Soussy. 1995. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob. Agents Chemother. 39:1667-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouabdesselam, S., J. Tankovic, and C. J. Soussy. 1996. Quinolone resistance mutations in the gyrA gene of clinical isolates of Salmonella. Microb. Drug Resist. 2:299-302. [DOI] [PubMed] [Google Scholar]
- 35.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 36.Piddock, L. J. 2002. Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol. Rev. 26:3-16. [DOI] [PubMed] [Google Scholar]
- 37.Piddock, L. J. V., K. Whale, and R. Wise. 1990. Quinolone resistance in salmonella: clinical experience. Lancet 335:1459. [DOI] [PubMed] [Google Scholar]
- 38.Piddock, L. J. V., V. Ricci, I. McLaren, and D. J. Griggs. 1998. Role of mutation in the gyrA and parC genes of nalidixic acid-resistant salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41:635-641. [DOI] [PubMed] [Google Scholar]
- 39.Reyna, F., M. Huesca, V. Gonalez, and L. Y. Fuchs. 1995. Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob. Agents Chemother. 39:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Springer, A. L., and M. B. Schmig. 1993. Molecular characterization of the Salmonella typhimurium parE gene. Nucleic Acids Res. 21:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tankovic, J., R. Bachoual, S. Ouabdesselam, A. Boudjadja, and C. J. Soussy. 1999. In vitro activity of moxifloxacin against fluoroquinolone-resistant strains of aerobic gram-negative bacilli and Enterococcus faecalis. J. Antimicrob. Chemother. 43:19-23. [DOI] [PubMed] [Google Scholar]
- 42.Threlfall, E. J., L. R. Ward, and B. Rowe. 1999. Resistance to ciprofloxacin in non-typhoidal salmonellas from humans in England and Wales—the current situation. Clin. Microbiol. Infect. 5:130-134. [DOI] [PubMed] [Google Scholar]
- 43.Troung, Q. C., J.-C. N. Van, D. Shlaes, L. Gutmann, and N. J. Moreau. 1997. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob. Agents Chemother. 41:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasallo, F. J., P. Martin-Rabadan, L. Alcala, J. M. Garcia-Lechuz, M. Rodriguez-Creixems, and E. Bouza. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin. Infect. Dis. 26:535-536. [DOI] [PubMed] [Google Scholar]
- 45.Vila, J., J. Ruiz, P. Goni, and M. T. J. de Anta. 1996. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:491-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wain, J., N. T. Hoa, N. T. Chin, H. Vinh, M. J. Everett, T. S. Diep, N. P. J. Day, T. Solomon, N. J. White, L. J. V. Piddock, and C. M. Parry. 1997. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin. Infect. Dis. 25:1404-1410. [DOI] [PubMed] [Google Scholar]
- 47.Walker, R. A., A. J. Lawson, E. A. Lindsay, L. R. Ward, P. A. Wright, P. A. Bolton, F. J. Wareing, D. R. Corkish, R. H. Davies, and E. J. Threlfall. 2000. Decreased susceptibility to ciprofloxacin in outbreak associated multiresistant Salmonella typhimurium DT104. Vet. Rec. 147:395-396. [DOI] [PubMed] [Google Scholar]
- 48.Walker, R. A., N. Saunders, A. J. Lawson, E. A. Lindsay, M. Dassama, L. R. Ward, M. J. Woodward, R. H. Davies, E. Liebana, and E. J. Threlfall. 2001. Use of a LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype Typhimurium DT104 isolates. J. Clin. Microbiol. 39:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weigel, L. M., C. D. Steward, and F. C. Tenover. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weigel, L. M., G. J. Anderson, R. R. Facklam, and F. C. Tenover. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:3517-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiuff, C., M. Madsen, D. L. Baggesen, and F. M. Aarestrup. 2000. Quinolone resistance among Salmonella enterica from cattle broilers, and swine in Denmark. Microb. Drug Resist. 6:11-17. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, J., Y. Dong, X. Zhao, S. Lee, A. Amin, S. Ramaswamy, J. Domagala, J. M. Musser, and K. Drlica. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517-525. [DOI] [PubMed] [Google Scholar]


