Abstract
Using HeLa cells, we have developed methods to determine 1) the number of RNA polymerases that are active at any moment, 2) the number of transcription sites, and 3) the number of polymerases associated with one transcription unit. To count engaged polymerases, cells were encapsulated in agarose, permeabilized, treated with ribonuclease, and the now-truncated transcripts extended in [32P]uridine triphosphate; then, the number of growing transcripts was calculated from the total number of nucleotides incorporated and the average increment in length of the transcripts. Approximately 15,000 transcripts were elongated by polymerase I, and ∼75,000 were elongated by polymerases II and III. Transcription sites were detected after the cells were grown in bromouridine for <2.5 min, after which the resulting bromo-RNA was labeled with gold particles; electron microscopy showed that most extranucleolar transcripts were concentrated in ∼2400 sites with diameters of ∼80 nm. The number of polymerases associated with a transcription unit was counted after templates were spread over a large area; most extranucleolar units were associated with one elongating complex. These results suggest that many templates are attached in a “cloud” of loops around a site; each site, or transcription “factory,” would contain ∼30 active polymerases and associated transcripts.
INTRODUCTION
The human genome probably contains ∼70,000 genes, and there are also reasonably accurate estimates of the steady-state numbers of mRNA, hnRNA, and rRNA molecules in a mammalian cell (for reviews see Lewin, 1975; Miklos and Rubin, 1996). However, it has proved difficult to determine how many polymerases are engaged in transcription at any moment in each cell. (See Iyer and Struhl [1996] for measurements of transcription rates in yeast.) Three main approaches have been used with cell extracts; one involves a comparison of the rate of RNA synthesis with the rate given by a known number of pure enzyme molecules (Sugden and Keller, 1973), another the binding of radiolabeled γ-methyl-amanitin to polymerase II (Chambon, 1974), and a third the incorporation of [3H]uridine triphosphate (UTP) into the 3′-termini of nascent RNA (Cox, 1976). While these approaches have drawbacks, they gave roughly similar results, namely that 20,000–100,000 polymerases are active. (Sollner-Webb and Tower [1986], Geiduschek and Tocchini-Valentini [1988], and Zawel and Reinberg [1995] review different polymerases.)
These results have been brought into focus by the finding that nascent transcripts and polymerases in extranucleolar regions are concentrated in discrete sites ∼80 nm in diameter (Jackson et al., 1993; Wansink et al., 1993; Bregman et al., 1995; Iborra et al., 1996a; Fay et al., 1997). The number of sites is at least one order of magnitude less than the above estimates of the number of active polymerases. This raises several questions. Does each site contain many active polymerases engaged on one transcription unit? Then, only several thousand transcription units can be active at any moment, and tens of thousands of different messages can only accumulate if many previously active genes are switched off, as other genes are switched on. In other words, many so-called “active” genes must spend most of their time not being transcribed (e.g., Lewin, 1975; Wijgerde et al., 1995). Or, at the other extreme, are many transcription units each associated with one polymerase in each site? In this case, what confines those transcription units and polymerases to sites that occupy <0.5% nuclear volume, a volume that is much less than that occupied by active chromatin? One possibility is that active polymerases are bound to the site, but this is inconsistent with the idea that polymerases track along the template.
As these possibilities are at variance with commonly accepted views, we developed a suite of methods to determine the number of active polymerases and sites. We find ∼75,000 nascent transcripts are concentrated in only about 2400 extranucleolar sites, and that a typical transcription unit is associated with one active polymerase.
MATERIALS AND METHODS
Cell Growth, Encapsulation, Lysis, and Buffers
HeLa cells in suspension were grown, encapsulated in agarose, and lysed with saponin (100 μg/ml; 3 min; Sigma, Poole, Dorset) in a modified “physiological” buffer (PB; Jackson et al., 1988). PB is 100 mM potassium acetate, 30 mM KCl, 10 mM Na2HPO4, 1 mM MgCl2, 1 mM Na2ATP, 1 mM DTT, and 0.2 mM PMSF (pH 7.4). As the acidity of ATP batches varies, 100 mM KH2PO4 (≤1/100th volume) can be added to adjust the pH. PB* is PB plus human placental ribonuclease inhibitor (10 U/ml; Amersham Pharmacia, Little Chalfont, Bucks, United Kingdom). PB** is PB plus 10 mM β-glycerophosphate, 2.5 mM potassium pyrophosphate, 10 mM NaF, 0.1 mM Na3V04, 1 μg/ml aprotinin, 10 μg/ml bestatin, 2 μg/ml E-54, 0.5 μg/ml leupeptin and 0.5 μg/ml pepstatin, and KCl reduced to 17.5 mM. PBS+ is 1% acetylated-BSA (Sigma), 0.2% Tween 20 (Sigma), tRNA (5 μg/ml), and 25 U/ml ribonuclease inhibitor in PBS. All buffers used during/after lysis were ice-cold, unless stated otherwise.
Numbers of Nascent Transcripts
Cells were grown in [methyl-3H]thymidine (0.05 μCi/ml; ∼80 Ci/mmol; Amersham Pharmacia) for 20 h to label DNA uniformly and allow accurate quantification of cell number, and then encapsulated (107/ml), regrown (2 h), washed twice in PBS, and once in PB. Next, 107 cells in 10 ml PB were lysed, washed twice in PB, resuspended in 2.5 ml PB ± ribonuclease A (RNase A) (150 Kunitz U/ml; deoxyribonuclease [DNase]-free; Sigma), incubated (30 min at 4°C; 5 min at 25°C), washed once in PB* plus 200 μg/ml saponin, and washed five times with PB*. Then, 250 μl packed beads were resuspended in 450 μl PB*, various concentrations of unlabeled UTP (±inhibitors) were added, and the mixture was incubated for 10 min at 4°C, and then for 3 min at 33°C, before reactions (500 μl final volume) were initiated by addition of a mixture of triphosphates to give 100 μM ATP, cytidine triphosphate (CTP), and guanosine triphosphate (GTP), [32P]UTP (100 μCi/ml; ∼800 Ci/mmol), and a molarity of MgCl2 equal to that of added triphosphates (PB contains 1 mM MgCl2). (No RNase activity remained during elongation, as after RNase treatment, washing, 30 min elongation, and addition of 2.5 mM EDTA, transcript length remained unchanged for 30 min.) After various times at 33°C, the amount of radiolabel incorporated into acid-insoluble material was determined by scintillation counting (Jackson et al., 1988). Other samples were washed six times in PB*, diluted four times with 1 mM MgCl2, and incubated (10 min; 33°C) with 500 U/ml RNase-free DNase and 25 U/ml ribonuclease inhibitor. After addition of SDS to 0.2%, beads were melted (10 min; 75°C), and RNA was prepared using RNAzol B (BioGenesis, Poole, Dorset, United Kingdom). Dried RNA was dissolved in RNase-free water plus 10 U/ml ribonuclease inhibitor, and RNA from equal numbers of cells run on 6% polyacrylamide “denaturing” gels (Sambrook et al., 1989), before autoradiographic images were collected using a PhosphorImager (Molecular Dynamics; Chesham, Bucks, United Kingdom) and data exported into Microsoft Excel. Short transcripts were not lost preferentially when RNA was purified, as transcripts had similar sizes when samples were applied to gels immediately after melting without purifying RNA (our unpublished results). Direct application was not used routinely as the large volume means that fewer transcripts can be loaded per lane, and this gives lower autoradiographic intensities.
Each transcript has an unlabeled and labeled part, but only the latter contributes to intensity; therefore, weight averages were not directly converted to number averages (e.g., Igo-Kemenes and Zachau, 1977). Instead, we assume transcripts contain 15 unlabeled nucleotides (the minimum seen; e.g. Figure 1C); more complicated corrections were not deemed necessary, as values obtained using the method of Igo-Kemenes and Zachau (1977) differed by ≤20%. Then, numbers of molecules in each of ∼1000 divisions from bottom to top of the gel were corrected for the effect of unlabeled nucleotides, and the number average was calculated by integration. The total number of nascent transcripts/cell was calculated from cell number, nucleotides incorporated, and increments in chain length taking into account: cell viability and lysis (<5% and >97% cells stained with trypan blue respectively); reduction in polymerizing activity by RNase treatment (<10%; e.g. Figure 1B); ribosomal transcripts (containing 16% U; GenEmbl accession number U13369) constituted 35% of total, while other transcripts (30% U; Lewin, 1974) constituted the remainder (Figure 2A; mitochondrial transcripts neglected). Lines in Figure 1D and 2C were drawn using linear regression. Slopes at 90% confidence intervals had values of ±25% (Figure 1D), ±30% (pol II,III; Figure 2C), and ±40% (pol I; Figure 2C) of slopes drawn. Lines deviated from linearity in the reproducible way shown, presumably because some transcripts terminated (leading to overestimates in the range shown).
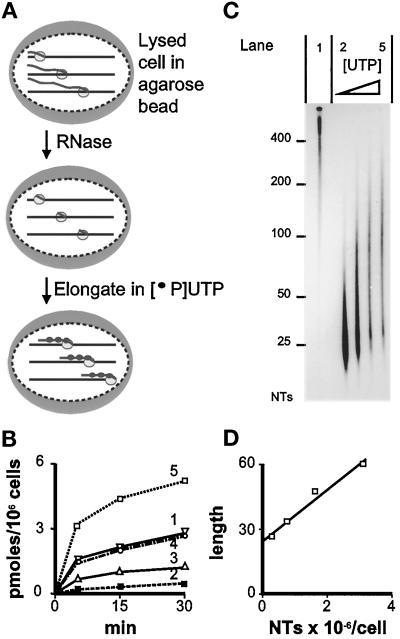
Figure 1.
The number of nascent transcripts in a HeLa cell. (A) Strategy. Cells are encapsulated in agarose (large gray oval) and lysed; three polymerases (small ovals) are shown extending nascent transcripts (wavy lines) in one cell. After RNase treatment, polymerases extend truncated transcripts in [32P]UTP plus various concentrations of UTP. The number of nascent transcripts is calculated from the total number of labeled nucleotides incorporated into all transcripts (determined as in Figure 1B), and the average increase in length of each transcript (determined as in Figure 1C). (B) Incorporation rates. Curve 1: No RNase treatment, extension in 1.125 μM UTP. Curves 2–5: RNase treated, extension in 0.125, 0.375, 1.125, or 2.625 μM UTP, respectively. (C) Autoradiograph of gel containing [32P]RNA. NTs: length in nucleotides. Lanes 1–5: 30 min samples from panel B, curves 1–5, respectively. RNA from equal numbers of cells is loaded in lanes 1–5; lanes 2–5 contain progressively fewer counts as the effects of reduced specific activity outweigh those of increased elongation. (D) The relationship between transcript length (number average in nucleotides) and the number of nucleotides (NTs) incorporated (from Figure 1, B and C).
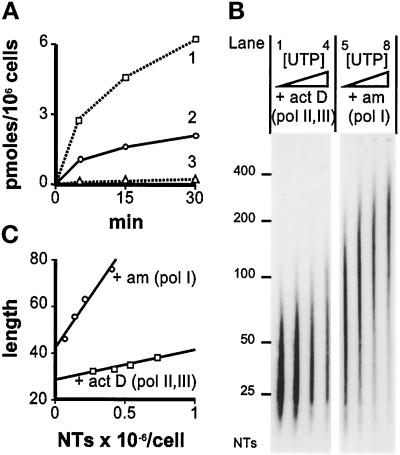
Figure 2.
Polymerase I transcripts. (A) Inhibitors affect incorporation (measured as in Figure 1B, using 2.5 μM UTP and no RNase treatment). Curve 1: no inhibitors added. Curve 2: 200 μg/ml α-amanitin (polymerases II + III inhibited). Curve 3: cells grown in actinomycin D (0.2 μg/ml; 15 min) before lysis, extension in 200 μg/ml α-amanitin (polymerases I, II, + III inhibited). (B) Autoradiographs of gels containing [32P]RNA from panel A. NTs: length in nucleotides. Lanes 1–4. Cells grown in actinomycin D (act D; 0.2 μg/ml; 15 min) before lysis, RNase treatment, and extension (5 min) in 0.225, 0.375, 0.625, or 1.125 μM UTP. Lanes 5–8. After RNase treatment, transcripts were extended (5 min) in 200 μg/ml α-amanitin (am) and 0.125, 0.225, 0.375 or 0.625 μM UTP; longer exposure than lanes 1–4. (C) The relationship between transcript length (number average in nucleotides) and the number of nucleotides (NTs) incorporated (from panels A and B).
A calculation used for Figure 1 is now given. Transcripts were elongated in different UTP concentrations for one time, as this reduces possible effects due to residual pools, DNA sequence, and transcript termination; similar results were obtained using one UTP concentration and different times (our unpublished results). Thus, cell numbers and radioactivity were counted; 100 μl cells contained 47,000 3H cpm, equivalent to 0.21 cpm/cell. After 30 min elongation in 0.125 μM [32P]UTP (100 μCi/ml), 100 μl elongation mixture contained 9900 and 9800 acid-insoluble 3H and 32P cpm, respectively, or 0.21 32P cpm/cell (normalized using 3H cpm/cell), equivalent to 0.12 pmol UMP/106 cells. As 65% transcripts are nucleoplasmic (with 30% U) and 35% are nucleolar (with 16% U), the average U content is 25%; therefore, 0.47 pmol nucleotide monophosphates/106 cells are incorporated, equivalent to 0.28 × 106 nucleotide monophosphates/cell (Figure 1B, curve 2, 30 min). The number-average length of such labeled transcripts (Figure 1C, lane 2) was 26.5 nucleotides. This length and nucleotide monophosphates/cell give the left-hand point in Figure 1D. After other points were obtained similarly, the slope of the resulting line shows that individual transcripts grow by 12 nucleotides as 106 nucleotides are incorporated/cell. Therefore, there are 83,000 (i.e. ∼80,000) nascent transcripts/cell.
The numbers of different polymerases were determined using drugs. Actinomycin D inhibits all polymerases to some degree; ≤0.1 μg/ml is commonly used to inhibit polymerase I by ≤90% (Perry and Kelley, 1970; Chambon, 1974). We require complete inhibition, as even slowly elongating polymerases generate labeled transcripts. Therefore, the minimum drug concentration required to eliminate nucleolar synthesis was determined by immunofluorescence. Cells were permeabilized, allowed to incorporate bromo-UTP (Br-UTP), and extracted to improve antibody access, and bromo-RNA (Br-RNA) was indirectly immunolabeled. Growth for 15 min in 0.2 μg/ml actinomycin D before permeabilization eliminated nucleolar fluorescence without detectable effect on nucleoplasmic labeling (our unpublished results). α-Amanitin (200 μg/ml) eliminated nucleoplasmic fluorescence without detectable effect on nucleolar labeling (our unpublished results; see also Weinmann et al., 1975).
Endogenous, native, nascent transcripts were sized, in experiments in which RNase treatment was omitted, after extension in 1 μM [32P]UTP (100 μCi/ml; 10 min), and electrophoresis on 0.8% agarose/formaldehyde (Sambrook et al., 1989).
Numbers of Transcription Sites
General procedures for postembedment labeling, stereology—and values for major/minor axes, nuclear/nucleolar axes, mean diameters and volumes—have been described (Iborra et al., 1996a). Cells were grown in bromouridine (Br-U), prefixed (10 min; 0°C) with 4% paraformaldehyde in 250 mM HEPES (pH 7.4), fixed (50 min; 20°C) with 8% paraformaldehyde, dehydrated, embedded, and Br-RNA on sections indirectly immunolabeled using protein A absorbed on to gold particles. After blocking nonspecific binding, sections were incubated successively with 1) monoclonal antibromodeoxyuridine (2 h; 10 μg/ml in PBS + Tween + BSA; Boehringer, Lewes, East Sussex, United Kingdom) that reacts with Br-RNA, 2) rabbit anti-mouse IgG (1 h; 1:50 dilution; Stratech, Luton, Bedfordshire, United Kingdom), 3) protein A absorbed on to 5/9 nm gold particles (1 h), and fixed with 2.5% glutaraldehyde. After uranyl acetate staining, sections were observed in a Zeiss 912 electron microscope (Carl Zeiss, Thornwood, NY).
Labeling in clusters marked newly made RNA as labeling was abolished by pretreating sections with RNase A (Iborra et al., 1996a). It was not due to incorporation into DNA as it was seen without acid denaturation, and in nonreplicating cells in G1 phase. Incubation in 20 μg/ml α-amanitin for 30 min before, and during, Br-U incorporation reduced numbers of particles/μm2 over the nucleoplasm from 9.1 ± 2.7 to 1.2 ± 0.7, and reduced numbers of clusters/μm2 from 0.88 ± 0.2 to 0.31 ± 0.2, confirming that most labeling reflected polymerase II incorporation. Exposure to Br-U has little effect initially on transcription rates; for example, when G1 cells are grown in [3H]adenosine + 2.5 mM Br-U, 3H is incorporated for 1 h into acid-insoluble material at ≥95% rate of controls grown in U (our unpublished results).
Sites containing Br- and biotin-RNA were analyzed as follows. Cells were encapsulated, grown (2 h), labeled with Br-U, and lysed, and transcription reactions were performed as above with 100 μM biotin-CTP (bio-CTP; Life Technologies, BRL, Paisley, Scotland), instead of CTP, and 100 μM UTP. After fixation and sectioning, samples were immunolabeled with gold particles of different sizes as described above (smallest particles applied first to minimize steric hindrance). In Figure 3B, bio-RNA was labeled using goat anti-biotin (5 μg/ml; Jackson Laboratories, West Grove, PA) and rabbit anti-goat IgG conjugated with 5 nm gold particles (1:50 dilution; British BioCell International, Cardiff, Wales); then, samples were fixed (1% glutaraldehyde; 15 min), incubated with 50 mM NH4Cl (1 h) and 10% FCS (30 min), and Br-RNA was labeled with 9 nm particles. Br-RNA was also labeled before bio-RNA, with similar results.
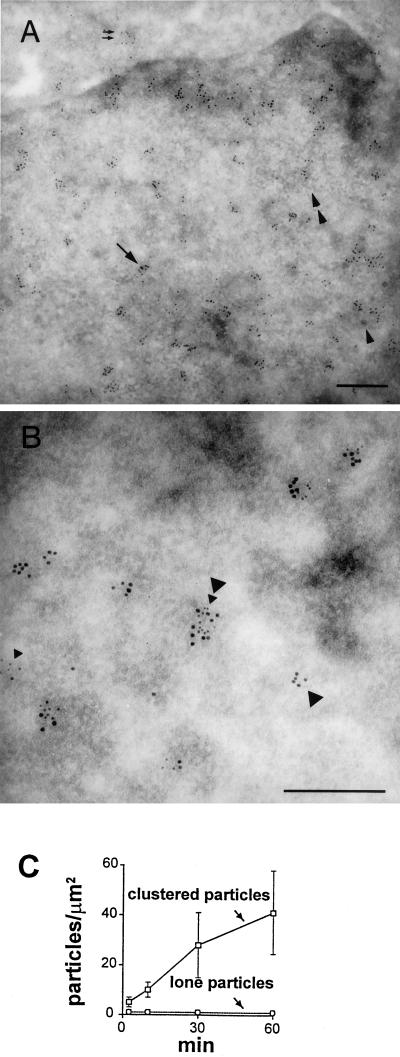
Figure 3.
Immunogold detection of newly-made RNA. (A) Low-power view. Cells were grown (1 h) in 2.5 mM Br-U and fixed, and sites containing Br-RNA were indirectly immunolabeled with 9 nm gold particles. Most particles are found in clusters (arrow marks one) over euchromatin, especially at the edge of heterochromatin (see also Puvion and Moyne, 1978); these clusters mark Br-RNA at synthetic sites and other places on the transport/processing pathway, including some that has reached the cytoplasm (2 small arrows). Interchromatin granule clusters (2 arrowheads mark one) are weakly labeled. Perichromatin granules (single arrowhead marks one) are unlabeled, although they are frequently surrounded by clusters. Bar, 300 nm. (B) High-power view. Cells were grown in Br-U (2.5 mM; 10 min), permeabilized, allowed to extend nascent transcripts in bio-CTP for 15 min, fixed, and indirectly immunolabeled with gold particles of 9 and 5 nm (marking Br- and bio-RNA, respectively). Most particles are found in clusters, some of which contain both Br- and bio-RNA (large and small arrowheads, respectively). Bar, 300 nm. (C) Clustered particles mark transcription sites. Cells were grown for various times in 2.5 mM Br-U and fixed, and sites containing Br-RNA were immunolabeled with 9-nm gold particles. Particles over the nucleoplasm were counted, and numbers normalized per unit area (error bars: ± SD).
Numbers of Polymerases per Transcription Unit
Cells (103 cells in 2 μl) were spotted onto a 3×1 inch glass slide and 5 μl 0.375% sarkosyl, 25 U/ml ribonuclease inhibitor, 10 mM EDTA, and 100 mM Tris-HCl (pH 7.4) were added; after 10 min at 20°C, tilting the slide allowed the drop to run down over 10 min. For light microscopy, samples were air-dried and fixed in 3:1 methanol/acetic acid for 10 min, and Br-RNA distributions were monitored by indirect immunolabeling using the general procedures of Iborra et al. (1996a) and successive additions of a monoclonal anti-Br antibody, anti-mouse Fab conjugated with digoxigenin, monoclonal anti-digoxigenin (all from Boehringer), and anti-mouse Ig conjugated with Cy3 (Jackson Laboratories). For electron microscopy, DNA was picked up on “sticky” nickel grids (coated with formvar and carbon, then 30 μg/ml ethidium bromide; Sogo and Thoma, 1989), air-dried, fixed (4% paraformaldehyde, 20 min), and stained with phosphotungstic acid and uranyl acetate (Osheim and Beyer, 1989). Sometimes, cells were grown in 2.5 mM Br-U (10 min) before lysis, and Br-RNA on grids was indirectly immunolabeled with 9-nm gold particles.
Two controls show that sarkosyl removes little transcriptional activity or nascent RNA. Polymerases elongate on naked DNA faster than on histone-covered DNA, so the first control was conducted in the presence of 4 mM cordycepin triphosphate to limit elongation; then, sarkosyl reduced [32P]UTP incorporation by 4%. Second, >95% acid-insoluble 32P remained in beads after extending nascent chains by ∼50 nucleotides in [32P]UTP and washing twice in 100 vol 0.25% sarkosyl.
Immunoblotting
For Figure 5, cells were encapsulated (107/ml), grown (2 h), washed twice in PBS and 1× in PB**, and divided into three. One sample was diluted four times with 1 mM MgCl2 and 0.2% Triton X-100 and incubated with DNase as above; other samples were treated (15 min, 4°C) with PB** + saponin or 0.25% sarkosyl, washed five times with PB**, diluted four times with 1 mM MgCl2, and incubated with DNase. Proteins in the three samples were dissolved in sample buffer containing SDS, resolved on 6% SDS-polyacrylamide gels, blotted on to nitrocellulose, and the largest subunit of RNA polymerase II was detected by autoradiography (Harlow and Lane, 1988) using the ECL detection kit (Amersham Pharmacia) and monoclonal antibodies 8WG16 (Promega, Southampton, United Kingdom) and H5 (supplied by Dr. S.L. Warren, Yale University, New Haven, CT). Digital autoradiographic images were analyzed using ImageQuant (Molecular Dynamics, Sunnyvale, CA).
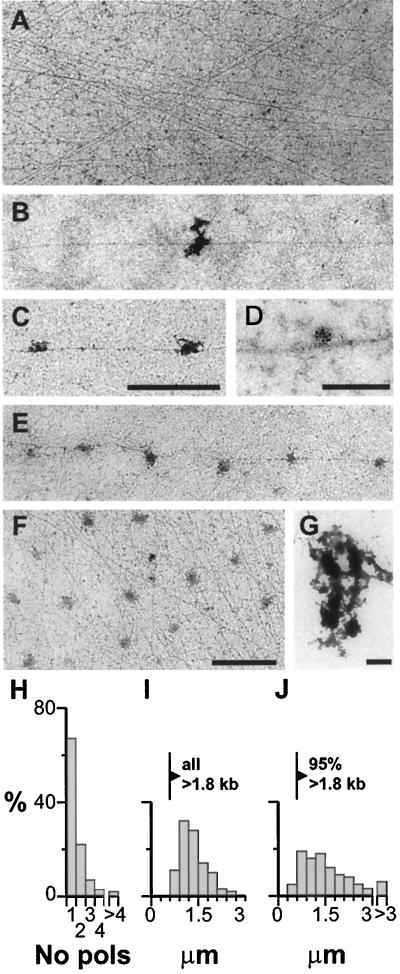
Figure 5.
Most active transcription units contain one engaged polymerase. After cells are lysed with sarkosyl, templates are spread and imaged in the electron microscope. (A–C) and (E–F) are at the same magnification. (A) Transcript-free region typical of most of the spread. (B, C, and E) DNA fibers from well-spread regions associated with 1, 2, and 6 transcripts. Bar, 1 μm (∼3 kb). (D) Immunogold labeling shows transcript zones contain nascent RNA. Cells were grown (2.5 mM Br-U; 10 min), and nascent RNA in spreads immunolabeled with 9-nm gold particles. Bar, 200 nm (∼0.6 kb). (F) An unusual region containing the highest density of extranucleolar transcripts seen. Bar, 1 μm (∼3 kb). (G) A nucleolar region with few DNA fibers, and a cluster of densely staining material with the dimensions and appearance of fibrillar centers. Bar, 1 μm (∼3 kb). (H and I) Most transcription units in well-spread regions are associated with one polymerase. One hundred fibers were selected for analysis if they could be traced without crossing any others for 4 μm on each side of a transcript (i.e., like those shown in panels B, C, and E). The number of transcripts/fiber is expressed as a percentage in H, and the distance between the 34 fibers associated with ≥2 transcripts is shown in panel I. (J) Densely-packed transcription units. The minimum intertranscript spacing in transcription units like those in panel F was determined as follows: 265 transcripts were selected at random, and the minimum distance along connecting DNA fiber(s) to a neighbor was measured (even if it involved apparently switching from fiber to fiber at crossovers in the tangle).
Selecting Br-RNA and bio-RNA
Cells were synchronized 2 h post mitosis (Hozák et al., 1993), encapsulated (5 × 106/ml), and grown for 2 h and then for 2.5 min in [3H]cytidine (250 μCi/ml; NEN, Hounslow, Middlesex, United Kingdom) ± 2.5 mM Br-U. After washing in ice-cold medium (1×), PBS (2×), and PB (1×), cells were lysed and washed, before “run-on” transcription reactions were performed (as above) using 20 μM UTP and 50 μCi [32P]UTP, and biotin-14-CTP instead of CTP. RNA was purified as above, except that samples were extracted twice with phenol to remove agarose before RNA purification using RNAzol B. Dried RNA from reactions was dissolved in 500 μl PBS+. Bio-RNA was selected using streptavidin-magnetic beads (Dynal, Bromborough, Cheshire, United Kingdom). Beads (50 μl) were washed in 0.5 ml PBS+ over 1 h, rewashed, mixed (5 h, 4°C) with 100 μl RNA, separated magnetically, and rewashed in PBS+. Supernatants were reextracted with fresh beads as above; after beads or supernatants were combined, the amount of 3H and 32P was determined by scintillation counting as above. Br-RNA was selected using the anti-BUdR antibody and Sepharose-protein G (Amersham Pharmacia). Sepharose-protein G (25 μl) was washed in PBS+, allowed to bind (2 h) the anti-bromodeoxyuridine antibody (5 μg in 0.5 ml PBS+), washed three times with PBS+, mixed with 100 μl RNA, separated by centrifugation, and rewashed in PBS+. Supernatants were reextracted with Sepharose-protein G and, after beads or supernatants were combined, the amount of 3H and 32P was determined as before.
RESULTS
The Total Number of Nascent Transcripts in a HeLa Cell
Figure 1A illustrates our strategy for counting the number of nascent transcripts. Cells are encapsulated in agarose microbeads to protect them during subsequent manipulation, lysed, and washed to remove endogenous nucleoside triphosphates. Then transcripts are trimmed with RNase A to leave a few nucleotides attached to the still-engaged polymerase, which is allowed to extend the truncated transcript in a limiting concentration of [32P]UTP. The number of radiolabeled nucleotides incorporated during this extension period is monitored (Figure 1B). Next, RNA is run on a gel, and the labeled transcripts are visualized by autoradiography so that their lengths can be determined (Figure 1C). As the incorporation of a known number of labeled nucleotides into all transcripts is associated with a known increase in length of each one (Figure 1D), the number of growing transcripts can be calculated.
This strategy was chosen for several reasons. First, labeling in vivo with [3H]U is impracticable, as endogenous pools prevent an accurate estimate of the specific activity of the immediate precursor, [3H]UTP. Moreover, the rate of elongation is so rapid (i.e., ∼1200 nucleotides/min; Shermoen and O’Farrell, 1991; O’Brien and Lis, 1993) that many transcripts will be completed during the several minutes required to give detectable labeling. Therefore, we label in vitro after permeabilizing cells with saponin in a physiological buffer. We show below that essentially all RNA polymerases active in vivo remain active in vitro. Under our conditions, few, if any, transcripts are initiated, but some elongating polymerases terminate. Second, RNase treatment increases the accuracy of sizing transcripts, as addition of even a few nucleotides to truncated transcripts can easily be seen on a gel. Under similar conditions, the growth of non-RNase–trimmed transcripts (average length, 7600 nucleotides; see below) would go undetected. (Extending untreated transcripts by more than ∼100 nucleotides defeats the approach, as so many terminate during incubation.) Fortunately, RNase treatment reduces incorporation only marginally (Figure 1B; compare curves 1 and 4). Third, elongation in different UTP concentrations minimizes any effects due to residual pools, DNA sequence, and transcript termination.
In 0.125 μM UTP, RNase-treated cells extend truncated transcripts slowly (Figure 1B, curve 2), and the rate increases as the UTP concentration is raised (Figure 1B, curves 3–5); this increase is reflected by an increase in chain length (Figure 1C, lanes 2–5). Extrapolation back to 0 μM UTP shows that RNase prunes nascent transcripts back to ∼25 nucleotides; the extent of such truncation varies from experiment to experiment. (Polymerase I transcripts prove more resistant to truncation [Figure 2B].) Ideally, the number of nucleotides incorporated should be related linearly to chain length, but asymmetry in base sequence and chain termination may contribute to the slight deviations seen (Figure 1D). Here, ∼2,000,000 nucleotides are incorporated into all transcripts as each typically grows by ∼25, so there must be ∼80,000 ± 20,000 growing transcripts/cell. The average of three experiments gave a value of 88,000 (range 80,000–95,000) active polymerases/cell (our unpublished results).
The Number of Polymerase I Transcripts
Mammalian cells contain two major RNA-polymerizing activities (i.e., I and II) and two minor activities (i.e., III and the mitochondrial enzyme; Chambon, 1974). Polymerase I in the nucleolus is resistant to high concentrations of α-amanitin (Figure 2A, curve 2), but not actinomycin D (Figure 2A, curve 3; residual activity is presumably due to mitochondrial enzyme). Assuming that the contribution of the mitochondrial enzyme is negligible, and α-amanitin completely inhibits polymerases II and III, we can estimate the numbers of transcripts made by polymerase I. Results from a typical experiment (Figure 2) show that 12,800 ± 5000 (polymerase I) complexes are resistant to α-amanitin. Averages from three experiments give 14,700 (range 12,800–18,600; our unpublished results) polymerase I complexes/cell. Then, the number of polymerase II/III complexes can be obtained by subtraction from the total (i.e., 88,000 − 14,700 = 73,300). Polymerase I numbers can also be estimated using actinomycin D, assuming it has no effect on polymerase II/III; 76,000 ± 23,000 complexes were resistant to actinomycin D (Figure 2). (Polymerase I elongates in vitro faster than polymerase II [Figure 2C]; this enhancement varies from ∼2× to ∼3.5× in 0.125 and 2.625 μM UTP, respectively [our unpublished results].) We conclude that each cell contains ∼75,000 polymerase II/III complexes and ∼15,000 polymerase I complexes.
The Size of Nascent Transcripts
We next determined the average length of (non-RNase–trimmed) nascent RNA. Permeabilized cells were allowed to extend transcripts by ∼25 nucleotides, before the now end-labeled transcripts were sized. The number average molecular weight of all transcripts was 7600 nucleotides; corresponding values for transcripts made by polymerase I and II/III (i.e., measured in α-amanitin and actinomycin D, respectively) were 6600 and 8400 nucleotides (our unpublished results). These values are similar to those obtained earlier (Lewin, 1975).
Labeling Extranucleolar Transcription Sites in Vivo Using Br-U
Previously, we and others have used independent methods to show that nascent transcripts extended in vitro are concentrated in ∼2000 sites (Iborra et al., 1996a; Fay et al., 1997). One method utilizing biotin-CTP (bio-CTP) was sensitive enough to detect most sites, as no more sites could be seen when 10× more biotin was incorporated. However, it remained possible that bio-CTP was incorporated by permeabilized cells into a fraction of sites active in vivo, or that sites aggregated on lysis. Therefore, we developed a method to label sites in vivo. Cells were grown in 2.5 mM Br-U for a short period, before the resulting Br-RNA was detected by immunogold labeling (Figure 3, A and B). Gold particles tended to be clustered, and the numbers of such particles (normalized per unit area) increased both with incubation time (Figure 3C) and Br-U concentration (our unpublished results); this suggests that clusters mark sites containing newly made RNA. (A cluster is defined as >1 particle lying within 40 nm of another [center-to-center distance].) There were 1.1 ± 0.5 clusters/μm2 (our unpublished results). A background of lone particles was scattered over both nucleus and cytoplasm; their number (i.e., ∼0.9/μm2) did not change with concentration (our unpublished results) or incorporation time (Figure 3C). Such exposures to Br-U are not toxic, and labeling in clusters marks newly-made RNA; for example, labeling was abolished by pretreatment with RNase A (see MATERIALS AND METHODS).
As newly made RNA moves quickly away from synthetic sites, many clusters seen after labeling for 1 h mark transcripts at later stages on the way to the cytoplasm. (Although complete substitution of U by Br-U prevents splicing in vitro [Sierakowska et al., 1989; Wansink et al., 1994], Br-RNA made in vivo moves to the cytoplasm [Figure 3A; Iborra, Jackson, and Cook, submitted].) Therefore, it is important to determine which clusters mark synthetic sites, and which mark downstream sites. The synthetic fraction was identified by double-labeling (Table 1). Cells were grown in Br-U for 2.5–60 min and permeabilized, and nascent transcripts were extended by ∼250 nucleotides in bio-CTP; then, sites containing Br-RNA and bio-RNA were labeled with 9 and 5 nm gold particles (Figure 3B). Here, no new transcripts are initiated in vitro, and bio-RNA is unable to move away from synthetic sites (Iborra et al., 1996a). Therefore, clusters of small gold particles mark biotin in still-growing transcripts at synthetic sites. At all times, there was approximately one such “bio-cluster”/μm2 in extranucleolar regions (Table 1, column B). After growth for 2.5 min in Br-U, there was an equivalent density of clusters of large particles (i.e., “Br-clusters”; column A); this shows that little Br-RNA had yet moved away from synthetic sites, and suggests that the same sites are labeled by the two precursors. However, when cells were grown in Br-U for longer, the density of Br-clusters increased progressively (column A); this is consistent with steady-state synthesis in a few sites, followed by a steady flux of products into/through a larger number of downstream sites.
Table 1.
Distinguishing transcription from downstream sites
| Labeling conditions | Colocalization
|
|||
|---|---|---|---|---|
| Clusters/μm2
|
% Br-clusters associated with bio-clusters (C) | % Bio-Clusters associated with Br-clusters (D) | ||
| Br-clusters (A) | Bio-clusters (B) | |||
| 1. 2.5 min, 5 mM Br-U; 15 min bio-CTP | 1.1 ± 0.5 | 1.1 ± 0.5 | 85 | 85 |
| 2. 10 min, 2.5 mM Br-U; 15 min bio-CTP | 2.0 ± 0.5 | 1.0 ± 0.4 | 58 | 85 |
| 3. 30 min, 2.5 mM Br-U; 15 min bio-CTP | 4.6 ± 1.6 | 1.1 ± 0.3 | 28 | 84 |
| 4. 60 min, 2.5 mM Br-U; 15 min bio-CTP | 7.1 ± 2.2 | 0.9 ± 0.6 | 18 | 85 |
Cells were grown in Br-U, permeabilized, and allowed to make RNA in the presence of bio-CTP, and clusters of gold particles marking nucleoplasmic sites containing Br- and bio-RNA were counted. Br- and bio-clusters had average diameters of 82 and 78 nm, respectively; neither size changed with labeling time. Clusters were considered to be colocalized if a gold particle in one lay within 40 nm of gold particle in another.
Such a movement of Br-RNA away from synthetic sites was confirmed by analysis of doubly-labeled sites. At all times, ∼85% bio-clusters colocalized with “Br-clusters” (Table 1, column D); essentially all transcription sites active after lysis were also active in vivo a few moments earlier. However, with longer growth in Br-U, the percentage of Br-clusters that colocalized with bio-clusters fell (column C); progressively fewer Br-clusters labeled in vivo are transcriptionally active in vitro, as more Br-RNA leaves synthetic sites. We conclude that 1) transcription and downstream sites can be distinguished, 2) transcription sites labeled in vivo and in vitro have the same size (Table 1, legend), and 3) lysis does not aggregate sites.
Single-labeling with Br-U proves sufficiently sensitive to allow detection of most transcription sites. We showed this by growing cells for short periods in high concentrations of Br-U. If only a fraction of sites were detected after 1.25 min, then doubling the Br-U concentration or incubation time should allow previously undetected sites to be detected; however, neither had any effect on cluster density (Table 2), despite an increase in labeling in each cluster (Figure 3C; Iborra, Jackson, and Cook, manuscript submitted for publication). These treatments had no effect on the doubling time of cells (our unpublished results).
Table 2.
Most transcription sites can be detected
| Br-U concentration (mM) | Clusters/μm2 (± SD)
|
|
|---|---|---|
| 1.25 min | 2.5 min | |
| 2.5 | 0.9 ± 0.2 | 1.1 ± 0.4 |
| 5 | 1.1 ± 0.3 | 1.1 ± 0.5 |
| 10 | 1.0 ± 0.2 | 1.2 ± 0.3 |
The Number of Extranucleolar Transcription Sites
The total number of transcription sites in three-dimensional space can be calculated (using standard stereological procedures) from the numbers and diameters of clusters seen in two-dimensional sections, with knowledge of nucleoplasmic volume (e.g., Iborra et al., 1996a); one cluster/μm2 corresponds to 2400 extranucleolar sites/cell. (We are currently analyzing the relative numbers of polymerase II and III sites [Pombo, Jackson, Hollingshead, and Cook, manuscript in preparation].)
Most Transcription Units Are Associated with Only One Polymerase
The number of active polymerases on a typical transcription unit has been determined using “Miller” spreads; chromatin is spread and imaged in the electron microscope, and the number of closely spaced transcripts are counted (reviewed by Osheim and Beyer, 1989). Most analyses have concerned highly active units such as rDNA in amphibian oocytes or nonribosomal units in insect embryos. Only a few mammalian nonribosomal units have been analyzed, because the higher complexity makes identification so difficult. Even so, nonribosomal units seem to be associated with only one to two transcripts (e.g., Laird and Chooi, 1976; McKnight and Miller, 1979; Beyer et al., 1981; Fakan et al., 1986). Therefore, we modified the method of Parra and Windle (1993) to enable us to visualize all transcription units in a nucleus (and not just highly selected examples). Then, we went on to analyze systematically several hundreds of nonribosomal units in detail.
One thousand cells are spotted on one end of a glass slide, and the strong detergent, sarkosyl, is added. This disassembles nuclei, strips histones from the template, and spreads templates over a wide area to improve resolution; >95% active polymerases that have formed the first phosphodiester bond in the transcript remain associated with the now-naked DNA (Hawley and Roeder, 1987; see MATERIALS AND METHODS). Then, DNA and associated transcripts are spread as they flow down the slide.
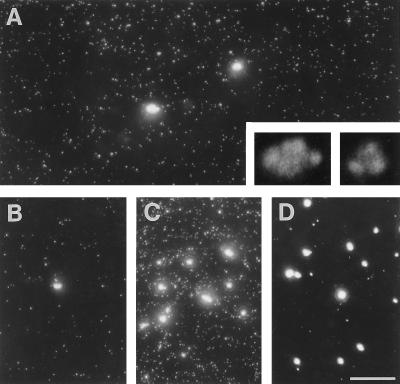
We first monitored the distribution of nascent transcripts in whole spreads by light microscopy; permeabilized cells were allowed to extend nascent transcripts in Br-UTP by ∼250 nucleotides, before spreading and indirect immunolabeling of nascent Br-RNA. Many small faint foci and fewer large bright ones are seen (Figure 4A); few bright ones are found at the origin of the spread (Figure 4B), but more are found at the leading edge (Figure 4C). Few faint foci are seen after extension in 250 μg/ml α-amanitin (Figure 4D), suggesting that the bright ones are generated by polymerase I and faint ones are generated by polymerase II/III. (The few faint foci represent background as the same numbers are seen when Br-UTP is omitted.) After counting the number of faint foci in 25 randomly selected regions (25 μm2) of several slides (with background subtraction), and knowing the area covered by the spread, we calculate that ∼55,000 faint foci are derived from each cell. If each cell contains ∼75,000 active polymerases II/III (see above), most foci must contain one transcript. Similar counts of bright foci in 25 regions of 250 μm2 indicate ∼30 bright foci are derived from each cell. Under higher power, bright foci are typically seen to be aggregates (Figure 4A, inset) of approximately four subfoci (98% foci had ≤10 subfoci; our unpublished results). Therefore, each cell contains 120 subfoci, which is close to the number of ribosomal cistrons active in a HeLa nucleus (i.e., ∼150; reviewed by Jackson et al., 1993). If ∼15,000 transcripts are generated by polymerase I (see above), each subfocus would contain ∼125 nascent transcripts. Again, this is close to the 100–120 expected, if each subfocus contains one active cistron (Osheim and Beyer, 1989).
Figure 4.
Imaging nascent transcripts in spreads by immunofluorescence. Permeabilized cells were allowed to extend nascent transcripts in Br-UTP by ∼250 nucleotides, and then 1000 cells were applied to a slide, their was DNA spread, and (nascent) Br-RNA was indirectly immunolabeled. Bar, 10 μm. (A) A view of a typical field illustrating the two kinds of fluorescent foci marking Br-RNA. Insets: 1/32 exposure of the two bright foci at 5× magnification. (B and C) Regions containing few and many bright foci, from the origin and the leading edge of the spread, respectively. (D) Extension in 250 μg/ml α-amanitin; bright foci and some background foci are seen in the leading edge.
We next analyzed the spreads by electron microscopy after picking up DNA from the surface onto a grid. Most of the grid is covered by transcript-free DNA fibers (Figure 5A). When transcripts are seen associated with fibers, they are not extended as they are in Miller spreads. They are usually alone, although sometimes several are strung along one fiber (Figure 5, B, C, and E). An area containing RNA such as that illustrated in Figure 5B probably contains one transcript, as such areas typically obscured 101 ± 60 nm of the underlying fiber, if it were fully extended (n = 200; our unpublished results); since polymerases are maximally packed on rDNA every 75 base pairs (bp) (McKnight and Miller, 1976), it is unlikely that more than one could be associated with the obscured DNA. Essentially all transcripts can be immunolabeled with gold particles after growth in Br-U for 10 min (Figure 5D); <10% transcripts were in dense DNA tangles; the highest density seen is shown in Figure 5F. Regions at the front of the spread contain poorly spread material containing few DNA fibers and clusters of dense bodies that resemble fibrillar centers of nucleoli (Figure 5G); these were not analyzed further. As the distributions of two kinds of foci seen by light microscopy correlate with distributions of individual transcripts and disrupted nucleolar material seen by electron microscopy (compare Figure 4, A–C, with Figure 5, B–G), we can be confident that no group of transcripts goes undetected by electron microscopy.
Numerical analysis showed that 66% transcripts in well-spread regions (like those in Figure 5, B–F) lay >4 μm (i.e., >12 kilobases [kb]) away from another (Figure 5H). As 66% nascent RNA is <12,000 nucleotides (our unpublished results), most of these transcription units must be associated with one engaged polymerase. Where ≥2 transcripts are attached to one stretch of DNA, the intertranscript distance was >0.6 μm (equivalent to ∼1.8 kb; Figure 5I), so we cannot exclude the possibility that each is associated with a different transcription unit in a tandem array. Only occasionally was a fiber associated with >4 transcripts; these were often uniformly spaced (e.g. Figure 5E). The maximum seen was 13 transcripts, spaced every 12 ± 3 kb (our unpublished results).
We also estimated minimum intertranscript spacings in the rare dense tangles (e.g., Figure 5F). A transcript was selected at random, and the minimum distance along connecting DNA fiber(s) to a neighbor was measured (even if it involved tracing a fiber around several sides of a polygon, apparently switching from fiber to fiber at crossovers in the tangle). Although most transcripts in a pair are usually from different transcription units, this enables us to put a lower limit on the distance between polymerases; 77% transcripts lay ≥0.9 μm (≥2.7 kb) from another (Figure 5J). Therefore, even in these regions, which we stress are exceptional, a typical transcription unit of 8400 bp is associated with ≤3 polymerases. All results are consistent with there being only one engaged polymerase on most extranucleolar units.
The Relative Proportions of Different Forms of Polymerase II
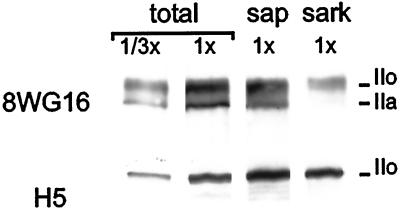
We next used sarkosyl to estimate the relative proportions of different phosphorylated forms of the largest subunit of polymerase II. This contains a C-terminal domain that becomes hyperphosphorylated when elongation begins (Dahmus, 1996). Hypo- and hyperphosphorylated forms (IIa and IIo) have apparent molecular masses on gels of ∼220 and ∼240 kDa and are both recognized by monoclonal antibody 8WG16 (Thompson et al., 1989). Saponin extracts little protein detected by 8WG16, whereas sarkosyl removes 85% IIa and 62% IIo (Figure 6, legend). As few radiolabeled transcripts or engaged polymerases are extracted by sarkosyl (see above), probably only this resistant fraction is active. Quantitative analysis of five such blots shows that ≤27% of total reactivity resists extraction (Figure 6, legend), and it is this fraction that is probably active.
Figure 6.
Sarkosyl removes most of large subunit of RNA polymerase II. Different numbers (1x or 1/3x) of encapsulated cells were either not extracted (total), or extracted with saponin (sap) or sarkosyl (sark). After electrophoresis and immunoblotting with 8WG16 or H5 antibodies, autoradiographs were prepared; the ∼220–240 kDa region is shown. Quantitative analysis (our unpublished results) of five pairs of blots shows the following. 8WG16 reactivity: 54% is due to IIo, saponin has little effect, and sarkosyl removes 85% IIa and 62% IIo, to leave 27% (range 22–34%) of total. H5 reactivity: saponin has little effect, but sarkosyl extracts 13% of major band.
This conclusion was confirmed using the H5 antibody that mainly recognizes epitopes on the highly phosphorylated forms of IIo (Bregman et al., 1995; Kim et al., 1997). Now, sarkosyl extracts ∼13% of the major band (Figure 6). (Incubation with saponin has a curious effect; new bands appear, probably due to phosphatases that remain active, despite the presence of inhibitors; sarkosyl prevents the appearance of [or removes] most of the new bands.)
Most Transcripts Are Labeled with Br-U (in Vivo) or bio-CTP (in Vitro)
As we were concerned that some polymerases might be inactivated by lysis, or could not incorporate the analogues, we confirmed that most polymerases active in vivo could incorporate both [32P]UTP and bio-CTP in vitro. Cells were grown in [3H]cytidine and lysed, and then nascent chains were elongated in both [32P]UTP and bio-CTP. (G1 cells were used to minimize incorporation of [3H]cytidine into DNA.) Then, biotin-labeled transcripts were selected using magnetic beads coated with streptavidin, and the 3H:32P ratio was measured before and after selection. We might expect three types of nascent transcript: singly labeled ones containing 3H (made if some engaged polymerases terminated during labeling, or were inactivated by lysis), doubly labeled ones containing 3H and 32P (made if some could not incorporate biotin), and triply labeled ones containing 3H, 32P, and biotin (made if all polymerases remained fully functional and able to incorporate the analogues). If there were many more singly or doubly labeled transcripts than triply labeled ones, selection should reduce the ratio accordingly; however, the ratio remained essentially the same (Table 3; minor 3H losses are consistent with some termination occurring naturally in vivo). This shows that most, if not all, polymerases able to incorporate 3H in vivo can go on to incorporate bio-CTP in vitro.
Table 3.
Most transcripts labeled with [3H]cytidine can be labeled with Br-U and bio-CTP
| Experiment | cpm
|
3H/32P ratio | |
|---|---|---|---|
| 3H | 32P | ||
| 1. [3H]C in vivo, then [32P]UTP + bio-CTP in vitro | |||
| Before selectiona | 14,151 | 6,796 | 2.1 |
| After selecting bio-RNAb | 5,943 | 3,138 | 1.9 |
| 2. [3H]C + Br-U in vivo, then [32P]UTP + bio-CTP in vitro | |||
| Before selectiona | 2,522 | 2,898 | 0.87 |
| After selecting bio-RNAb | 1,396 | 2,410 | 0.58 |
| After selecting Br-RNAc | 1,789 | 1,980 | 0.90 |
Selection procedures worked efficiently, as in experiments 1 and 2, 40 and 67% of all 3H cpm in cells were recovered in pure RNA, respectively,a <1% [32P]RNA made in the absence of bio-CTP bound to streptavidin-beads,b and <2% [32P]RNA made in the absence of Br-U bound to beads coated with anti-Br antibody.c
We extended this approach to determine whether Br-U was incorporated into a subset of sites. Cells were now grown in [3H]cytidine supplemented with Br-U and lysed, and the nascent chains were elongated in [32P]UTP and bio-CTP as before, before Br-labeled transcripts were selected (Table 3). Again, the ratio was essentially the same before and after selection, showing that there was no bias against Br-U incorporation. Selection for bio-RNA confirmed that most, if not all, polymerases can incorporate bio-CTP.
DISCUSSION
Methods for Counting Active Polymerases and Transcription Sites
Recent findings suggest that nascent transcripts and active RNA polymerases are confined within mammalian nuclei to a limited number of sites (Jackson et al., 1993; Wansink et al., 1993; Bregman et al., 1995; Iborra et al., 1996a). This raises the issue of how many transcripts each site contains. Therefore, we devised a suite of methods to count numbers of nascent transcripts and sites in HeLa cells. All counting methods have associated errors, but we are concerned here with reconciling differences of more than an order of magnitude, and not twofold differences or less.
One method allows the total number of transcripts being made at any moment to be estimated. Cells are encapsulated in agarose, permeabilized, and treated with RNase, and still-engaged polymerases are allowed to extend the now-truncated transcripts in [32P]UTP; then, the number of growing transcripts can be calculated from the total number of nucleotides incorporated into all transcripts, and the average increment in length of each one (Figure 1). We find that ∼90,000 transcripts are being made by all polymerases, ∼15,000 by RNA polymerase I, and ∼75,000 by polymerases II and III. These figures lie within the ranges found previously (see INTRODUCTION). Two potential sources of error would lead to underestimates. 1) Some polymerases might disengage on lysis. We cannot formally monitor such losses because the rate of transcription in vivo is unknown, but we have shown that most, if not all, transcripts that can be labeled with [3H]cytidine (±Br-U) in vivo can also be labeled with [32P]UTP and bio-CTP in vitro (Table 3). This means that although lysis may change elongation rates, it disengages few, if any, enzymes. 2) Some polymerases might disengage, terminate, or pause during incubation in vitro, but we minimize such effects using different UTP concentrations.
A second method enables the number of nucleoplasmic transcription sites to be counted. Cells are grown for different periods in Br-U, before sites containing Br-RNA are marked with gold particles; Br-RNA is made in transcription sites and then moves away down the transport/processing pathway. After growth for 2.5 min or less in Br-U, gold particles are concentrated in ∼2400 discrete clusters. The identification of these as transcription sites was confirmed after lysis by extending nascent RNA chains in bio-CTP; then, essentially all sites containing Br-U also contained bio-RNA (Table 1). This shows that sites labeled in vivo do not aggregate on lysis. After growth in Br-U for longer, additional downstream sites become labeled that are functionally distinct, as they cannot incorporate bio-CTP.
How accurate is this estimate that transcription is confined to only ∼2400 sites? Do we fail to detect many less active sites? The experiments summarized in Table 3 eliminate the possibility that Br-U is incorporated into a fraction of transcripts. It is also unlikely that many less active sites go undetected as increasing the incorporation time (from 1.25 to 2.5 min) and Br-U concentration (from 2.5 to 10 mM) should raise more sites above the threshold of detection, but it does not (Table 2). We might also expect different detection methods to have different thresholds (Figure 7A), but three others gave similar numbers of sites (i.e., 2000–2700; Iborra et al., 1996a; see also, Fay et al., 1997). One involved incubating lysed cells with Br-UTP and light microscopy; a second involved bio-CTP incorporation and electron microscopy; in the latter case, the number remained unchanged despite a 10× increase in incorporation. A third was indirect; antibodies against polymerase II labeled 2500 sites that partially overlapped transcription sites, and lysis had no effect on numbers. Therefore, four independent approaches now give similar results. They involve precursors that are incorporated with different efficiencies, and different fixation, embedding, immunolabeling, imaging and stereological procedures. Moreover, many sites cannot aggregate on lysis, as similar counts are obtained with unlysed cells.
Figure 7.
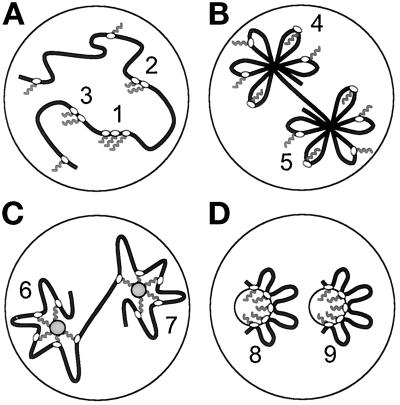
Different models for the organization of extranucleolar transcription. (A) Active polymerases (ovals) and associated transcripts (wavy lines) are spread throughout euchromatin. An insensitive method might only allow transcripts in site 1 to be detected; then, increased Br-U incorporation should allow detection of less-active sites 2 and 3. A more sensitive method would allow initial detection of sites 1–3, and other sites after increased incorporation. All sites can only be seen using a method that enables single transcripts to be detected efficiently. This model cannot apply because most sites are detected, there is one polymerase on most transcription units, and there are 30× more nascent transcripts than sites. However, all sites (i.e., 4–9) can be detected in models B–D (even using insensitive methods) as each site contains so many transcripts. (B) Polymerases track around chromatin loops tied into a rosette. Here, sites contain most of the length of the templates. However, ∼30 polymerases, associated transcripts and splicing factors, and 20–30 complete transcription units cannot be fitted into a site (diameter ∼80 nm). This is only feasible if most of the length of each transcription unit is excluded (as in panels C and D). (C) Nascent transcripts are tethered to a splicing complex (gray circles in sites 6 and 7) during synthesis. (D) Polymerases are attached to the surface of two sites (i.e. 8 and 9); nascent transcripts are extruded into each site, which is surrounded by a cloud of transcription units.
The third method permits rough estimates of 1) the minimum number of nascent transcripts in a cell (by light microscopy), and 2) the average number of polymerases on individual transcription units (by electron microscopy). It allows visualization of all transcription units in a nucleus, and not just selected examples. One thousand cells are spotted on to a glass slide, and the strong detergent, sarkosyl, is added; this disassembles nuclei and strips histones from the template, but leaves engaged polymerases and transcripts still associated with now-naked DNA (Parra and Windle, 1993). Then, DNA and associated transcripts are spread as they flow down the slide.
For light microscopy, permeabilized cells were allowed to extend nascent transcripts in Br-UTP, before spreading; then Br-RNA was indirectly immunolabeled (Figure 4). Each cell gave rise to ∼55,000 small faint foci and ∼120 larger brighter subfoci (generated by polymerase II/III and I, respectively). As each focus could contain more than one transcript, this allows us to put a lower limit on the number of nascent transcripts/cell. Moreover, if each cell contains ∼75,000 transcripts generated by polymerases II/III (see above), most faint foci must contain 1. Similarly, if each cell contains ∼15,000 transcripts generated by polymerase I (see above), each bright subfocus would contain ∼125, which is close to the 100–120 expected if each contained one active rDNA cistron (Osheim and Beyer, 1989).
Knowledge of the numbers and distribution of nascent transcripts obtained by light microscopy enabled us to identify the different foci by electron microscopy and allowed us to be sure that no group of transcripts had gone undetected. Analysis of 100 individual (nonnucleolar) transcription units of >24 kb confirmed that 66% were associated with only one transcript (Figure 5H). As 65% nascent RNA is ≤12,000 nucleotides, most active units must be associated with one polymerase. Again, this is consistent with earlier results; even the active major late unit of adenovirus 2 is associated with only one transcript every 7.5 kb (Beyer et al., 1981; Wolgemuth et al., 1981).
The fourth method allows purification of nascent RNA. (See Bonner et al., [1975] and Huang et al. [1978] for others.) After extension in Br-U in vivo or biotin-CTP in vitro, transcripts are selected using beads coated with antibodies or streptavidin. We used this method to show that under our conditions essentially all polymerases active in vivo remain active after lysis (Table 3).
Numbers of Active Polymerases and Sites in Extranucleolar Regions
Our results indicate that ∼75,000 nascent transcripts are concentrated in only ∼2400 extranucleolar sites. Even if we take the lowest estimate of transcript number (i.e., 69,000), and the highest estimate of site number (i.e., 2700; Iborra et al., 1996a), the two still differ by more than an order of magnitude. This makes the model depicted in Figure 7A unlikely. The simplest interpretation of these results is that a typical site contains ∼30 nascent transcripts, and so active polymerases. Then, as two-thirds or more transcription units are transcribed by approximately one polymerase at any moment, each site would contain transcripts from 20–30 different units (Figure 7,B–D).
These numbers of polymerases can sustain ribosomes and messages at known levels (e.g., Cox, 1976). For example, if polymerase I takes ∼5 min to make a primary transcript, ∼15,000 enzymes can double rRNA numbers in 21 h to ensure that each daughter receives ∼4 × 106 rRNAs when the cell divides. Moreover, polymerase II elongating at ∼1200 nucleotides/min (e.g. Shermoen and O’Farrell, 1991) can make a typical transcript of 8400 nucleotides in ∼7 min. In 500 min, the average half-life of a message (Cox, 1976), 75,000 such complexes would make 5× more transcripts than the number needed to sustain a steady-state level of ∼106 messages/cell. After correcting for cell doubling, this means that three out of four primary transcripts could be completely degraded, as processing removes four-fifths of the length of the remainder (calculated from the length difference between the primary transcript and mature message), so that ∼5% RNA reaches the cytoplasm as message of ∼1500 nucleotides.
The Organization of Extranucleolar Transcription
These results impose constraints on models of how extranucleolar polymerases are organized, as some mechanism must confine ∼30 polymerases (engaged on ≥20 templates) so that the ∼30 nascent transcripts become concentrated in a site only ∼80 nm across. How might this be achieved? Templates might be tied into a rosette of short loops, as polymerases track around the loops (Figure 7B). However, it is impossible to pack all the components into one site, as the nucleosomes associated with only ∼10 typical transcription units of 8.6 kb completely fill a sphere of 80 nm, leaving no room for anything else. Moreover, cutting such loops with restriction enzymes should detach polymerases, transcripts, and transcription units, but it does not (Jackson and Cook, 1985; Jackson et al., 1996). Alternatively, nascent transcripts might be captured by a neighboring splicing complex, as they are being made (Figure 7C). If such a site contains little DNA, it becomes feasible to pack ∼30 nascent transcripts and splicing components into ∼80 nm. However, treatment with a restriction endonuclease and RNase (to sever links with the complex) should release polymerases, but, again, it does not (e.g., Jackson and Cook, 1985). Another possibility involves attaching polymerases to a structure that we call a transcription factory as it contains so many polymerizing machines (Figure 7D; Jackson et al., 1981; Iborra et al., 1996b). Then, a cloud of templates would surround the factory, as transcripts are extruded into the factory. Here, templates move past fixed polymerases, rather than vice versa. Packing ∼30 nascent transcripts into ∼80 nm is again feasible and gives about half the density found in the dense fibrillar component of the nucleolus, the site where nascent rRNA is found (Hozák et al., 1994; Shaw and Jordan, 1995). This density is also roughly equivalent to that found in a ribosome (diameter ∼25 nm); each ribosome contains ∼7000 nucleotides rRNA, and ∼30 ribosomes can be packed into a sphere with a diameter of ∼80 nm. Only this model is consistent with all the results described here and with others showing that still-engaged polymerases and transcribed DNA remain after nuclease treatment (Jackson and Cook, 1985; Jackson et al., 1996; Iborra et al., 1996a). Then, a tripartite factory contains spatially contiguous template, polymerase, and transcript zones; as in the nucleolus, the transcription machinery need not occupy much of the transcript zone.
ACKNOWLEDGMENTS
We thank Drs. M. Hollingshead, S.L. Warren, and J. Renau-Piqueras for kindly supplying reagents or software, and A. Pombo, J. Sanderson, and J. Bartlett for their help. This work was supported by the Cancer Research Campaign and Wellcome Trust.
REFERENCES
- Beyer AL, Bouton AH, Hodge LD, Miller OL. Visualization of the major late R strand transcription unit of adenovirus serotype 2. J Mol Biol. 1981;147:269–295. doi: 10.1016/0022-2836(81)90441-1. [DOI] [PubMed] [Google Scholar]
- Bonner J, Gottesfeld J, Garrard W, Billing R, Uphouse L. Isolation of template active and inactive regions of chromatin. Methods Enzymol. 1975;40:97–102. doi: 10.1016/s0076-6879(75)40010-6. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. The Enzymes. P.D. Boyer, New York: Academic Press; 1974. RNA polymerases; pp. 261–331. [Google Scholar]
- Cox RF. Quantitation of elongating form A and B RNA polymerases in chick oviduct nuclei and effects of estradiol. Cell. 1976;7:455–465. doi: 10.1016/0092-8674(76)90176-8. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Fakan S, Leser G, Martin TE. Immunoelectron microscope visualization of nuclear ribonucleoprotein antigens spread within transcription complexes. J Cell Biol. 1986;103:1153–1157. doi: 10.1083/jcb.103.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Tocchini-Valentini GP. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hawley DK, Roeder RG. Functional steps in transcriptional initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- Hozák P, Cook PR, Schöfer C, Mosgöller W, Wachtler F. Site of transcription of ribosomal RNA and intra-nucleolar structure in HeLa cells. J Cell Sci. 1994;107:639–648. doi: 10.1242/jcs.107.2.639. [DOI] [PubMed] [Google Scholar]
- Hozák P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to a nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Huang RC, Smith MM, Reeve AE. Studies on gene transcription in vitro by analysis of the primary transcripts. Cold Spring Harb Symp Quant Biol. 1978;42:589–596. doi: 10.1101/sqb.1978.042.01.061. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J Cell Sci. 1996a;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Iborra FJ, Pombo A, McManus J, Jackson DA, Cook PR. The topology of transcription by immobilized polymerases. Exp Cell Res. 1996b;229:167–173. doi: 10.1006/excr.1996.0355. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T, Zachau HG. Domains in chromatin structure. Cold Spring Harbor Symp Quant Biol. 1977;42:109–118. doi: 10.1101/sqb.1978.042.01.012. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Bartlett J, Cook PR. Sequences attaching loops of nuclear and mitochondrial DNA to underlying structures in human cells: the role of transcription units. Nucleic Acids Res. 1996;24:1212–1219. doi: 10.1093/nar/24.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Transcription occurs at a nucleoskeleton. EMBO J. 1985;4:919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, McCready SJ, Cook PR. RNA is synthesised at the nuclear cage. Nature. 1981;292:552–555. doi: 10.1038/292552a0. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Yuan J, Cook PR. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J Cell Sci. 1988;90:365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird CD, Chooi WY. Morphology of transcription units in Drosophila melanogaster. Chromosoma. 1976;58:193–218. doi: 10.1007/BF00701359. [DOI] [PubMed] [Google Scholar]
- Lewin B. Gene Expression-2: Eukaryotic Chromosomes. II. London: Wiley; 1974. [Google Scholar]
- Lewin B. Units of transcription and translation: sequence components of heterogeneous nuclear RNA and messenger RNA. Cell. 1975;4:77–93. doi: 10.1016/0092-8674(75)90113-0. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Miller OL. Ultrastructural patterns of RNA synthesis during early embryogenesis in Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Miller OL. Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell. 1979;17:551–563. doi: 10.1016/0092-8674(79)90263-0. [DOI] [PubMed] [Google Scholar]
- Miklos GLG, Rubin GM. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, Beyer AL. Electron microscopy of ribonucleoprotein complexes on nascent RNA using Miller chromatin spreading method. Methods Enzymol. 1989;180:481–509. doi: 10.1016/0076-6879(89)80119-3. [DOI] [PubMed] [Google Scholar]
- Parra I, Windle B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nat Genet. 1993;5:17–21. doi: 10.1038/ng0993-17. [DOI] [PubMed] [Google Scholar]
- Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol. 1970;76:127–140. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- Puvion E, Moyne G. Intranuclear migration of newly synthesized extranucleolar ribonucleoproteins. Exp Cell Res. 1978;115:79–88. doi: 10.1016/0014-4827(78)90404-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowska H, Shuklas RR, Dominski Z, Kole R. Inhibition of pre-mRNA splicing by 5-fluoro-, 5-chloro-, and 5-bromouridine. J Biol Chem. 1989;264:19185–19191. [PubMed] [Google Scholar]
- Sogo JM, Thoma F. Electron microscopy of chromatin. Methods Enzymol. 1989;170:142–179. doi: 10.1016/0076-6879(89)70045-8. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B, Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Sugden B, Keller W. Mammalian deoxyribonucleic acid-dependent ribonucleic acid polymerases, I; purification and properties of an α-amanitin-sensitive ribonucleic acid polymerase and stimulatory factors from HeLa and KB cells. J Biol Chem. 1973;248:3777–3788. [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Wansink DG, Nelissen RL, de Jong L. In vitro splicing of pre-mRNA containing bromouridine. Mol Biol Rep. 1994;19:109–113. doi: 10.1007/BF00997156. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labelling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann R, Raskas HJ, Roeder RG. The transcriptional role of host DNA-dependent RNA polymerases in adenovirus-infected KB cells. Cold Spring Harb Symp Quant Biol. 1975;34:495–500. doi: 10.1101/sqb.1974.039.01.061. [DOI] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- Wolgemuth DJ, Hsu M-T. Visualization of nascent RNA transcripts and simultaneous transcription and replication in viral nucleoprotein complexes from adenovirus 2-infected HeLa cells. J Mol Biol. 1981;147:247–268. doi: 10.1016/0022-2836(81)90440-x. [DOI] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]