Abstract
SCCmec is a mobile genetic element that carries the gene (mecA) mediating methicillin resistance in staphylococci. For Staphylococcus aureus, four SCCmec types have been described, one (type IV) of which has been associated with newly identified community-acquired methicillin-resistant S. aureus. However, the distribution of SCCmec types among S. epidermidis is not known. SCCmec typing of a collection of 44 methicillin-resistant Staphylococcus epidermidis (MRSE) isolates recovered between 1973 and 1983 from the blood of patients with prosthetic valve endocarditis (PVE) was performed by PCR amplification of key genetic elements (mecA, mecI, IS1272, and ccrAB). Of the 44 isolates, 1 (2%) harbored SCCmec type I, 15 (34%) harbored type II, 12 (28%) harbored type III, and 16 (36%) harbored type IV. The complete nucleotide sequence of SCCmec type IV was determined for 16 isolates and found to be identical in size (24 kb) and 98% homologous to DNA sequences published for S. aureus. Type IV SCCmec was also common (5 of 10 isolates) among a geographically dispersed collection of 10 recent (1998 to 2001) S. epidermidis bloodstream isolates. Multilocus sequence typing (MLST) (using the same seven genes presently employed for S. aureus MLST) of these MRSE isolates and of 10 additional recent geographically dispersed methicillin-susceptible isolates demonstrated that all 16 PVE isolates and 2 of 5 recent isolates harboring type IV SCCmec were in three related clonal groups. All three MSSE PVE isolates recovered from patients between 1976 and 1979 were in the same clonal groups as type IV SCCmec MRSE isolates. These data support the hypothesis of intra- and interspecies transfer of type IV SCCmec and suggest that there are clonal associations in S. epidermidis that correlate with SCCmec type.
Resistance of staphylococci to methicillin and other beta-lactam antibiotics is mediated by a specific penicillin binding protein (PBP) that has a reduced affinity for beta-lactam antibiotics (PBP 2a) (6, 15, 29). PBP 2a is encoded by the mecA gene, which is carried on a chromosomal genetic element designated staphylococcus chromosomal cassette mec (SCCmec). Four types of SCCmec, differing in DNA sequences and gene content, have been identified in Staphylococcus aureus (18, 19, 21), and the first methicillin-resistant (MR) S. aureus (MRSA) isolates (isolated in the early 1960s) are thought to have emerged from an epidemic methicillin-susceptible (MS) S. aureus (MSSA) lineage upon acquisition of SCCmec type I from coagulase-negative staphylococci (CoNS) (7, 12).
MRSA isolates recovered from patients who have had little or no contact with hospital-based care have recently been described from the community (1, 13, 14, 16, 23, 24, 26, 28). Most of these isolates have genetic backgrounds similar to those of MSSA isolates present in the community and most have a novel SCCmec type (type IV) rarely found in S. aureus isolates recovered before 1990 (22, 25). Type IV SCCmec is smaller than the SCCmec types previously reported (21 to 24 kb versus 34 to 67 kb) (18, 19, 21). These data suggest that type IV SCCmec is mobile and has been recently acquired by S. aureus, possibly from Staphylococcus epidermidis, the CoNS that is the most common MR species.
This study was performed to assess the degree of similarity of SCCmec between S. epidermidis and S. aureus clinical isolates and to determine whether S. epidermidis is a likely reservoir of this resistance determinant. The clinical S. epidermidis isolates used were recovered from the blood of patients with documented prosthetic valve endocarditis (PVE) diagnosed between 1973 and 1983, before the first reported MRSA isolates shown to carry type IV SCCmec were diagnosed, and from the blood of patients without PVE, as part of a Surveillance and Control of Pathogens of Epidemiologic Importance (SCOPE) study (10). SCOPE collects bacteria isolated from the blood of patients in 52 hospitals representing all geographic regions of the United States. We also performed multilocus sequence typing (MLST) to investigate the distribution of SCCmec types among S. epidermidis genotypes.
MATERIALS AND METHODS
Bacterial strains and media.
S. epidermidis isolates used in this study were from two sources. First, there were 47 isolates collected between 1973 and 1983 from the blood of patients with PVE in Richmond, Va., Boston, Mass., Birmingham, Ala., and Winnipeg, Manitoba, Canada (Table 1). A total of 44 of these isolates were MR and three were MS. The clinical criteria for PVE and characterization of many of the infections have been previously published (5, 20). Second, there were 20 non-PVE isolates (10 MR and 10 MS) from the blood of patients with bloodstream infection (BSI), collected through the SCOPE study (10, 30). They were classified as true bacteremic isolates on the basis of previously published criteria (10). They were from four different geographic areas in the United States (the southeast, northeast, northwest, and southwest regions) and were recovered between 1998 and 2001 (Table 1). S. aureus strains COL (SCCmec type I), N315 (SCCmec type II), ANS46 (SCCmec type III), and MW2 (SCCmec type IV) were used as SCCmec standards. Brain heart infusion broth and brain heart infusion agar (Becton Dickinson, Sparks, Md.) were used for cultivation of S. epidermidis and S. aureus strains. All strains have been identified to species level according to NCCLS criteria in the original studies. Identification of S. epidermidis was confirmed by demonstrating that the isolates produced catalase, did not produce coagulase, and could not use trehalose. Resistance to oxacillin was confirmed when strains grew on Mueller-Hinton agar plates containing 6 μg of oxacillin/ml after incubation for 72 h at 30°C.
TABLE 1.
Characteristics of S. epidermidis study isolates
| Yr of isolation | U.S. or Canadian locationa | No. of strains | Source | Studyb |
|---|---|---|---|---|
| 1976-1983 | Richmond, Va. | 12 | Blood-valved | PVE |
| 1973-1975 | Winnipegc | 5 | Blood-valve | PVE |
| 1976-1982 | Boston, Mass. | 20 | Blood-valve | PVE |
| 1976-1980 | Birmingham, Ala. | 10 | Blood-valve | PVE |
| 1998-2001 | NE | 3 | Blood | SCOPE |
| 1998-2001 | NW | 9 | Blood | SCOPE |
| 1998-2001 | SW | 2 | Blood | SCOPE |
| 1998-2001 | SE | 6 | Blood | SCOPE |
NE, northeastern United States; NW, northwestern United States; SW, southwestern United States; SE, southeastern United States.
Studies (reference[s]): PVE (5, 20); SCOPE (10).
Manitoba, Canada.
Valve, prosthetic valve.
Determination of SCCmec types.
Genomic DNA of S. epidermidis strains was extracted with a QIAGEN plasmid minikit (QIAGEN, Hilden, Germany) as recommended by the manufacturer. SCCmec types were determined with the use of specific primers for amplification of the key genetic elements mecA (primers mecA1 and mecA2), mecI (primers 214 and 215), IS1272 (primers F3 and mA2 and primers F1 and B3), and ccrA/B (primers α1, α2, α3, and β2). PCR was performed using a Taq PCR MasterMix kit (QIAGEN) with a 50-μl reaction volume in a MiniCycler thermocycler (MJ Research, Boston, Mass.). The overall size of SCCmec was determined (using Expand Taq [Roche Diagnostics, Mannheim, Germany] according to the procedure recommended by the manufacturer) by long-range PCR with five sets of overlapping primers (18, 19, 21). All PCR products were purified with a QIAquick PCR purification kit (QIAGEN). The nucleotide sequences of all primers are shown in Table 2 or were reported previously (18, 19, 21). Nucleotide sequences of SCCmec for all strains designated type IV were established (utilizing an ABI Prism 377 DNA sequencer [Applied Biosystems, Foster City, Calif.] with BigDye fluorescent terminators) by automated sequencing of PCR products obtained with several sets of overlapping primers. For most isolates the nucleotide sequence was determined on a single strand. However, for several strains most of the sequence was confirmed on both strands.
TABLE 2.
Primers used for SCCmec typing
| Primer | Sequence |
|---|---|
| F3 | TTGGGTTTCACTCGGATG |
| mA2 | AACGTTGTAACCACCCCAAGA |
| F1 | CACAATCTGTATTCTCAGGTCG |
| B3 | ATTAGTGCTCGTCTCCACG |
| α2 | AACCTATATCATCAATCAGTACGT |
| α3 | TAAACGCATCAATGCACAAACACT |
| α4 | AGCTCAAAAGCAAGCAATAGAAT |
| β2 | ATTGCCTTGATAATAGCCTCT |
| mecA1 | GGAGGATATTGATGAAAAAG |
| mecA2 | GCTTCACTGTTTTGTTATTC |
| 214 | CGGATCCGAAATGGAATTAATATAATG |
| 215 | CGGAATTCGACTTGATTGTTTCCTC |
Computer analysis of nucleotide sequences.
All analysis was carried out using Vector NTI suite 7.1 software for Windows (InforMax, Bethesda, Md.). A homology search was performed with the BLAST program for the EMBL and GenBank databases. Sequences were compared to the published sequences of CA05 (JCSC1968) and 8/6-3P (JCSC1978) (GenBank accession numbers AB063172 and AB063173, respectively) (21).
MLST.
Housekeeping genes were selected on the basis of the published MLST scheme for S. aureus (11). The DNA sequences of the housekeeping genes of S. epidermidis RP62A were from the unpublished genome sequence found on The Institute for Genomic Research website (www.tigr.org). Primers were designed by using the sequences of highly conserved regions flanking more variable regions. Each primer pair amplified an internal fragment of the housekeeping gene (∼500 bp) and allowed accurate sequencing of fragments of approximately 450 bp of each gene on both strands. The following seven housekeeping genes were used in the final MLST scheme (the fragments were amplified by using the primers shown in Table 3): carbamate kinase (arcC), shikimate 5-dehydrogenase (aroE), glycerol kinase (glpK), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpiA), and acetyl coenzyme A acetyltransferase (yqi). Chromosomal DNA was extracted using a bacterial genomic DNA purification kit (Edge BioSystems, Gaithersburg, Md.) according to the instructions of the manufacturer, with the addition of lysostaphin at a final concentration of 50 μg/ml. For each isolate, the allele at each of the seven loci gave an allelic profile that was used to define its sequence type (ST). Polymorphic sites were displayed by using Sequence Output, a Macintosh program available from the MLST website (http://www.mlst.net). Clonality was assessed using BURST, a web-implemented clustering algorithm designed for use on MLST data sets from bacterial pathogens. Groups with at least five out of seven identical alleles were defined as clonal groups. The program was developed on the basis of the epidemic model of population structure devised by Smith et al. (27) and specifically examines the relationships between very closely related genotypes within clonal complexes, thereby bypassing the difficulties resulting from the frequent recombination that overwhelms the deep-rooted phylogenetic signal (8).
TABLE 3.
Genes and primers used in MLST for S. epidermidis
| Gene
|
Gene size (bp) | Primer
|
||
|---|---|---|---|---|
| Name | Symbol | Name | Sequence | |
| Carbamate kinase | arcC | 468 | arcC-F | TGTGATGAGCACGCTACCGTTAG |
| arcC-R | TCCAAGTAAACCCATCGGTCTG | |||
| Shikimate 5-dehydrogenase | aroE | 423 | aroE-F | CATTGGATTACCTCTTTGTTCAGC |
| aroE-R | CAAGCGAAATCTGTTGGGG | |||
| Glycerol kinase | glpK | 471 | glpK-F | CATCACCACGGTCAAAACATGC |
| glpK-R | CAGGTCGTCCAATCTATCACGC | |||
| Phosphate acetyltransferase | pta | 477 | pta-F | TACTGCATCGTATCCACCTAAACG |
| pta-R | TGGTGCTGCACATTCTACTGGAG | |||
| Triosephosphate isomerase | tpiA | 411 | tpiA-F | CCACCATATTGAATACGTGTAGCG |
| tpiA-R | GCTTACTTTGAAGAAAGCGGTG | |||
| Guanylate kinase | gmk | 465 | gmk-F | TCGATTCTTAGCGAGTTCAACC |
| gmk-R | CCTTCAGGTGTTGGAAAGGG | |||
| Acetyl-CoA acetyltransferase | yqi | 474 | yqi-F | TGCTGGACGGAGTTGTGCTAAC |
| yqi-R | ATCCTGCTCGTATTGCTGCG | |||
RESULTS
SCCmec typing.
Among the 44 S. epidermidis PVE strains, the SCCmec types were as follows: one (2%) was SCCmec type I, 15 (34%) were type II, 12 (28%) were type III, and 16 (36%) were type IV. Among the 10 SCOPE isolates, 5 (50%) were type IV, 4 (40%) were type III, and 1 (10%) was type II. Long-range PCR analysis with overlapping primers of the SCCmec region of the type IV isolates revealed a size of 24.2 kb for all strains. No significant variation in the sizes of the individual PCR products was observed among strains.
Sequencing.
The entire nucleotide sequence was determined for each SCCmec IV strain. From the left inverted repeat to the right inverted repeat, all elements were 24,248 bp in size. Comparison to the published sequences of S. aureus CA05 (JCSC1968) revealed a homology of 98 to 99%, with no insertions or deletions. Differences were usually single-base mismatches. Most of the sequence was only determined on one strand, but this was felt to be sufficient because of the high homology of the single strand to the published sequence.
MLST.
Seven housekeeping gene fragments (between 411 and 477 bp in size) (Table 3) were sequenced from each of 44 MR S. epidermidis (MRSE) isolates from patients with PVE (isolated from 1973 to 1983), three MSSE isolates were sequenced from isolates from patients with PVE (isolated in 1976, 1979, and 1979), 10 MRSE isolates were sequenced from isolates from patients with confirmed BSI (isolated for the SCOPE study [1998 to 2001]) and 10 MSSE isolates were sequenced from isolates from patients with confirmed BSI (SCOPE, 1998 to 2001). For each isolate, the sequences obtained at each of the seven loci were compared with those of every other isolate and the alleles were numbered consecutively. Sequences were assigned as distinct alleles even when they differed at a single nucleotide. No weighting was applied to reflect the number of nucleotide differences between alleles. Between 5 (yqi) and 10 (arcC) alleles were present at each locus (Table 4), with a mean of 8.1 alleles per locus, which allowed 32 STs to be distinguished. Most of the S. epidermidis genes were relatively uniform, with the number of polymorphic (variable) nucleotide sites at the seven loci ranging between 10 in tpiA and 34 in glpK (Fig. 1).
TABLE 4.
MLST results
| ST | Date(s) of isolation | No. of locations | SCCmec type(s) | No. of strains | MLST profilea |
|---|---|---|---|---|---|
| 1 | 1979 | 1 | MS | 1 | 1-1-2-1-1-1-1 |
| 2 | 1973-1980 | 4 | II, IV, MS | 7 | 1-1-3-1-1-1-1 |
| 3 | 2000-2001 | 2 | IV, MS | 3 | 1-1-3-1-2-1-1 |
| 4 | 1976 | 1 | IV | 1 | 1-1-3-1-5-1-1 |
| 5 | 1976-1979 | 1 | IV | 3 | 1-1-3-2-1-1-1 |
| 6 | 1976 | 1 | IV | 1 | 1-1-3-3-5-1-4 |
| 7 | 1980s | 1 | II, III | 2 | 1-1-3-6-2-1-1 |
| 8 | 1973-1978 | 4 | I-IV | 13 | 1-2-3-1-1-1-1 |
| 9 | 1979 | 1 | IV | 1 | 1-2-3-5-1-1-1 |
| 10 | 1979 | 1 | II | 1 | 1-2-8-1-1-1-1 |
| 11 | 2000 | 1 | MS | 1 | 1-5-3-1-1-8-5 |
| 12 | 1978-1983 | 2 | II, III | 3 | 1-6-3-1-1-1-1 |
| 13 | 1979 | 1 | III | 1 | 1-6-4-1-6-1-1 |
| 14 | 2001 | 1 | MS | 1 | 1-7-3-8-4-9-5 |
| 15 | 1998 | 1 | II | 1 | 1-7-7-1-1-1-1 |
| 16 | 1978 | 1 | III | 1 | 10-2-3-1-1-6-4 |
| 17 | 1973 | 1 | III | 1 | 2-1-10-1-1-1-1 |
| 18 | 1973-2000 | 3 | II, IV | 3 | 2-1-3-1-1-1-1 |
| 19 | 1976 | 1 | III | 1 | 2-6-6-1-1-1-1 |
| 20 | 2000 | 1 | MS | 1 | 3-1-5-6-2-5-3 |
| 21 | 2000 | 1 | MS | 1 | 3-3-5-6-2-5-3 |
| 22 | 2000 | 1 | MS | 1 | 3-3-9-7-3-2-2 |
| 23 | 2001 | 1 | MS | 1 | 3-4-5-6-2-5-3 |
| 24 | 1978 | 1 | III | 1 | 4-8-3-4-1-7-4 |
| 25 | 2000 | 1 | MS | 1 | 5-1-3-6-1-5-1 |
| 26 | 1976 | 1 | III | 1 | 6-6-3-1-1-1-1 |
| 27 | 1976-2000 | 6 | II, III | 9 | 7-1-3-1-1-1-1 |
| 28 | 2000 | 1 | III | 1 | 7-1-3-1-1-3-1 |
| 29 | 1998 | 1 | IV | 1 | 7-1-3-6-2-4-1 |
| 30 | 2000 | 1 | IV | 1 | 8-1-3-1-2-1-1 |
| 31 | 2001 | 1 | MS | 1 | 9-5-1-6-1-5-1 |
| 32 | 2001 | 1 | MS | 1 | 9-5-3-1-1-1-5 |
Allele numbers. The genes corresponding to the numbers are (from left to right) arcC, aroE, glpK, gmk, pta, tpiA, and yqi.
FIG. 1.

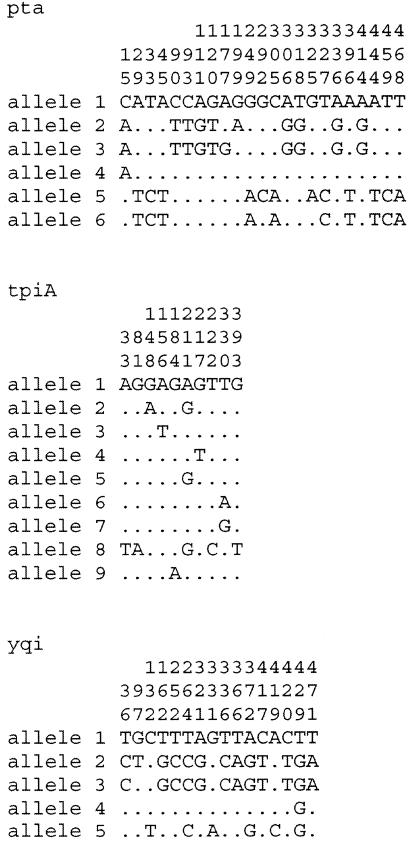
Polymorphic sites of all alleles (generated with Sequence Output software [www.mlst.net]). The numbers above the nucleotide designations (rows 1 to 3 of each allele data set) show the nucleotide numbers (to be read from top to bottom). The nucleotide at each numbered position is displayed in cases in which nucleotides differ. The differences define the alleles. Dots indicate that the nucleotide is the same as that of allele 1.
Association of MLST and SCCmec.
Figure 2 shows the clonal associations (derived from the matrix of pair-wise differences between the allelic profiles) of the 67 isolates, as depicted using the BURST program (www.mlst.net). A total of 52 of the 54 MRSE isolates clustered into three related clonal groups. In addition, the three MSSEs isolated in the 1970s had STs that were identical to those of MRSE PVE isolates from the same decade. However, while all of the contemporary (1998 to 2001) MRSE blood isolates were in the same clonal groups as the MRSE PVE isolates from the 1970s, 9 of the 10 contemporary MSSE isolates were unrelated to all of the MRSE isolates. All of the type IV SCCmec-containing isolates, whether contemporary or from the 1970s, were clustered into three clonal groups on the basis of the presence of ST 2, 3, or 8. However, isolates with either type II or type III SCCmec were also in these clonal groups, and isolates with identical MLSTs carried more than one SCCmec type (ST 2 with SCCmec types II and IV and ST 8 with SCCmec types I, II, III, and IV).
FIG. 2.
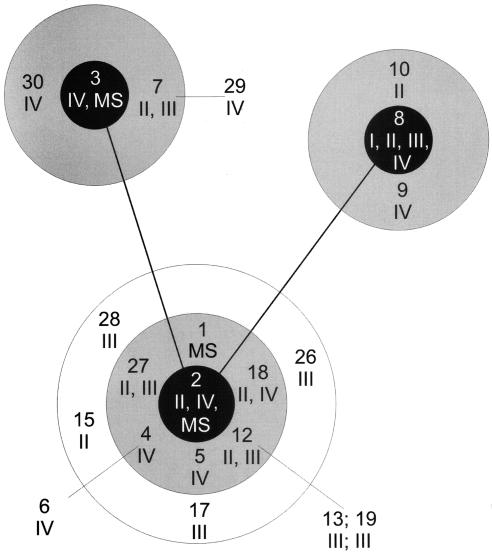
Clustering of STs in a graphic display of the results of BURST analysis (www.mlst.net). Arabic numbers 1 to 30, STs (see Table 4 for details); roman numerals I to IV, SCCmec types of isolates. STs differ by one allele within a circle and from one circle to the next (i.e., from the black- to gray-shaded circle portions and from the gray- to white-shaded circle portions). Therefore, the gray-shaded circle portions show the single-locus variants of the respective ancestral strains and the white circle portion shows the double-locus variants of that ancestral strain. Also, STs differ by one allele between connected black circle portions (representing ST ancestor types 2, 3, and 8). STs outside of any circle differ by two alleles from the ST to which they are connected (e.g., ST 6 to ST 4).
DISCUSSION
In the present study we found that 36% of S. epidermidis isolates recovered in the 1970s from patients with documented PVE contained mecA within a type IV SCCmec resistance island. The DNA sequence of S. epidermidis SCCmec type IV was identical to the element found in community-onset MRSA strains of S. aureus (21). Evaluation of MLST data has suggested that transfer of SCCmec into MSSA strains prevalent in a given region produced MRSA. Enright et al. (12) proposed that pandemic clones of hospital-associated MRSA emerged as a result of the insertion of SCCmec into five separate sensitive epidemic lineages. Similarly, strains of MRSA have arisen in Native American communities that have little association with the health care system. These isolates have pulsed-field gel electrophoresis patterns that are identical to those of prevalent MSSA isolates except for the presence of type IV SCCmec. These isolates are completely distinct from hospital-associated isolates (14, 22, 25).
The hypothesis of the transfer of SCCmec between S. epidermidis and S. aureus is supported by several lines of evidence. First, the nucleotide sequence of the entire 24 kb of type IV SCCmec in the 16 isolates of S. epidermidis that we examined was identical to the published sequence of type IV SCCmec in S. aureus strains acquired in the community. This is in contrast to findings for the rest of the S. epidermidis genome, in which only 17% of the open reading frames have at least 80% identity to S. aureus over their entire length (S. Gill, The Institute for Genome Research, personal communication). This suggests that interspecies exchange of this DNA has occurred.
Second, we previously documented that the insertion junction of IS1272 into the mecR1 gene of SCCmec type I from S. aureus COL was identical to the junction in S. epidermidis. On the basis of the finding that IS1272 is present throughout the genomes of S. haemolyticus and S. epidermidis and that it is less frequently found in the genome of S. aureus, we postulated that the insertion event occurred in CoNS, with subsequent transfer of IS1272-containing SCCmec to S. aureus (4). In the present study we also found the IS1272 insertion junctions of S. epidermidis and S. aureus SCCmec type IV to be identical. Thus, not only does this provide additional evidence for the interspecies transfer of the SCCmec element, but it also suggests that recombination occurred in CoNS between type I sequences and additional sequences and generated type IV SCCmec. The type IV sequences were subsequently transferred into S. aureus.
Third, MR is highly prevalent among S. epidermidis isolates and is less common among S. aureus isolates. Over 80% of S. epidermidis isolates from documented infections are MR, MRSE is the predominant skin flora of hospitalized patients, and colonizing MRSE can persist on the skin for months after discharge (2, 3, 5). In contrast, MRSA infections comprise from 30 to 50% of hospital-associated S. aureus cases, after discharge MRSA is usually carried in the nose only, and only a small percentage of carriers harbor MRSA. Thus, the reservoir of SCCmec is likely to be larger in S. epidermidis both in the hospital and in the community.
Finally, as we documented in the present study, type IV SCCmec was highly prevalent among S. epidermidis isolates causing infections in the 1970s (being found in 36% of isolates) and was not found among MRSA isolates recovered during that time. Ito et al. found that only 2 of 38 MRSA isolates recovered from patients in Europe, Israel, and Japan between 1961 and 1993 carried SCCmec type IV, and these were from 1981 and 1985 (18). Among 15 MRSA isolates in our collection of isolates from the United States in which IS1272 was found to have been inserted into mecR1, 9 were isolated between 1961 and 1983 and 6 were isolated between 1983 and 1990. None of nine (0%) of isolates from the early group contained type IV SCCmec; all were type I. In contrast, five of six isolates recovered between 1983 and 1990 (83%) carried type IV SCCmec (H. Wisplinghoff and G. Archer, unpublished data). Thus, the first S. aureus isolates carrying type IV SCCmec were recovered from patients in the early 1980s and S. epidermidis isolates carrying this element were prevalent in the early 1970s.
As mentioned above, MLST has been an excellent tool for assessing the movement of SCCmec among S. aureus strains (11, 12). Identical or closely related STs have been found to harbor different SCCmec types, including type IV. We used the same MLST scheme for S. epidermidis that was used for S. aureus and found that it provided excellent clonal discrimination. There were from 5 to 10 alleles of each gene and 32 STs among 67 isolates. We found that identical or closely related (i.e., differing by only one allele) STs carried each of the SCCmec types (I, II, III, or IV) and that the same SCCmec type could be found within multiple, often unrelated STs. However, among the S. epidermidis isolates that carried type IV SCCmec, all 16 recovered from patients between 1973 and 1983 and two of the five SCOPE isolates from the late 1990s were in three clonal groups. In addition, all three MSSE isolates recovered from patients with PVE in the 1970s were in the same clonal group as the MRSE isolates containing type IV SCCmec. These data suggest that there is transfer of SCCmec among S. epidermidis strains and that certain clones are both preferential recipients for type IV SCCmec and more successful at environmental competition, having survived for almost 30 years.
MLST and SCCmec typing have proven invaluable for epidemiological and evolutionary investigations of S. aureus, and here we demonstrated the utility of these techniques for studies of S. epidermidis. The theory that CoNS represent a large genetic pool of resistance genes (available to the more virulent species S. aureus) has been confirmed in this report, and future work applying the methods used here will be useful in quantifying the extent of gene flow in the staphylococci.
Acknowledgments
This work was supported in part by NIH grant R-37 AI 35705.
REFERENCES
- 1.Adcock, P. M., P. Pastor, F. Medley, J. E. Patterson, and T. V. Murphy. 1998. Methicillin-resistant Staphylococcus aureus in two child care centers. J. Infect. Dis. 178:577-580. [DOI] [PubMed] [Google Scholar]
- 2.Archer, G. L., and B. C. Armstrong. 1983. Alteration of staphylococcal flora in cardiac surgery patients receiving antibiotic prophylaxis. J. Infect. Dis. 147:642-649. [DOI] [PubMed] [Google Scholar]
- 3.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer, G. L., J. A. Thanassi, D. M. Niemeyer, and M. J. Pucci. 1996. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 40:924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archer, G. L., N. Vishniavsky, and H. G. Stiver. 1982. Plasmid pattern analysis of Staphylococcal epidermidis isolates from patients with prosthetic valve endocarditis. Infect. Immun. 35:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, D. F., and P. E. Reynolds. 1980. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 122:275-278. [DOI] [PubMed] [Google Scholar]
- 7.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 9.Dickinson, T. M., and G. L. Archer. 2000. Phenotypic expression of oxacillin resistance in Staphylococcus epidermidis: roles of mecA transcriptional regulation and resistant-subpopulation selection. Antimicrob. Agents Chemother. 44:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, A. L., J. F. Marcinak, P. D. Mangat, and P. C. Schreckenberger. 1999. Community-acquired and clindamycin-susceptible methicillin-resistant Staphylococcus aureus in children. Pediatr. Infect. Dis. J. 18:993-1000. [DOI] [PubMed] [Google Scholar]
- 14.Groom, A. V., D. H. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201-1205. [DOI] [PubMed] [Google Scholar]
- 15.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu, K., Y. Katayama, H. Yuzawa, and T. Ito. 2002. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 292:67-74. [DOI] [PubMed] [Google Scholar]
- 18.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karchmer, A. W., G. L. Archer, and W. E. Dismukes. 1983. Staphylococcus epidermidis causing prosthetic valve endocarditis: microbiologic and clinical observations as guides to therapy. Ann. Intern. Med. 98:447-455. [DOI] [PubMed] [Google Scholar]
- 21.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O'Boyle, R. N. Danila, J. E. Cheek, M. T. Osterholm, K. A. Moore, and K. E. Smith. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996-1998. Clin. Infect. Dis. 33:990-996. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien, F. G., J. W. Pearman, M. Gracey, T. V. Riley, and W. B. Grubb. 1999. Community strain of methicillin-resistant Staphylococcus aureus involved in a hospital outbreak. J. Clin. Microbiol. 37:2858-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 26.Shahin, R., I. L. Johnson, F. Jamieson, A. McGeer, J. Tolkin, E. L. Ford-Jones, and Toronto Child Care Center Study Group. 1999. Methicillin-resistant Staphylococcus aureus carriage in a child care center following a case of disease. Arch. Pediatr. Adolesc. Med. 153:864-868. [DOI] [PubMed] [Google Scholar]
- 27.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suggs, A. H., M. C. Maranan, S. Boyle-Vavra, and R. S. Daum. 1999. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. Pediatr. Infect. Dis. J. 18:410-414. [DOI] [PubMed] [Google Scholar]
- 29.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voelker, R. 1996. New group tracks hospitals' drug-resistant bugs. JAMA 275:177-178. [PubMed] [Google Scholar]


