Abstract
The presence of integrons in 85 multiresistant German isolates of the predominating Salmonella enterica subsp. enterica serovar Paratyphi B dT+ clone was investigated. All isolates possessed a chromosomally located Tn7-like class 2 integron carrying the same dfrA1-sat1-aadA1 array of gene cassettes. Only four isolates (4.7%) revealed an additional class 1 integron with two strains each containing the aadA1 or dfrA1-aadA1 gene cassettes.
During the last decade multidrug-resistant d-tartrate-positive Salmonella enterica subsp. enterica serovar Paratyphi B (Salmonella serovar Paratyphi B dT+) isolates (formerly called Salmonella serovar Java) have increasingly been isolated from poultry and poultry products in Germany and The Netherlands (7, 18, 29). Recent studies by Brown et al. (1) strongly suggest that the same multiresistant clone found in German and Dutch poultry was responsible for 10 human cases of Salmonella serovar Paratyphi B dT+ infections in Scotland.
By studying the phenotypic and molecular features of contemporary and old Salmonella serovar Paratyphi B dT+ isolates from poultry, Miko et al. (18) could show that since the mid-1990s the majority of contemporary isolates have been multidrug resistant and belong to a particular clone defined by a distinct pulsed-field gel electrophoresis (PFGE) and IS200 profile. Despite the multiple-drug resistance, class 1 integrons were found in only 1 of the 39 contemporary isolates, suggesting the minor importance of this integron class in the acquisition of multidrug resistance by the predominating Salmonella serovar Paratyphi B dT+ clone.
The role of integrons and gene cassettes in the evolution and dissemination of multidrug resistance in gram-negative bacteria is well established (22, 23). Integrons are characterized by their ability to integrate and excise genes that are part of gene cassettes via a site-specific recombination event (2, 3, 5, 22). Integrons harbor a gene, intI, which encodes a site-specific recombinase (IntI integrase), and an attachment site, attI, into which individual gene cassettes are inserted (24). To date, five distinct integron classes associated with gene cassettes that contain antibiotic resistance genes have been described (4). The best-characterized group is class 1 integrons. They are the most widely disseminated ones among the members of the family Enterobacteriaceae, including many of the Salmonella enterica subsp. enterica serovars (10, 11, 15, 25, 26). For class 1 integrons, intI1, attI1, and promoter Pc are found within potentially mobile elements that are transposons, e.g., Tn402, or defective transposon derivatives. Class 2 integrons have been shown to contribute to the spread of antibiotic resistance genes in the family Enterobacteriaceae as well (9, 31). In Salmonella they have been identified so far only in the serovar Typhimurium (21). For class 2 integrons, the typical intI2* gene, which includes a termination codon, and the attI2 and Pc are found within transposons such as Tn7 (27, 28). The remaining three integron classes are not well characterized, since to date only a single example of each of them has been detected.
The aim of this study was to determine whether class 2 integrons are common mediators of multidrug resistance in Salmonella serovar Paratyphi B dT+ isolates of the predominating clone in Germany.
Characterization of Salmonella serovar Paratyphi B dT+ isolates.
A total of 85 contemporary multidrug-resistant strains isolated in Germany between 1995 and 2001 were included in the study. Thirty-nine of the isolates had been described in reference 18 and were already known to belong to the predominating clone (group 3 strains). The other 46 independent isolates from 2001 were selected by the same criteria given in reference 18. They originated from food products of poultry origin (n = 31) and from poultry (n = 12), cattle (n = 1), and feed (n = 2). Fourteen comparative strains described as group 1 and 2 strains in reference 18 were also analyzed. They did not belong to the contemporary clone (Table 1).
TABLE 1.
Integrons in Salmonella serovar Paratyphi B dT+ strains
| No. of strains | Yrs of isolation | Antibiotic resistancea | Integron (cassettes/locationb)
|
Profilec
|
||
|---|---|---|---|---|---|---|
| Class 1 | Class 2 | PFGE | IS200 | |||
| Multiresistant strains of the predominating clone | ||||||
| 81 | 1995-2001 | TMP, SPE, STR, SMX, NAL, AMP, KAN-NEO | dfrA1-sat1-aadA1/C | X8 | ISP9 | |
| 4 | 1995-2001 | TMP, SPE, STR, SMX, TET, NAL, AMP, GEN | dfrA1-aadA1/P or aadA1/P | dfrA1-sat1-aadA1/C | X8 | ISP9 |
| Comparative strains | ||||||
| 5 | 1994-1999 | TMP, SPE, STR, SMX, TET, NAL, CHL, KAN-NEO | dfrA12-aadA2/P | X7 | ISP8 | |
| 9 | 1960-1993 | Sensitive | X1-X6 | ISP1-ISP7 | ||
Boldface indicates resistance exhibited by all strains, and lightface indicates resistance exhibited by only some strains. Abbreviations: TMP, trimethoprim; SPE, spectinomycin; STR, streptomycin; SMX, sulfamethoxazole; NAL, nalidixic acid; AMP, ampicillin; KAN-NEO, kanamycin-neomycin; TET, tetracycline; GEN, gentamicin; CHL, chloramphenicol.
P, high-molecular-weight plasmid; C, chromosome.
Assignment according to reference 18. X, XbaI; ISP, IS200 profile.
Serotyping, d-tartrate fermentation, plasmid profile typing, PFGE, and IS200 profiling were done as described previously (18). All isolates from 2001 revealed the PFGE profile X8 and the IS200 profile ISP9 as well and could consequently be assigned to the predominating German Salmonella serovar Paratyphi B dT+ clone. Susceptibilities to a panel of antimicrobial agents (Table 1) were assessed by determining the MIC by the NCCLS broth microdilution method (19). Breakpoints given by the NCCLS and DANMAP (6) were used. All 85 isolates exhibited a core spectrum of antibiotic resistance determinants for trimethoprim, spectinomycin, and streptomycin. Additional resistances were found to sulfamethoxazole (in 71% of the strains), nalidixic acid (62%), ampicillin (51%), tetracycline (7%), kanamycin-neomycin (3%), and gentamicin (3%).
Identification of integrons.
Integrons were detected in all 85 multiresistant isolates by using PCR with degenerate primers to amplify conserved regions of the integron integrase genes intI1, intI2, and intI3 (30). In order to distinguish class 1 and class 2 integrons, PCR amplifications were carried out with intI1 (20)- and intI2 (17)-specific primers. All isolates carried class 2 integrons, and four of them revealed a class 1 integron as well (Table 1).
To detect inserted gene cassettes, the variable regions of class 1 integrons, carried by the four intI1-positive isolates, were amplified using primers 5′ CS and 3′ CS, which annealed with the DNA regions flanking the recombination site attI (16). Direct sequencing of the purified PCR products revealed the dfrA1 and aadA1 gene cassettes or the aadA1 gene cassette alone in two isolates each.
Class 2 integron cassette arrays, carried by the 85 intI2-positive isolates, were amplified using primers hep74, which binds to attI2, and hep51, which binds to orfX, situated at the right end of the cassette region within transposon Tn7 (12, 31). All of these isolates and two Tn7-containing positive control strains, Escherichia coli K-12 J62 (ColE1::Tn7) and E. coli K-12 χ6149 S17-1 (RP4-2), carried a class 2 integron characterized by a PCR product of about 2.2 kb. By contrast, no PCR product was observed in the comparative strains (Table 1).
The sequence similarity among the 2.2-kb amplicons was investigated and confirmed by restriction analysis with the restriction enzyme HincII, HinfI, or AvaI. All amplicons gave identical restriction patterns, and the sum of the sizes of the restriction fragments was consistent with the size of the undigested amplicon (Fig. 1 and data not shown).
FIG. 1.
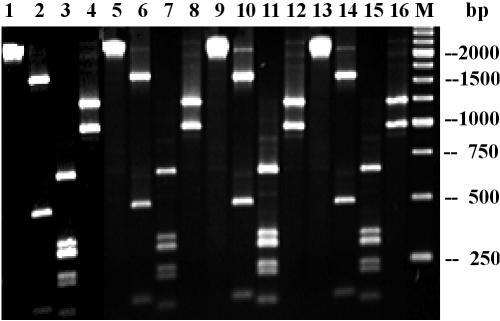
Results of the restriction analysis of the 2.2-kb amplicons of ColE1::Tn7, RP4-2, and two representative class 2 integron-positive Salmonella serovar Paratyphi B dT+ strains with HincII (lanes 2, 6, 10, and 14), HinfI (lanes 3, 7, 11, and 15), and AvaI (lanes 4, 8, 12, and 16). Lanes 1, 5, 9, and 13 contain the undigested amplicons. Lane M, 250-bp ladder (Roche Diagnostics, Mannheim, Germany).
Tn7 has been shown to carry three integrated resistance gene cassettes, namely, dfrA1, sat1, and aadA1, encoding resistance to trimethoprim, streptothricin, and spectinomycin-streptomycin, respectively (22, 27, 28). Recently, a fourth gene cassette, ORFX, coding for a protein of unknown function was described by Hansson et al. (12).
We detected the Tn7-borne antibiotic resistance genes in all Salmonella serovar Paratyphi B dT+ isolates carrying class 2 integrons by PCR analysis. The gene-specific primers P3/P4 for dfrA1 (8); sat1-F/B (GAAACATTGGATGCTGAG/GAACCAGTACCAGTACAT), designed for this work; and aadA-F/B (26) were used. To confirm the cassette content and the specific gene arrangement in the integron, PCR amplifications using primers flanking adjacent gene regions (hep74/P4, P3/sat1-B, sat1-F/aadA-B, and aadA-F/hep51) were performed. Partial sequencing of the amplicons corroborated the presence of the same array of cassettes, dfrA1-sat1-aadA1, in all class 2 integrons.
Localization of the class 2 integrons.
In order to map the class 2 integrons, PFGE after digestion of genomic DNAs with XbaI and subsequent Southern blot analysis with intI2-, dfrA1-, sat1-, and aadA1-specific probes (which were not cut by XbaI) was carried out. Plasmid DNA extracted by the method of Kado and Liu (14) was hybridized with these probes as well, in order to map these genes on the plasmids. Results of the experiments revealed that the intI2-, dfrA1-, sat1-, and aadA1-specific probes hybridized to the same two chromosomal bands of about 78 and about 410 kb in all Salmonella serovar Paratyphi B dT+ isolates carrying class 2 integrons (Fig. 2). This suggests the presence of two copies of the integrase 2 gene and the other integron-specific genes on the bacterial chromosome. In contrast to class 1 integrons, which could be localized on high-molecular-weight plasmids (data not shown), none of the class 2 integron genes was plasmid encoded.
FIG. 2.
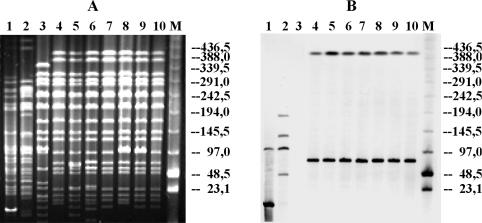
Localization of class 2 integrons by PFGE mapping. Shown are PFGE patterns of representative Salmonella serovar Paratyphi B dT+ strains (A) and results of a Southern blot analysis performed on the same gel with the intI2- (or dfrA1-, sat1-, or aadA1-) specific probe (B). Lanes 1, ColE1::Tn7; lanes 2, RP4-2; lanes 3, class 2 integron-negative comparative strain; lanes 4 to 10, representative class 2 integron-positive Salmonella serovar Paratyphi B dT+ strains; lanes M, low-range PFGE markers (New England Biolabs, Frankfurt, Germany). The control strain ColE1::Tn7 revealed the expected strong reactivity with the 20-kb plasmid fragment and with at least one chromosomal fragment (13). In the control strain RP4-2 the probes recognized four chromosomal fragments of different sizes, indicating the presence of four copies of the integron-specific genes.
In conclusion, this study shows that all isolates of the predominating German Salmonella serovar Paratyphi B dT+ clone carry two Tn7-like class 2 integrons and that these integron structures are located on the bacterial chromosome. The chromosomal occurrence of class 2 integrons in this Salmonella serovar is a novel finding and in agreement with the hypothesis that the multiresistance in Salmonella serovar Paratyphi B dT+ has evolved during the past decade by insertion of the transposon Tn7 into the bacterial chromosome. The subsequent vertical transmission within the serotype explains why the resistance has become so widespread and persistent in European Salmonella serovar Paratyphi B dT+ isolates.
Acknowledgments
We thank the staff of the National Salmonella Reference Laboratory for skilled technical assistance.
REFERENCES
- 1.Brown, D. J., H. Mather, L. M. Browning, and J. E. Coia. 2003. Investigation of human infections with Salmonella enterica serovar Java in Scotland and possible association with imported poultry. Eurosurveillance 8:35-40. [DOI] [PubMed] [Google Scholar]
- 2.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 3.Collis, C. M., and R. M. Hall. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875-2885. [DOI] [PubMed] [Google Scholar]
- 4.Collis, C. M., M. J. Kim, S. R. Partridge, H. W. Stokes, and R. M. Hall. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 184:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis, C. M., M. J. Kim, H. W. Stokes, and R. M. Hall. 2002. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol. Microbiol. 46:1415-1427. [DOI] [PubMed] [Google Scholar]
- 6.DANMAP. 2001. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. Danish Veterinary Institute, Copenhagen, Denmark.
- 7.Dorn, C., A. Schroeter, A. Miko, D. Protz, and R. Helmuth. 2001. Increasing number of Salmonella Paratyphi B isolations from broilers received at the National Salmonella Reference Laboratory. Berl. Münch. Tierärztl. Wochenschr. 114:179-183. [PubMed] [Google Scholar]
- 8.Gibreel, A., and O. Skold. 1998. High-level resistance to trimethoprim in clinical isolates of Campylobacter jejuni by acquisition of foreign genes (dfr1 and dfr9) expressing drug-insensitive dihydrofolate reductases. Antimicrob. Agents Chemother. 42:3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein, C., M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerra, B., S. Soto, S. Cal, and M. C. Mendoza. 2000. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob. Agents Chemother. 44:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra, B., S. M. Soto, J. M. Argüelles, and M. C. Mendoza. 2001. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4, 5, 12:i:−]. Antimicrob. Agents Chemother. 45:1305-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansson, K., L. Sundstrom, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heikkila, E., L. Sundstrom, M. Skurnik, and P. Huovinen. 1991. Analysis of genetic localization of the type I trimethoprim resistance gene from Escherichia coli isolated in Finland. Antimicrob. Agents Chemother. 35:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon, H., T. Kim, S. Cho, J. Seol, B. Kim, J. Hyun, K. Park, S. Kim, and H. Yoo. 2002. Distribution and characterization of class 1 integrons in Salmonella enterica serotype Gallinarum biotype Gallinarum. Vet. Microbiol. 89:303-309. [DOI] [PubMed] [Google Scholar]
- 16.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 18.Miko, A., B. Guerra, A. Schroeter, C. Dorn, and R. Helmuth. 2002. Molecular characterization of multiresistant d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates. J. Clin. Microbiol. 40:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 5th ed. NCCLS M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Ng, L. K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orman, B. E., S. A. Pineiro, S. Arduino, M. Galas, R. Melano, M. I. Caffer, D. O. Sordelli, and D. Centron. 2002. Evolution of multiresistance in nontyphoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 46:3963-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 23.Recchia, G. D., and R. M. Hall. 1997. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 5:389-394. [DOI] [PubMed] [Google Scholar]
- 24.Recchia, G. D., H. W. Stokes, and R. M. Hall. 1994. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 22:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridley, A., and E. J. Threlfall. 1998. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb. Drug Resist. 4:113-118. [DOI] [PubMed] [Google Scholar]
- 26.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 27.Sundstrom, L., P. H. Roy, and O. Skold. 1991. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tietze, E., and J. Brevet. 1991. The trimethoprim resistance transposon Tn7 contains a cryptic streptothricin resistance gene. Plasmid 25:217-220. [DOI] [PubMed] [Google Scholar]
- 29.van Pelt, W., H. van der Zee, W. J. B. Wannet, A. W. van de Giessen, D. J. Mevius, N. M. Bolder, R. E. Komijn, and Y. T. H. P. van Duynhoven. 2002. Explosive increase in Salmonella Java in poultry. Consequences for public health. Tijdschr. Diergeneeskd. 127:625-629. [PubMed] [Google Scholar]
- 30.White, P. A., C. J. McIver, Y. M. Deng, and W. D. Rawlinson. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 31.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]


