Abstract
Five sequential Cryptococcus neoformans isolates recovered from an AIDS patient with recurrent meningitis were analyzed. Four isolates were fluconazole susceptible, while the fifth isolate developed fluconazole resistance. Analysis of the 14-α lanosterol demethylase gene (ERG11) showed a point mutation in the resistant strain responsible for the amino acid substitution G484S.
Mechanisms of azole resistance already described for yeasts include altered affinity of lanosterol 14-α demethylase (ERG11) to azole drugs due to target site mutation or its overexpression and decreased accumulation of drugs due to enhanced energy-dependent drug efflux (5, 13). Changes in the azole affinity of the lanosterol 14-α demethylase have already been related to low-level fluconazole resistance in Cryptococcus neoformans isolates (15). In addition, the decreased affinity of lanosterol 14-α demethylase for azole derivatives due to mutations that contributes to the increase in the MICs of fluconazole has been described for sequential clinical isolates of Candida albicans (5, 6). To elucidate if this mechanism could also be implicated in the resistance of C. neoformans to azole, we compared the ERG11 genomic sequence in five sequential isolates recovered from recurrent episodes of cryptococcal meningitis.
Clinical case.
The five strains of C. neoformans were isolated from a 33-year-old male patient who had been positive for human immunodeficiency virus since 1990 and was presenting advanced AIDS (CD4+ count, <100 cells/mm3) and recurrent cryptococcosis. The first episode of cryptococcosis was diagnosed in August 1997 at Hospital Fernandez, Buenos Aires, Argentina. During the following 15 months, four more episodes of cryptococcal meningitis were detected and documented by cultures recovered from cerebrospinal fluid. In the first episode the patient was treated with amphotericin B (AMB), and for the rest of the episodes a fluconazole therapy at different doses was always established, reaching a cumulative dose of 336 g at the moment of the fifth episode. The five isolates from each episode were identified as C. neoformans var. grubii by the following parameters: morphology, assimilation and fermentation of carbon and nitrogen compounds, and molecular taxonomy.
Susceptibility testing was performed by microdilution and E-test methods. Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used as quality control strains throughout the experiments (7).
Microdilution method.
The susceptibility testing followed the NCCLS recommendations (7) but included some modifications as previously described (11). Briefly, the susceptibility testing included RPMI medium supplemented with 2% glucose as the assay medium (RPMI-2% glucose), an inoculum size of 105 CFU/ml, flat-bottomed trays, spectrophotometric reading at 530 nm, incubation at 30°C, and shaking at 350 rpm for 48 h (11). The antifungal agents used in the study were as follows: AMB (Sigma Aldrich Quimica S.A., Madrid, Spain), 5-flucytosine (5FC) (Sigma Aldrich Quimica), fluconazole (FCZ) (Pfizer S.A., Madrid, Spain), itraconazole (ITZ) (Janssen S.A., Madrid, Spain), and voriconazole (VOR) (Pfizer S.A.). For AMB the MIC endpoints were defined as the lowest drug concentration exhibiting reduction in growth of 90% or more compared with that of the control growth, while for flucytosine and azole drugs the MIC endpoint was defined as an inhibition of 50%.
E-test method.
Tests were performed according to the manufacturer's instructions. Plates containing RPMI 1640 medium without sodium bicarbonate and with l-glutamine (Sigma Aldrich Quimica), supplemented with 2% glucose and buffered to pH 7.0 with 0.165 M MOPS (morpholinepropanesulfonic acid) (Sigma Aldrich Quimica), were prepared. Bacto agar (Difco, Soria Melguizo, Madrid, Spain) was added at a final concentration of 1.5 g/100 ml (Etest technical guide 4b: antifungal susceptibility testing of yeasts, AB BIODISK, Piscataway, N.J.).
Strain typing.
The five strains were genotyped by using restriction fragment length polymorphisms generated by digesting total DNA samples to completion with the restriction enzyme SacI (Promega, Madison, Wis.) followed by hybridization with the CNRE-1 probe (kindly provided by S. Spitzer, New York, N.Y.) as previously described (1, 14). Three other C. neoformans isolates recovered from different patients were included as control strains. Hybridization patterns were analyzed visually. DNA fingerprinting using primers aimed to microsatellites M13 and (GACA)4, applied to all five strains, was performed to confirm their clonal origin.
Mutation detection.
For the sterol 14-α demethylase gene (ERG11) amplification, primers were designed on the basis of the sequence of the ERG11 gene from C. neoformans (GenBank accession no. AF225914). Primer CnERG11A (5′ TCGTCGAACCATCTTTCG 3′) was designed 83 bp upstream of the ATG initiation codon, and CnERG11B (5′ CGTCTATGACTTCATGACC 3′) was designed 73 bp downstream of the termination codon. The rest of the primers were designed to complete the full sequence of the gene. All the primers used in the present work were synthesized by Pharmacia (Madrid, Spain). The PCRs were carried out with a 50-μl volume containing 10 mM (NH4)2SO4, 10 mM KCl, 20 mM Tris-Cl (pH 8.8), 2 mM MgSO4, 10 ng of bovine serum albumin, 0.1% Triton X-100, a 250 μM concentration (each) of dATP, dGTP, dCTP, and dTTP (Applied Biosystem, Madrid, Spain), a 0.5 μM concentration of each primer, 2.5 U of Taq DNA polymerase (Applied Biosystem), and 50 ng of genomic DNA. Amplification was performed in a thermal cycler (Applied Biosystem) for 1 cycle of 5 min at 94°C, and then 30 cycles of 30 s at 94°C, 45 s at 48°C, and 2 min at 72°C, followed by one final cycle of 10 min at 72°C. The PCR products were analyzed by electrophoresis on 0.8% agarose gels and visualized by transillumination after being stained with ethidium bromide.
Cloning and plasmids.
C. neoformans isolates were grown at 30°C in YPD-Na (2% glucose, 1% yeast extract, 2% peptone, 3% NaCl), during 24 h in an orbital shaker at 150 rpm. DNA was extracted as previously described (14). Escherichia coli JM109 was grown in Luria-Bertani medium (12), supplemented with ampicillin (100 μg/ml), for propagation of plasmids for DNA extraction. PCR products were purified by Spin columns-200 (Clontech, Madrid, Spain) and cloned into pGEM-T easy vector system (Promega, Madrid, Spain). Insert DNAs of recombinant plasmids were sequenced by the BigDye terminator cycle sequencing ready reaction system (Applied Biosystem) according to the manufacturer's instructions. All the clones were sequenced on both strands. For each Cryptococcus strain, at least two inserts were analyzed. Sequence analysis was performed on an ABI prism 377 DNA sequencer (Applied Biosystem) by using the sequencing facilities available at the Biopolymers Unit at Instituto de Salud Carlos III, Majadahonda, Madrid, Spain.
Sequence analysis.
The amino acid sequences of sterol 14-α demethylase were deduced from nucleotide sequences and analyzed by using the MegAlign software package (DNAstar, Inc., Lasergene, Madison, Wis.). The multiple amino acids alignments were derived by CLUSTAL analysis (3).
Development of resistance to FCZ.
Recurrent episodes of cryptococcal meningitis are frequent events among patients with AIDS. Several authors have described strains of C. neoformans exhibiting resistance in vitro to FCZ (2, 8). Herein, we describe a case of cryptococcal meningitis in which resistance to FCZ is developed.
Table 1 shows the results of antifungal susceptibility testing. For isolates 1 to 4, the MICs were between 1 and 2 μg/ml but for isolate 5 the MICs were 32 μg/ml. No variations in the MICs of ITZ and VOR were detected in the sequential isolates. The strain exhibiting decreased susceptibility to FCZ (MIC, 16 μg/ml) was isolated after 15 months of treatment (after having received a cumulative dose of fluconazole of 336 g). A similar event has been described by Martinez et al. with C. albicans in a recurrent oropharyngeal candidiasis (6). In addition, Friese et al. reported a cryptococcal meningitis case in which the emergence of an FCZ-resistant strain (FCZ MIC, 64 μg/ml) was documented after three episodes of meningitis (2). The authors concluded that the development of resistance to FCZ was probably due to the FCZ maintenance therapy. It is interesting that our isolates and those studied by Friese et al. remained susceptible to other triazole agents (Table 1).
TABLE 1.
Development of in vitro fluconazole (FCZ) resistance in the five sequential clinical isolates of C. neoformans
| Episode (strain) | Time of follow-up (mo) | Cumulative dose of FCZ (g) | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|---|
| FCZa | FCZb | ITZa | VORa | 5FCa | AMBa | |||
| Initial (CN-1) | 0 | 0 | 2 | 2 | 0.06 | 0.5 | 16 | 0.12 |
| First relapse (CN-2) | 2 | 0 | 1.5 | 2 | 0.12 | 0.06 | 8 | 0.25 |
| Second relapse (CN-3) | 3 | 24 | 2 | 1 | 0.06 | 0.06 | 16 | 0.12 |
| Third relapse (CN-4) | 9 | 180 | 1.5 | 4 | 0.12 | 0.12 | 32 | 0.25 |
| Fourth relapse (CN-5) | 15 | 336 | 32 | 16 | 0.06 | 0.25 | 32 | 0.12 |
Determined by microdilution.
Determined by E-TEST.
The genotypic characterization of all strains by hybridization with CNRE-1 (Fig. 1) and DNA fingerprinting using primers aimed to microsatellites (data not shown) demonstrated that all isolates were isogenic and therefore clonal.
FIG. 1.
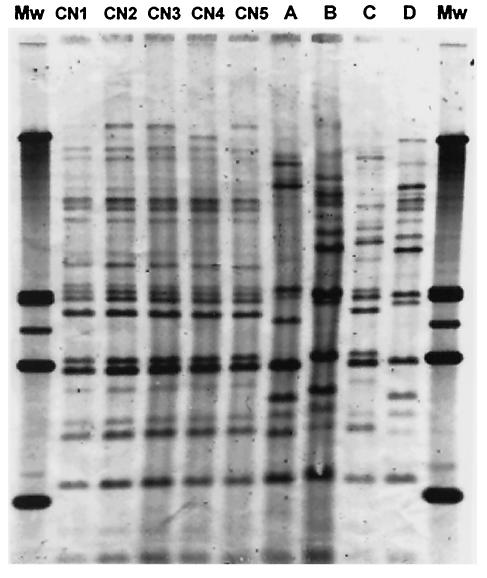
Southern hybridization analysis of C. neoformans total cellular DNAs digested with SacI and hybridized with the CNRE-1 probe. Lanes CN-1 to CN-5 show successive strains of recurrent episodes of cryptococcal meningitis. Lanes A, B, and C show DNAs of C. neoformans strains isolated from three patients from the same hospital, and lane D corresponds to C. neoformans ATCC 90112. Mw, molecular weight.
Little is known about mechanisms of azole resistance in C. neoformans (9, 15). Only recently, up-regulation of an ABC transporter-encoding gene (CnAFR1) causing an active drug efflux mechanism has been shown to be directly related to azole resistance in a C. neoformans strain (10). In this study, the sequences of the ERG11 genes of all isolates were analyzed in order to find any ERG11 point mutations (4, 6, 13) responsible for resistance to FCZ, as has been previously described for C. albicans strains (5).
Fragments of 2,147 bp containing the full ERG11 genomic sequence from all five isolates were obtained by PCR amplification. A point mutation (G1855T) in the ERG11 gene was detected in the FCZ-resistant isolate (CN-5) only. The experiments were repeated a second time to confirm this result. This mutation is responsible for the amino acid substitution glycine 484 for serine (G484S) in the ERG11 deduced protein sequence of C. neoformans. The G484 is a residue that forms part of the conserved hemo-binding domain and is conserved in all cytochrome P450 ERG11/Cyp51 of yeasts and filamentous fungi (Fig. 2). The amino acid substitution present in the fifth isolate, G484S, corresponds with the G464S of C. albicans ERG11 (Fig. 2). Several studies have demonstrated that this amino acid substitution detected in C. albicans ERG11 confers a change in the orientation of the P450 hemo-binding domain, leading to a decrease of azole binding and a decrease of the catalytic activity of the enzyme (4, 13). In sequential C. albicans strains recovered from oropharyngeal candidiasis, it has been demonstrated that this substitution appeared in isolates for which the MICs of FCZ were between 32 and 64 μg/ml (6). Similarly, for the C. neoformans isolates from the presented patient it was also clear that the increase of the MIC of FCZ (from 2 to 16 μg/ml) was matched to the G484S amino acid substitution. However, the possibility of involvement of another concomitant molecular mechanism of resistance cannot be disregarded.
FIG. 2.

Alignment of the 33 amino acid residues of ERG11/Cyp51p sequences from C. neoformans (CnCyp51; GenBank accession no. AAF35366), C. neoformans var. grubii (CnN1Cyp51; GenBank accession no. AY265353), Aspergillus fumigatus (AfCyp51A; GenBank accession no. AAK73659; and AfCyp51B; GenBank accession no. AAK73660), Leptosphaeria maculans (GenBank accession no. AAN28927), Aspergillus nidulans (AnCyp51; GenBank accession no. AAF79204), Penicillium italicum (PiCyp51; GenBank accession no. CAA89824), Botryotinia fuckeliana (BcCyp51; GenBank accession no. AAF85983), Erysiphe graminis (EgCyp51; GenBank accession no. AAC97606), Uncinula necator (UnCyp51; GenBank accession no. AAC49812), Saccharomyces cerevisiae (ScCyp51; GenBank accession no. AAA34546), Candida glabrata (CgErg11; GenBank accession no. AAB02329), Candida tropicalis (CtCyp51; GenBank accession no. AAA53284), C. albicans (CaErg11; GenBank accession no. AAF00598), and Ustilago maydis (UmCyp51; GenBank accession no. CAA88176). Residues that are identical among all the filamentous fungi and yeast are bolded. The same 33 amino acids from the FCZ-resistant strain of C. neoformans (CN-5) are at the bottom. The residue at position 484 is boxed.
Nucleotide sequence accession number.
The full nucleotide sequences of the ERG11 gene from C. neoformans var. grubii determined in this work appear in the GenBank nucleotide sequence database under accession number AY265353.
Acknowledgments
We thank the Biopolymers Unit at Instituto de Salud Carlos III, Majadahonda, Madrid, Spain, for DNA sequencing and Sylvia Spitzer for providing the CNRE-1 repetitive probe.
E.M. held a contract Ramon y Cajal from Ministry of Science and Technology. L.R. held a grant from PAHO and ISCIII.
L.R. and E.M. contributed equally to this work.
REFERENCES
- 1.Casadevall, A., and D. Spitzer. 1995. Involvement of multiple Cryptococcus neoformans strains in a single episode of cryptococcosis and reinfection with novel strains in recurrent infection demonstrated by random amplification of polymorphic DNA and DNA fingerprinting. J. Clin. Microbiol. 33:1682-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friese, G., T. Discher, R. Fussle, A. Schmalreck, and J. Lohmeyer. 2001. Development of azole resistance during fluconazole maintenance therapy for AIDS-associated cryptococcal disease. AIDS 15:2344-2345. [DOI] [PubMed] [Google Scholar]
- 3.Higgins, D. G., and P. M. Sharp. 1988. A package for performing multiple sequence alignments on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 4.Kelly, S. L., D. C. Lamb, J. Loeffler, H. Einsele, and D. E. Kelly. 1999. The G464S amino acid substitution in Candida albicans sterol 14alpha-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262:174-179. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Ribot, J. L., R. K. McAtee, S. Perea, W. R. Kirkpatrick, M. G. Rinaldi, and T. F. Patterson. 1999. Multiple resistant phenotypes of Candida albicans coexist during episodes of oropharyngeal candidiasis in human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 43:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez, M., J. L. Lopez-Ribot, W. R. Kirkpatrick, S. P. Bachmann, S. Perea, M. T. Ruesga, and T. F. Patterson. 2002. Heterogeneous mechanisms of azole resistance in Candida albicans clinical isolates from an HIV-infected patient on continuous fluconazole therapy for oropharyngeal candidosis. J. Antimicrob. Chemother. 49:515-524. [DOI] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. M27-A2. National Committee For Clinical Laboratory Standards, Wayne, Pa.
- 8.Paugam, A., J. Dupouy-Camet, P. Blanche, J. P. Gangneux, C. Tourte-Schaefer, and D. Sicard. 1994. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin. Infect. Dis. 19:975-976. [DOI] [PubMed] [Google Scholar]
- 9.Perfect, J. R., and G. M. Cox. 1999. Drug resistance in Cryptococcus neoformans. Drug Resist. Updates 2:259-269. [DOI] [PubMed] [Google Scholar]
- 10.Posteraro, B., M. Sanguinetti, D. Sanglard, M. La Sorda, S. Boccia, L. Romano, G. Morace, and G. Fadda. 2003. Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Mol. Microbiol. 47:357-371. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Tudela, J. L., F. Martinez, M. Cuenca-Estrella, L. Rodero, Y. Carpintero, and B. Gorgojo. 2000. Influence of shaking on antifungal susceptibility testing of Cryptococcus neoformans: a comparison of the NCCLS standard M27A medium, buffered yeast nitrogen base, and RPMI-2% glucose. Antimicrob. Agents Chemother. 44:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitzer, E. D., S. G. Spitzer, L. F. Freundlich, and A. Casadevall. 1993. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet 341:595-596. [DOI] [PubMed] [Google Scholar]
- 15.Venkateswarlu, K., M. Taylor, N. J. Manning, M. G. Rinaldi, and S. L. Kelly. 1997. Fluconazole tolerance in clinical isolates of Cryptococcus neoformans. Antimicrob. Agents Chemother. 41:748-751. [DOI] [PMC free article] [PubMed] [Google Scholar]


