Abstract
Pyrosequencing was used to detect rapidly and estimate the number of 23S rRNA genes with a G2576T mutation in 43 linezolid-resistant and -susceptible clinical isolates of enterococci. The method showed 100% concordance with PCR-restriction fragment length polymorphism for detecting isolates homozygous for either G2576 or T2576 or heterozygous for this mutation. A good correlation was found between linezolid MICs and the number of 23S rRNA gene copies carrying the mutation.
Enterococci, particularly Enterococcus faecium, are often resistant to multiple antibiotic classes, and therapy of serious infections caused by them may be problematic. In consequence, linezolid, a novel oxazolidinone (4), is increasingly used to treat enterococcal infections. Linezolid-resistant enterococci (LRE) are rare but may emerge during linezolid therapy. This resistance is associated with chromosomal mutations that affect the peptidyltransferase domain of 23S rRNA (9). In clinical LRE isolates, linezolid resistance is mediated by a G2576T single nucleotide polymorphism (SNP) in multiple alleles encoding 23S rRNA (1, 5, 9); other mutations, which have been reported in laboratory-generated linezolid-resistant mutants of enterococci (9), have not yet been detected in clinical isolates. This SNP was also present in the two reported linezolid-resistant isolates of methicillin-resistant Staphylococcus aureus (MRSA) (11, 12).
Previously, rapid detection of the G2576T polymorphism in enterococci by real-time PCR was described (13). That work, and the results of others (6, 12), indicated that some LRE and linezolid-resistant staphylococci were homozygous for T2576 but that more were heterozygous, containing some 23S rRNA gene copies with T2576 and others with G2576. Homozygosity for T2576, but not heterozygosity, has been associated with decreased fitness in vitro in the absence of linezolid selective pressure (W. Mazur, C. Knob, and H. S. Fraimow, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1607, 2002). We report here the use of pyrosequencing, which is an innovative technology suitable for the rapid detection of SNPs (http://www.pyrosequencing.com) (3, 7, 10), to detect and estimate the number of 23S rRNA genes containing T2576 mutations in clinical isolates of LRE.
Forty-three clinical isolates of E. faecium (n = 27, from 10 patients) or Enterococcus faecalis (n = 16, from 5 patients) were studied: 31 isolates were LRE (linezolid MICs, ≥8 μg/ml) known to contain the T2576 mutation (13); 12 isolates were susceptible to linezolid (MICs, ≤4 μg/ml).
Pyrosequencing was performed by using a PSQ 96 sample preparation kit and a PSQ 96MA analyzer (Pyrosequencing AB, Uppsala, Sweden) in accordance with the manufacturer's instructions. Briefly, a 96-bp fragment of 23S rRNA genes, spanning the G2576T SNP, was amplified from all isolates by using whole cells as the template and with previously published primers (13), except that the forward primer was labeled at the 5′ position with biotin. All the primers for pyrosequencing were purified by high-performance liquid chromatography (Sigma-Genosys Ltd., Pampisford, United Kingdom). The PCR product was captured, and the biotin-labeled (forward) strand was separated by using streptavidin-Sepharose beads (Amersham Biosciences, Little Chalfont, United Kingdom). The resulting single-stranded DNA was used as a template for pyrosequencing with a primer (5′-CGT TCT GAA CCC AGC-3′) located downstream of the G2576T SNP (Fig. 1). The pyrosequencing primer was designed to be complementary to the captured strand and was suitable for detecting this SNP in isolates of E. faecalis and E. faecium.
FIG. 1.
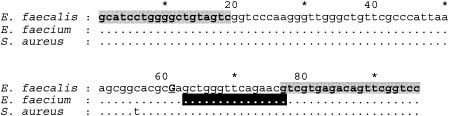
Pyrosequencing assay to detect the G2576T mutation in LRE and linezolid-resistant staphylococci. PCR primers used to amplify a 96-bp fragment of 23S rRNA genes spanning the mutation are shaded in grey; the forward primer was biotin labeled. The pyrosequencing primer (complementary to the forward strand) is shaded in black. Nucleotide G2576 is underlined and in upper case.
During the pyrosequencing reaction, the sequencing primer is hybridized to the single-stranded template and incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase, and apyrase and the substrates adenosine 5′ phosphosulfate and luciferin. Four nucleotides are added to the reaction sequentially. Incorporation of each nucleotide complementary to the template initiates a cascade and results in separate releases of light. These are detected and presented as peaks on a data trace called Pyrogram (for full details, see http://www.pyrosequencing.com). In the G2576T SNP assay described here, extension of the pyrosequencing primer resulted in the incorporation, first of T (in all isolates), followed either by C (complementary to G2576, wild type) or by A (complementary to T2576, mutant), followed by nucleotides complementary to those upstream of the SNP (Fig. 1). As the amount of light released at each extension step is directly proportional to the amount of nucleotide added, the relative numbers of T2576 and G2576 23S rRNA gene copies could be estimated from the relative peak heights on data traces and based on the assumption of six copies in E. faecium and four copies in E. faecalis (http://rrndb.cme.msu.edu). In parallel experiments, qualitative detection of the G2576T SNP was performed by PCR-restriction fragment length polymorphism analysis by using NheI digests of a 633-bp 23S rRNA gene fragment (13).
Qualitatively, the pyrosequencing data were in complete agreement with the PCR-restriction fragment length polymorphism results. Thus, the new method clearly identified isolates that were homozygous for either G2576 or T2576 or heterozygous at this position (Fig. 2). The estimation of the numbers of T2576 copies was based on averaging data from three pyrosequencing experiments (and expressing them as a whole number of gene copies); each experiment was performed with a separate batch of amplified template DNA. Estimates of copy number were reproducible between runs; they were identical in all three experiments for 19 isolates and varied by no more than one gene copy for other isolates. To allow for this variation, estimates for each isolate were considered to be a range of copies rather than a precise value (Table 1). There was a good correlation between the estimated number of T2576 copies and an increasing linezolid MIC, particularly for the E. faecium isolates (Table 1). Pyrosequencing confirmed the presence of the T2576 polymorphism in one to two 23S rRNA genes of an E. faecium isolate that was susceptible to linezolid (MIC, 4 μg/ml), thereby confirming the qualitative data obtained previously by real-time PCR (13).
FIG. 2.
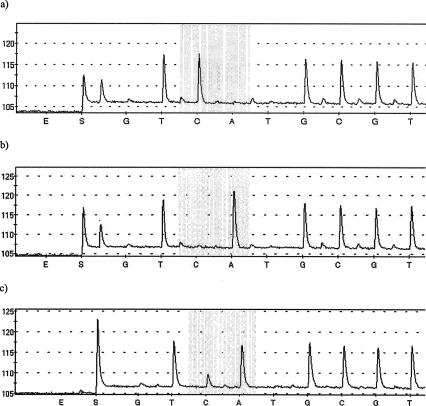
Detection by pyrosequencing of the G2576T mutation in 23S rRNA genes of linezolid-resistant and -susceptible enterococci. (a) A homozygous isolate with wild-type sequence (G2576) in all gene copies; (b) a homozygous isolate with a mutation (T2576) in all gene copies; (c) a heterozygous isolate with both mutant and wild-type copies of the gene. The shaded region shows the polymorphism at position 2576. The additions of enzymes and substrates to the reaction mix are indicated by E and S, respectively.
TABLE 1.
Correlation between linezolid MIC and the estimated number of 23S rRNA genes containing the T2576 mutation for clinical isolates of enterococci
| Linezolid MIC (μg/ml) | No. of isolates with indicated no. of T2576 gene copiesa
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
E. faecium (n = 27)
|
E. faecalis (n = 16)
|
|||||||||
| 0-1 | 1-2 | 2-3 | 3-4 | 4-5 | 5-6 | 0-1 | 1-2 | 2-3 | 3-4 | |
| 1 | ||||||||||
| 2 | 4 | |||||||||
| 4 | 3 | 1 | 4 | |||||||
| 8 | 2 | 1 | ||||||||
| 16 | 2 | |||||||||
| 32 | 4 | 3 | 2 | 2 | ||||||
| 64 | 2 | 2 | 3 | 2 | 3 | |||||
| 128 | 1 | 2 | ||||||||
Determined by averaging data from three pyrosequencing runs, each performed with a separate batch of amplified template DNA and based on six gene copies in E. faecium and four copies in E. faecalis (http://rrndb.cme.msu.edu).
In conclusion, pyrosequencing was an effective, rapid method for detecting and estimating copy numbers of the G2576T polymorphism in LRE. The assay reported here might also be useful for detecting this SNP in linezolid-resistant isolates of methicillin-resistant S. aureus. The manual setup of each experiment (for 43 isolates) took less than 90 min, and the actual pyrosequencing SNP analysis runs (of up to 96 samples) were completed in less than 15 min. Pyrosequencing was, therefore, slightly quicker than the real-time PCR assay reported previously (13) and had the additional benefit of being semiquantitative. Pyrosequencing setup time could be reduced further with automation (http://www.pyrosequencing.com) (2). Previous attempts to estimate the number of 23S rRNA gene copies containing the T2576 mutation in LRE or linezolid-resistant staphylococci have usually been time-consuming, involving either the cloning of pooled 23S ribosomal DNA amplicons, followed by analysis of multiple recombinants (6), Southern hybridization of suitable probes to NheI-digested genomic DNA (8), or specific amplification and sequencing of every 23S rRNA gene in the genome of the isolates investigated (8, 12). None of these approaches is convenient for the analysis of multiple isolates. Another approach, based on the densitometric analysis of digested PCR amplicons, has also been reported (Mazur et al., 42nd ICAAC). Pyrosequencing provided an extremely rapid alternative, and its application to other medical microbiological purposes deserves investigation.
Acknowledgments
We are grateful to Pyrosequencing AB for their support of this work.
REFERENCES
- 1.Auckland, C., L. Teare, F. Cooke, M. E. Kaufmann, M. Warner, G. Jones, K. Bamford, H. Ayles, and A. P. Johnson. 2002. Linezolid-resistant enterococci: report of the first isolates in the United Kingdom. J. Antimicrob. Chemother. 50:743-746. [DOI] [PubMed] [Google Scholar]
- 2.Diggle, M., and S. Clarke. 2003. A novel method for preparing single-stranded DNA for pyrosequencing. Mol. Biotechnol. 24:221-224. [DOI] [PubMed] [Google Scholar]
- 3.Elahi, E., N. Pourmand, R. Chaung, A. Rofoogaran, J. Boisver, K. Samimi-Rad, R. W. Davis, and M. Ronaghi. 2003. Determination of hepatitis C virus genotype by pyrosequencing. J. Virol. Methods 109:171-176. [DOI] [PubMed] [Google Scholar]
- 4.Ford, C. W., G. E. Zurenko, and M. R. Barbachyn. 2001. The discovery of linezolid, the first oxazolidinone antibacterial agent. Curr. Drug Targets Infect. Disord. 1:181-199. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, A. P., L. Tysall, M. W. Stockdale, N. Woodford, M. E. Kaufmann, M. Warner, D. M. Livermore, F. Asboth, and F. J. Allerberger. 2002. Emerging linezolid resistant Enterococcus faecalis and Enterococcus faecium isolated from two Austrian patients in the same intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 21:751-754. [DOI] [PubMed] [Google Scholar]
- 6.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacey-Miller, T., and R. Henry. 2003. Single-nucleotide polymorphism detection in plants using a single-stranded pyrosequencing protocol with a universal biotinylated primer. Anal. Biochem. 317:166-170. [DOI] [PubMed] [Google Scholar]
- 8.Pillai, S. K., G. Sakoulas, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., M. J. Ferraro, and H. S. Gold. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J. Infect. Dis. 186:1603-1607. [DOI] [PubMed] [Google Scholar]
- 9.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronaghi, M., M. Uhlen, and P. Nyren. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363-365. [DOI] [PubMed] [Google Scholar]
- 11.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 12.Wilson, P., J. A. Andrews, R. Charlesworth, R. Walesby, M. Singer, D. J. Farrell, and M. Robbins. 2003. Linezolid resistance in clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 51:186-188. [DOI] [PubMed] [Google Scholar]
- 13.Woodford, N., L. Tysall, C. Auckland, M. W. Stockdale, A. J. Lawson, R. A. Walker, and D. M. Livermore. 2002. Detection of oxazolidinone-resistant Enterococcus faecalis and Enterococcus faecium strains by real-time PCR and PCR-restriction fragment polymorphism analysis. J. Clin. Microbiol. 40:4298-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]


