Abstract
A series of ring expanded nucleoside (REN) analogues were synthesized and screened for inhibition of cellular RNA helicase activity and human immunodeficiency virus type 1 (HIV-1) replication. We identified two compounds 1 and 2 that inhibited the ATP dependent activity of human RNA helicase DDX3. Compounds 1 and 2 also suppressed HIV-1 replication in T cells and monocyte-derived macrophages. Neither compound at therapeutic doses was significantly toxic in ex vivo cell culture or in vivo in mice. Our findings provide proof-of-concept that a cellular factor, an RNA helicase, could be targeted for inhibiting HIV-1 replication.
Introduction
Human immune deficiency virus type 1 (HIV-1) is a retrovirus with an RNA genome of ~9 kilobases. HIV-1 is the etiological agent for the acquired immunodeficiency syndrome (AIDS). Since the first report of AIDS in 1981, HIV-1 has cumulatively caused 33 million deaths worldwide. Current HIV therapeutics target viral proteins such as reverse transcriptase, protease, integrase, and gp120. Because HIV-1 can mutate its genome rapidly, a consequence of this treatment approach has been the selection for, and the rapid emergence of, drug-resistant HIV-1’s (1). Thus, there is a compelling need to evolve therapeutic strategies that do not readily elicit resistant mutations.
One way to circumvent the mutability of the HIV-1 genome is to target cellular factors used by the virus for replication (2). Several such cellular factors have been characterized (3–8), including a cellular RNA-helicase reported as necessary for HIV-1 to replicate optimally in human cells (9, 10). Mammalian RNA helicases include proteins that have DEAD-, DEAH-, DExH-, and DExD –motifs (11, 12). These helicases participate in many aspects of cellular RNA metabolism including transcription, RNA splicing and stability, mRNA- export and translation, and mitochondrial gene expression (13–16). While all RNA helicases apparently conserve the same eight discrete protein motifs (12), there is some evidence that individual RNA helicases can be sufficiently different that each could be targeted with some degree of specificity (17–22). Thus, there are reports of compounds that inhibit with relative specificity the RNA-helicases encoded by viruses such as Hepatitis C, West Nile, Herpes Simplex, and Japanese encephalitis (18–22).
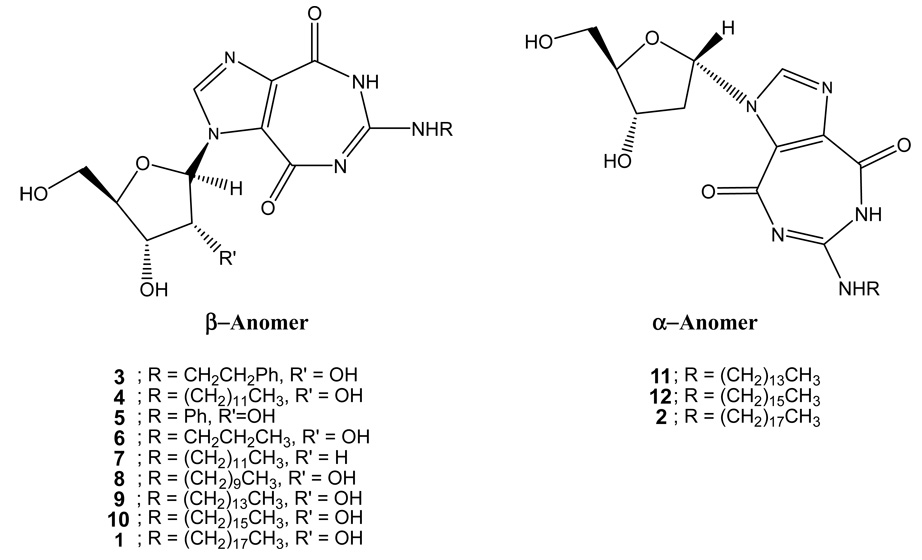
HIV-1 does not encode a viral helicase, but it does engage several cellular RNA-helicases for replication (23). In this study, we asked if novel compounds could be discovered that target cellular RNA helicases and whether such compounds would inhibit HIV-1 replication in cultured cells. Towards these goals, we screened several ring-expanded nucleoside (REN) analogs (Figure 1), some of which were previously described to be inhibitors of the NTPase/helicase of West Nile virus, Hepatitis C virus, and Japanese encephalitis virus (21, 22). We identified two RENs, 1 and 2, that inhibited the activity of human RNA helicase DDX3 in vitro and intracellular HIV-1 replication. Encouragingly, neither 1 nor 2 was toxic to cells or to mice at concentrations that were inhibitory for DDX3 and HIV-1. Our findings offer proof-of-principle for treating HIV-1 infection using small molecules that target a cellular protein.
Figure 1.
α- and β-anomers of ring-expanded nucleoside analogs.
Results
Compounds 1 and 2 inhibit productive HIV replication in T cells and macrophages
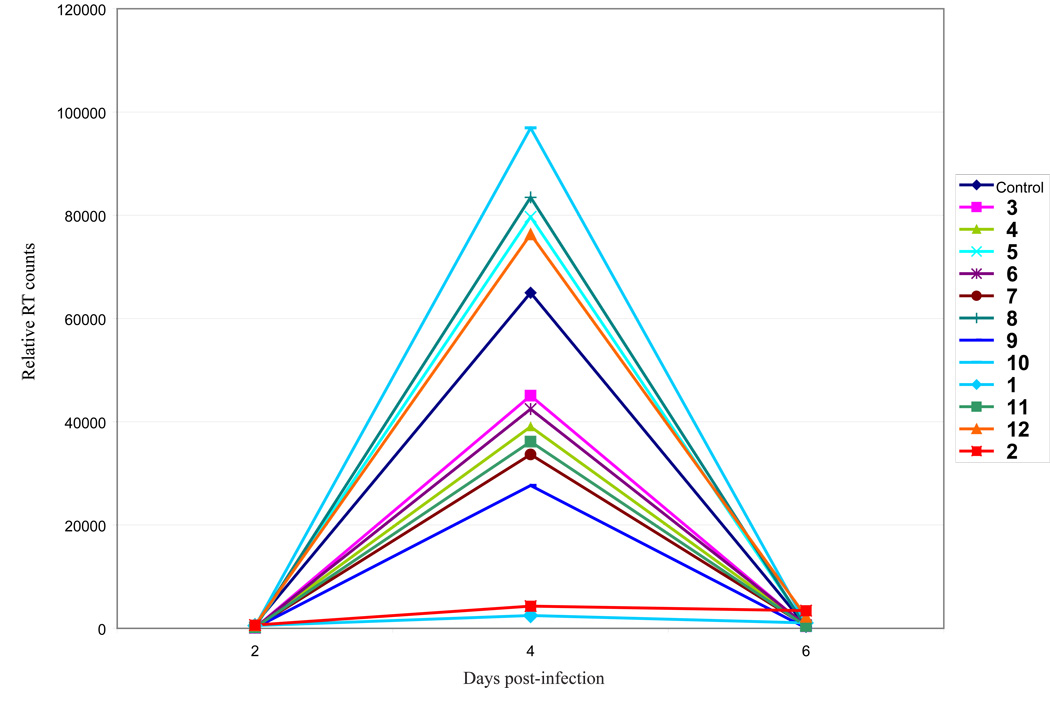
We tested a panel of RENs (Figure 1) for inhibition of HIV-1 replication in MT4 cells. Amongst twelve compounds, two strongly inhibited peak HIV-1 replication (10 µM concentration) on day 4 of infection in cultured cells (Figure 2). We selected 1 and 2 (Figure 3A), the two that were most potent, for further characterization of their virus-inhibitory activities.
Figure 2.
Screening of RENs for inhibition of HIV-1 replication. 12 REN compounds were synthesized and used at 10 µM concentration to analyze their effects on HIV-1 replication in T cell line (MT4 cells). MT4 cells were infected with 20,000 RT counts of virus and treated with the indicated compounds. Cell supernatants were collected every 2 days and assayed for RT activity. Compounds 1 and 2 were the most potent for inhibiting HIV-1 replication. Note that by day 8 because no new cells are being added to the tissue cultures, continuous HIV-1 replication in cells that are not inhibited by drugs results in cell killing and a reduction of RT production to base line.
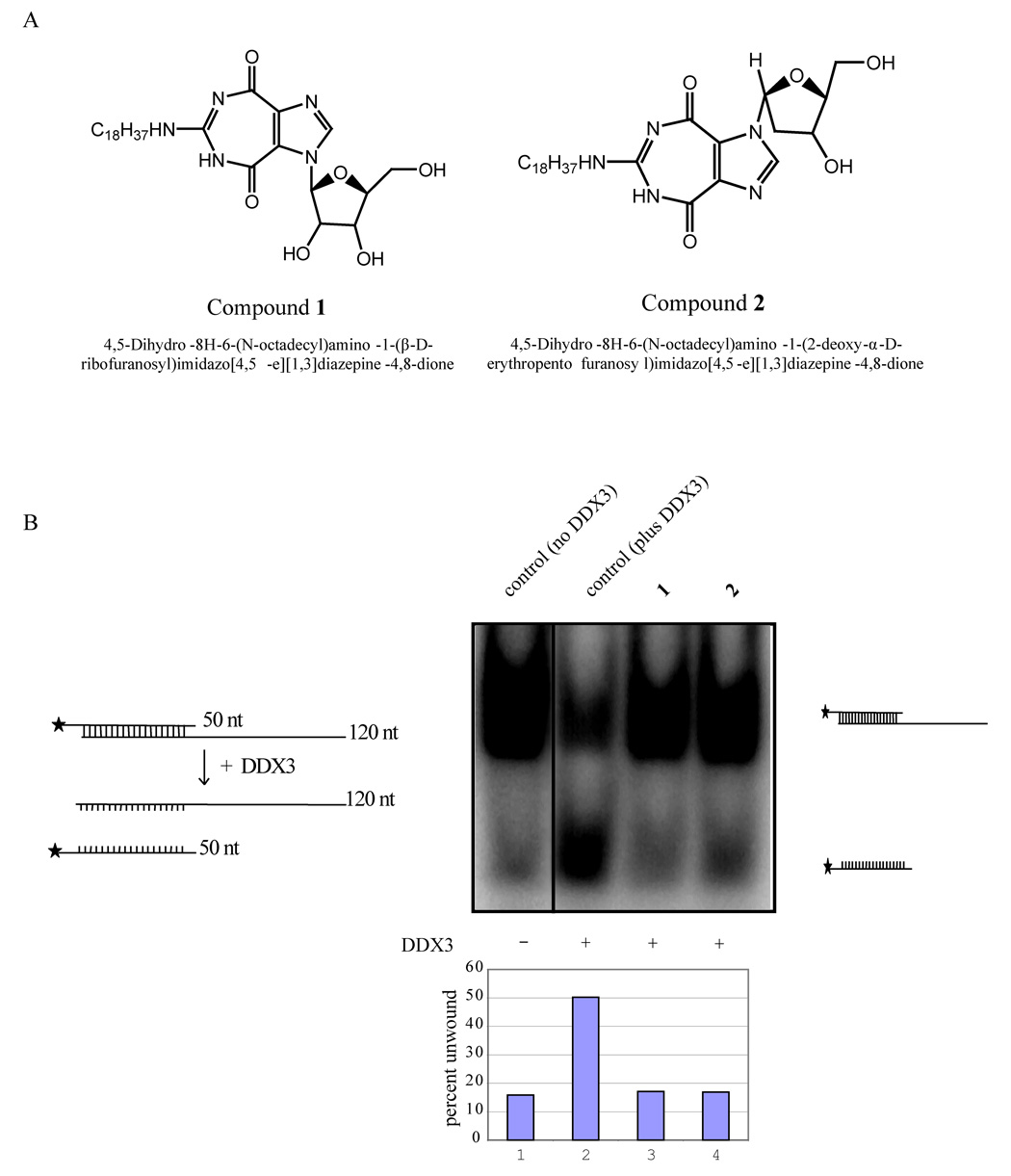
Figure 3.
Inhibition of helicase activity in vitro. A) Structures of 1 and 2. B) Inhibition of helicase activity of DDX3 by 1 and 2. The pBluescript vector was digested to completion with Kpn I and transcribed with T3 polymerase to generate a 120 base long transcript. The vector was also digested with EcoRI and transcribed with T7 polymerase in the presence of (α32p) UTP. The two complementary transcripts (50nt and 120 nt) were purified and hybridized at 95° C for 20 minutes. Note that the 120 nt is unlabeled while the 50 nt strand is 32P-labeled. Autoradiography of the slower migrating band is due to duplex formation between the labeled and unlabeled strands (lane 1). Duplexed RNA was unwound by DDX3 in the presence of ATP resulting in increased release of the labeled strand (50 nt; lane 2) and a reduction in intensity of the duplexed strands. Both 1 and 2 inhibited the ATP dependent RNA unwinding activity of DDX3 as measured by the reduced release of the 50 nt strand (lane 3 and 4). The bar diagram indicates percent duplex unwound by DDX3 in the presence and absence of 1 and 2.
RENs were previously found to inhibit virus encoded RNA helicases (21, 22). We asked if 1 or 2 could inhibit the activity of a cellular RNA helicase, DDX3. DDX3 was identified earlier to be important for HIV-1 replication in human cells(9). As an initial test, 1 and 2 were queried for their inhibition of DDX3’s ability to unwind in vitro a partially double stranded RNA. A partially double stranded RNA substrate was generated (see Methods), and this was incubated with purified WT DDX3 in the presence of ATP. We observed that the double stranded substrate could indeed be unwound by WT-DDX3 to release single stranded 32P labeled RNA (Figure 3B, lane 2). However, when either 1 or 2 was added into the reaction, DDX3’s unwinding activity was inhibited (Figure 3B, lanes 3 and 4).
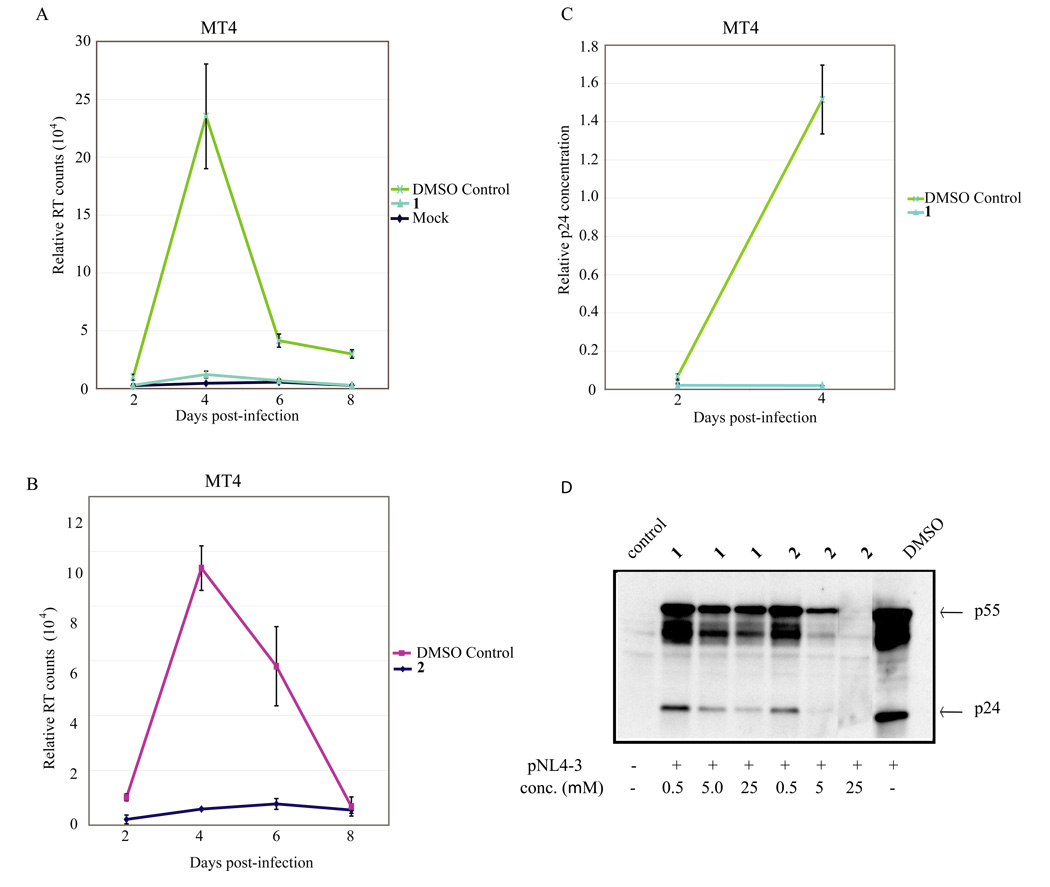
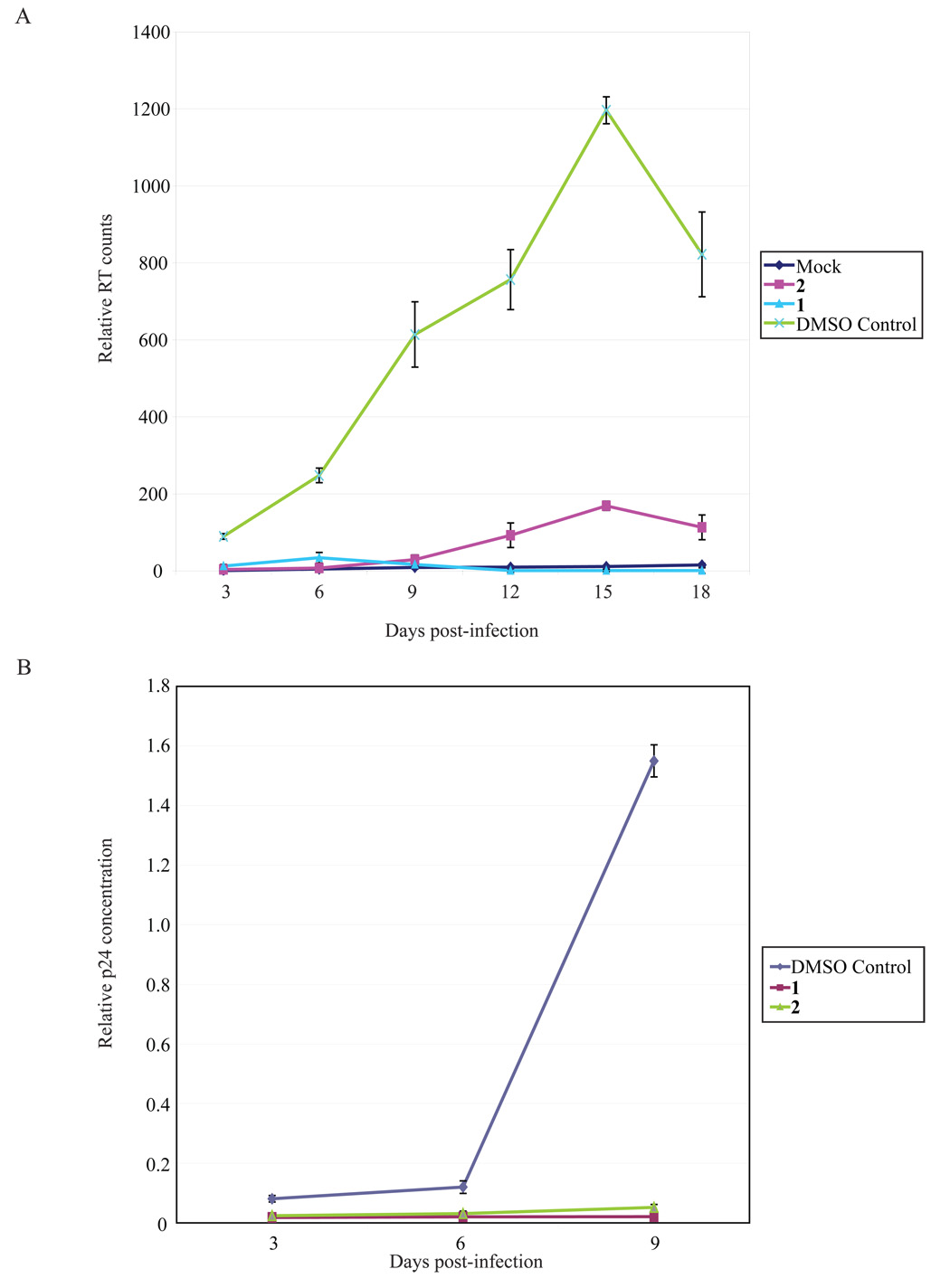
We next tested 1 and 2 for inhibition of HIV-replication in cultured cells. We first assayed 1 and 2 on HIV-1 infection of a suspension T cell line, MT4. In this setting over the time course of 8 days, an essentially complete suppression of HIV-1 replication was seen when 1 or 2 was used at 25 µM (Figure 4A and 4B).
Figure 4.
Measurement of 1 and 2 inhibition of HIV-1 replication using RT, p24 and Western blotting assays. MT4 cells were infected with 20,000 RT counts of virus and treated with the indicated compounds. The experiments were repeated three times. Cell supernatants were collected every 2 days and assayed for both RT activity (4A and 4B) and p24 ELISA (4C). ELISA shows a corresponding decrease in the amount of p24 present in supernatants collected from infected MT4 cultures similar to the results from the RT assays. D) 1 and 2 suppressed HIV-1 protein expression in HeLa cells transfected with HIV-1 molecular clone pNL4-3. HeLa cells were transfected with 2µg pNL4-3, and 3 hrs post-transfection the cells were treated with 25 µM concentration of 1 and 2. Cell lysates were prepared 24 hrs later and analyzed by Western blotting using anti-HIV-1 patient IgG.
Because the readout for HIV-1 replication in the MT4 cells was an in vitro assay of reverse transcriptase (RT) activity produced by the replicating virus and because 1 and 2 are nucleoside analogs which can potentially interfere with the RT assays, we were concerned that the observed reduction in RT activity could simply be an inhibition of the enzymatic assay and not an actual suppression of virus replication in the infected MT4 cells. To exclude this possibility, HIV-1 replication in MT4 cells was quantified using an enzyme-linked immunosorbent assay (ELISA) measuring the viral p24 capsid protein. Indeed, similar to the RT results, the p24 ELISA findings (Figure 4C) confirmed a significant reduction of virus production (p24) by 1 (similar results were achieved with 2, data not shown). We also asked whether 1 and 2 inhibited HIV-1 gene expression in transfected HeLa cells using a Western blotting assay (Figure 4D). Based on protein blotting using hyperimmune patient serum, we observed that both 1 and 2 greatly reduced the synthesis of viral proteins in the HIV-1 transfected HeLa cells (Figure 4D) consistent with a mechanism affecting a post-integration step in viral replication.
Macrophage- (M) tropic HIV-1 differs from T-cell tropic HIV-1 in that the former uses the CCR5 (not CXCR4) co-receptor and is the predominant viral genotype found in newly infected individuals. The above HIV-1/MT4 assays tested the effect of 1 and 2 on a T-tropic (CXCR4-utilizing) infection. Next, we queried whether 1 and 2 would also inhibit the replication of a M-tropic HIV-1 AD8 (a CCR5 using virus) in macrophages. We compared mock infected macrophages to macrophages infected with HIV-1 AD8 and found that virus replication in macrophages was indeed substantially suppressed by both 1 and 2 , respectively, as assayed by measuring RT (Figure 5A) or p24 (Figure 5B) production.
Figure 5.
Compounds 1 and 2 suppressed HIV-1 replication in monocyte-derived-macrophages (MDMs). MDMs were infected with M-tropic HIV-1 AD8 with 100,000 RT counts of virus on day 5 in culture and were treated with the indicated concentrations of 1 ad 2. Supernatants were collected every 3 days and assayed for virus replication by (A) RT activity and (B) p24 ELISA. RT assays and p24 ELISA showed that both 1 and 2 suppressed HIV-1 replication. The experiments were repeated three times.
Lack of toxicity of 1 and 2 in cultured cells and in mice
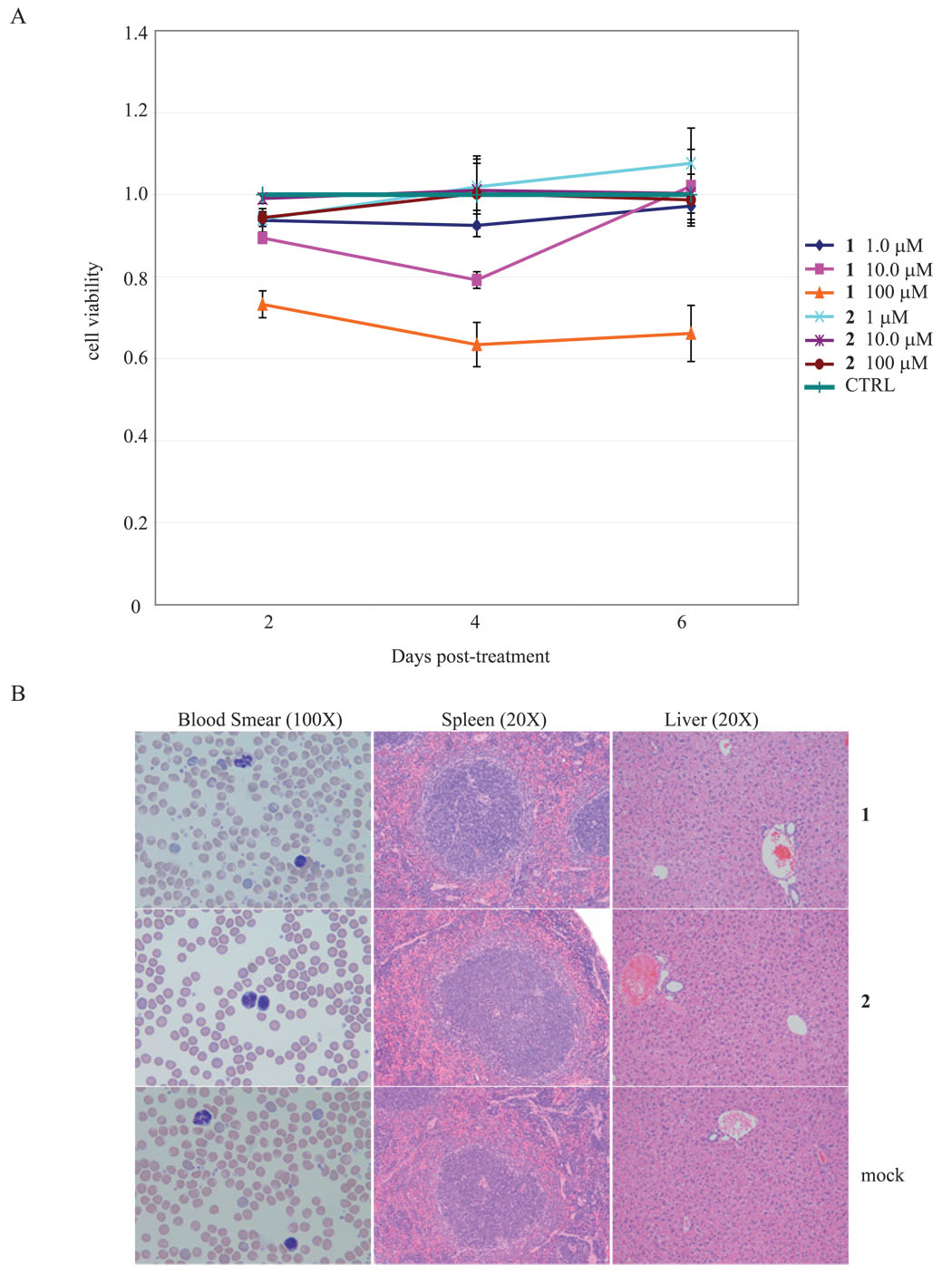
Potentially useful candidate anti-viral compounds should have acceptable toxicity profiles. To check 1 and 2 for cytotoxicity, we employed a colorimetric assay that monitors cell viability after treatment with the compounds. MT4 cells were incubated individually with 1 or 2 at 1, 10 or 100 µM concentrations. Over a 6 day period, we compared 1 or 2 treated cells to mock treated control cells and found no significant differences in cell growth and viability (Figure 6A).
Figure 6.
Lack of toxicity of 1 and 2 in cells and in mice. A) 4 × 106 MT4 cells in 2 ml were incubated in culture media containing 1, 10, and 100 µM concentrations of 1 and 2. 100 µl of each cell suspension were incubated with 10 µl of WST8 (water-soluble tetrazolium) for 2 hrs and then subjected to colorimetric measurements. Production of color is a measurement of the metabolism of viably dividing cells. B) In vivo toxicities of 1 and 2 were evaluated in 8–10 week old Balb-C mice. Mice, in groups of 4, were injected intraperitoneally twice weekly with drug boluses calculated to achieve final body 1 and 2 distributions of 0.2, 2, 20 and 200 µM, respectively for 4 weeks. None of the mice demonstrated any signs of toxicity from intra-peritoneal injections of 1 or 2. At the end of 5 weeks of drug injection and observation, all mice were sacrificed and histological examinations of tissues were performed. Examples of tissues from 1 and 2 injected mice compared to tissues from control mice are shown.
We next checked 1 and 2 for in vivo toxicity in mice. Mice, in groups of 4, were injected intraperitoneally twice weekly with drug boluses calculated to achieve final 1 and 2 body distributions of 0.2, 2, 20 and 200 µM, respectively. The mice were injected and monitored for 5 weeks. Control mice were injected with either normal saline or DMSO. At the end of the study period, none of the mice injected with 1 and 2 displayed any symptoms or showed any visible signs of toxicity. Detailed necropsies and histologies were performed on multiple organs from the 1, 2, and control mice. No difference was seen in blood smears, liver-, spleen- (Figure 6B), kidney-, heart-, and lung- histologies (data not shown). These results are consistent with a lack of in vivo toxicity elicited by 1 and 2 over the studied period.
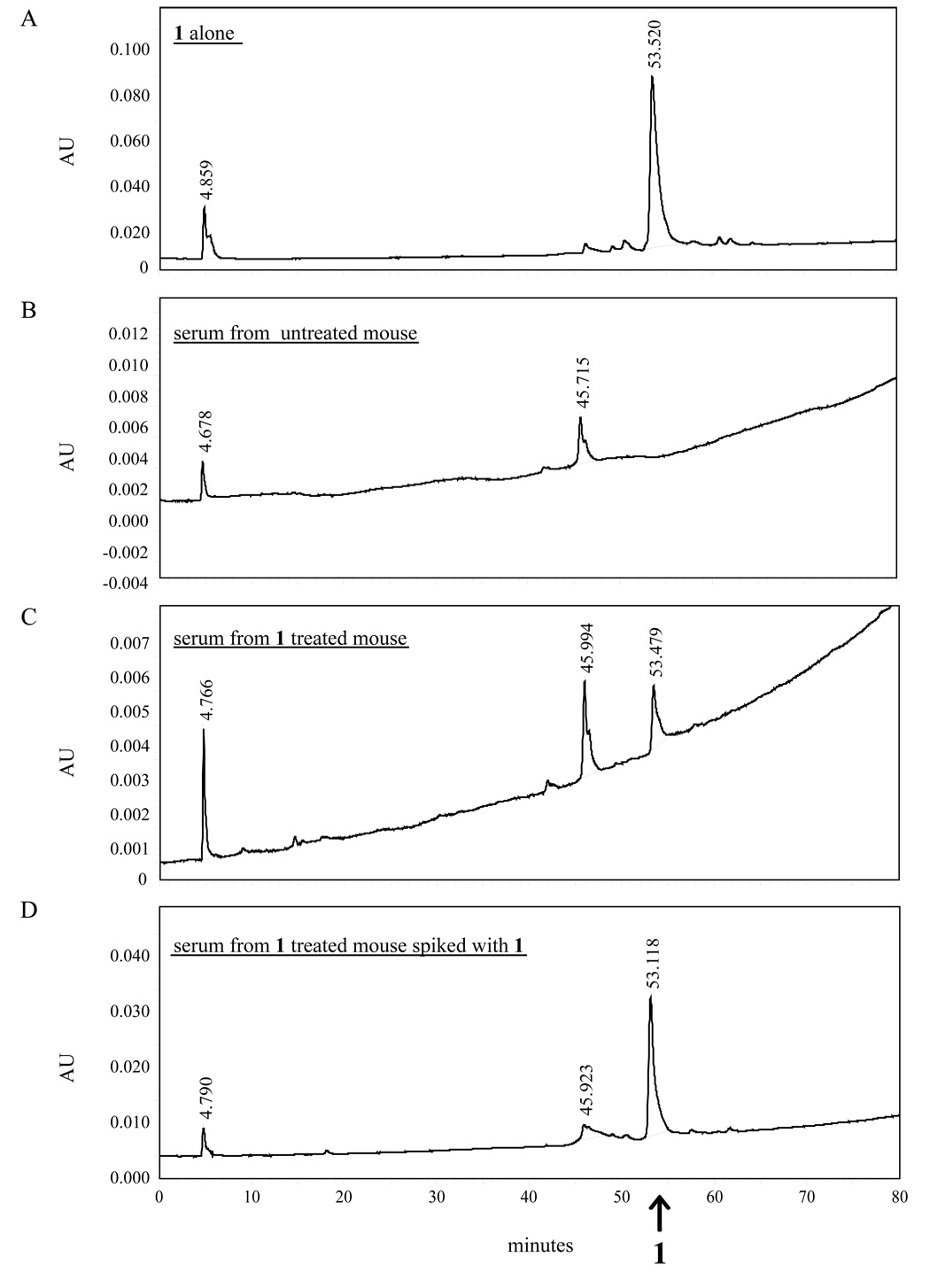
We further verified 1 for good in vivo systemic distribution in the injected mice. Absorption of the drug into the bloodstream and its distribution throughout the body are desirable properties for drug candidates. To measure if peritoneal-injection of 1 distributed well in the host, we collected sera from compound 1 injected mice and analyzed the samples by HPLC. The sera from mice injected with 200 µM of 1 (Figure 7C) were extracted with 3 volumes of ethyl acetate and passed through a C4 Vydac 214TP1010 column (10 × 250 mm), and then eluted with a gradient of 25% (H2O - 0.1%TFA) and 75% (CH3CN-0.1% TFA). This analysis revealed an absorption peak that corresponded to an 1-specific signal in the positive control (Figure 7A), and in 1-treated mice –“spiked” with additional 1 compound (Figure 7D). Conversely, the “compound 1-peak” was absent from the sera from DMSO-treated mice (Figure 7B).
Figure 7.
Systemic distribution of 1 in vivo. Sera collected from mice injected intraperitoneally with 1 were analyzed by HPLC for the presence of 1. In panel A and B HPLC was performed using 1 dissolved in MeOH:AcCN (1:1) and serum from an untreated mouse respectively to establish control profiles. The sera from mice injected with 1 (panel C) showed an absorption peak corresponding to the control 1 peak in panel A. A serum sample from a mouse injected with 1 was deliberately spiked with more 1 (panel D) and this produced an increase in the absorbance peak corresponding to the 1 peak in serum from a mouse injected with 1 (panel C) and the 1-alone control (panel A).
Discussion
The increased prevalence of HIV-1 resistant to current antiviral drugs highlights an urgent need to develop new treatment approaches. Existing therapies are targeted to virus-encoded proteins (e.g. reverse transcriptase, protease, integrase, and gp120). Because of the relaxed fidelity of HIV-1’s reverse transcriptase enzyme, virus-encoded proteins can rapidly mutate under drug selection (1, 24). Here, we have tested the concept of targeting HIV-1 replication using small molecules that inhibit the activity of a cellular RNA helicase, previously identified as important for virus replication in human cells.
We chose to target a cellular RNA helicase for two reasons. First, we and others (9, 25–28) have begun to broach the possibility that cellular proteins which do not mutate rapidly under drug selection could be bona fide targets for inhibiting virus replication. Second, targeting helicase activity is attractive because there is a body of literature that virus-encoded RNA helicases from hepatitis C virus, West Nile virus, Herpes simplex virus, and Japanese Encephalitis virus can be inhibited with small molecule drugs (18–22). While inhibiting a cellular protein risks cytotoxicity, we reasoned that there could be a therapeutic window of drug concentration which affects differentially the virus versus the host cell. We note that inhibition of cell-encoded enzymes in medical therapy is not an unprecedented strategy. Several clinical examples exist for the use of drugs to inhibit cellular enzymes (27, 29–33).
All RNA helicases share generally conserved motifs; however, recent findings have shown that small molecule inhibitors can work against some viral helicases but not others, suggesting that target discrimination between different helicases may be possible (18–22). For example, human herpesviruses encoded helicases have been shown in recent studies to be targeted by two new classes of drugs, amino-thiazolyphenyl-molecules (18) and thiazole amide derivatives (19), with little apparent toxicity on cellular helicases. Our current results extend these earlier anti-viral studies by testing the idea that a small molecule (e.g. 1) can target a cellular helicase and inhibit viral (HIV-1) replication. Indeed, our results suggest a therapeutic level for 1, which appears to be well-distributed systemically in vivo, that can inhibit HIV-1 replication without eliciting significant host toxicity. We note, however, that 1 and 2 could have additional complex intracellular effects; further studies are needed before one can conclusively determine that their anti-HIV-1 activities arise from direct inhibition of RNA-helicases.
The benefit of targeting a cellular protein versus a viral protein is that cellular genes do not mutate rapidly like viral genes. Hence in cells treated with 1 , HIV-1 cannot develop drug-resistance through mutation of its viral genome. Consistent with this proposed concept, a synthetic immunomodulator, Murabutide, was shown recently to suppress HIV-1 replication in macrophages and T cells (34). Intriguingly, Murabutide was found to inhibit the activity of a cellular RNA helicase, RH116, which works at the level of HIV-1 gene transcription rather than at post-transcriptional RNA processing like DDX3 (23). Collectively, the Murabutide results and our present work encourage further explorations of helicase inhibitors as anti-HIV-1 agents.
Experimental Section
Compounds
The chemical synthesis of ring-expanded nucleosides (RENs) with a β-configuration, including but not limited to 1, have been previously reported (21, 22). The procedure for the synthesis of RENs with an α-configuration is described below.
A general procedure for the synthesis of α-deoxy ring-expanded nucleoside (REN) analogs
An appropriate alkylamine (6 mmol) and 3, 5-dimethylpyrazole-1-carboxamidine nitrate (1.244 g, 6 mmol, 97%) was refluxed in methanol (20 ml) for 2 hours. The solvent was evaporated under vacuum, dried in a desiccator over P2O5 under vacuum for two nights. Without purification, it was dissolved in anhydrous methanol (6.0 mL), cooled to 0°C, and a solution of methanolic sodium methoxide (25% wt in methanol, 3.15 ml) was added drop-wise. The reaction mixture was kept in ice-bath for 0.5 hour. Solution from the reaction mixture was directly filtered into butyl 1-(2’-deoxy-3’,5’-di-O-p-toluoyl-α-D-erythropentofuranosyl)-4,5-imidazoledicarboxylate (21) (1.38–1.81 mmol) in anhydrous methanol (30 ml). The ring closure reaction went complete after stirring the reaction mixture overnight. The product was purified by silica gel chromatography, eluting successively with a mixture of chloroform:methanol (30:1), chloroform:methanol (10:1), and methanol. During solvent evaporation of the collected fractions, solid started to precipitate from solution. The precipitate was filtered and dried over P2O5 under vacuum.
4,5-Dihydro-8H-6-(N-tetradecyl)amino-1-(2’-deoxy-α-D-erythropentofuranosyl)imidazo[4,5-e]diazepine-4,8-dione (compound 11)
Yield: 92.6 %, mp > 250 °C; 1H NMR (DMSO-d6): δ 10.37 (brs, 1H, NH, exchangeable with D2O), 8.18 (s, 1H, imidazole), 7.10 (brs, 1H, NH, exchangeable with D2O), 6.62 (d, J= 5.9 Hz, 1H, 1’-H), 5.10 (m, 1H, 3’-OH, exchangeable with D2O), 4.87 (m, 1H, 5’-OH, exchangeable with D2O), 4.24 (m, 2H, 3’,4’-H), 3.42 (m, 2H, 5’-H1,2), 3.20 (m, 2H, CH2), 2.62 (m, 1H, 2’-H1), 2.01 (d, J= 14.3 Hz, 1H, 2’-H2), 1.46 (m, 2H, CH2), 1.22 (m, 22H, (CH2)11), 0.83 (t, J= 5.9 Hz, 3H, CH3); Anal. Calcd. for C25H41N5O5·H2O (509.64): C, 58.92; H, 8.50; N, 13.74. Found: C, 58.67; H, 8.29; N, 13.58.
4,5-Dihydro-8H-6-(N-hexadecyl)amino-1-(2’-deoxy-α-D-erythropentofuranosyl)imidazo[4,5-e]diazepine-4,8-dione (compound 12)
Yield: 99.0 %, mp > 250 °C; 1H NMR (DMSO-d6): δ 10.56 (brs, 1H, NH, exchangeable with D2O), 8.19 (s, 1H, imidazole), 7.11 (brs, 1H, NH, exchangeable with D2O), 6.63 (d, J= 6.6 Hz, 1H, 1’-H), 5.11 (m, 1H, 3’-OH, exchangeable with D2O), 4.87 (m, 1H, 5’-OH, exchangeable with D2O), 4.24 (m, 2H, 3’,4’-H), 3.42 (m, 2H, 5’-H1,2), 3.20 (m, 2H, CH2), 2.64 (m, 1H, 2’-H1), 2.01 (d, J= 13.9 Hz, 1H, 2’-H2), 1.47 (m, 2H, CH2), 1.22 (m, 26H, -(CH2)13), 0.83 (t, J= 6.0 Hz, 3H, CH3); Anal. Calcd. for C27H45N5O5·1.5H2O (546.70): C, 59.32; H, 8.85; N, 12.81. Found: C, 59.71; H, 8.60; N, 13.05.
4,5-Dihydro-8H-6-(N-octadecyl)amino-1-(2’-deoxy-α-D-erythropentofuranosyl)imidazo[4,5-e]diazepine-4,8-dione (compound 2)
Yield: 86.7 %, mp > 250 °C; 1H NMR (DMSO-d6): δ 10.30 (brs, 1H, NH, exchangeable with D2O), 8.19 (s, 1H, imidazole), 7.06 (brs, 1H, NH, exchangeable with D2O), 6.62 (d, J= 6.6 Hz, 1H, 1’-H), 5.10 (d, J= 2.2 H, 1H, 3’-OH, exchangeable with D2O), 4.87 (t, J= 5.1 Hz, 1H, 5’-OH, exchangeable with D2O), 4.24 (m, 2H, 3’,4’-H), 3.43 (m, 2H, 5’-H1,2), 3.21 (m, 2H, CH2), 2.64 (m, 1H, 2’-H1), 2.01 (d, J= 14.3 Hz, 1H, 2’-H2), 1.46 (m, 2H, CH2), 1.22 (m, 30H, (CH2)15), 0.84 (t, J= 6.4 Hz, 3H, CH3); Anal. Calcd. for C29H49N5O5·H2O (565.75): C, 61.57; H, 9.09; N, 12.38. Found: C, 61.88; H, 9.19; N, 12.18.
Antiviral assays
HIV-1 T-cell-tropic molecular clone (NL4-3) and macrophage-tropic molecular clone (AD8) were used. HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum. Infectious virus stocks were generated by transfecting HeLa cells with either pNL4-3 or pAD8 plasmids. Culture supernatant was collected 36 hrs post transfection, and the virus stock was quantified by RT assay (35). MT4 cells were maintained in RPMI supplemented with 10% fetal bovine serum. Usually 4 × 106 cells were infected with 20,000 normalized RT counts of NL4-3, and compounds were added 4 hrs post infection. Afterwards, 50% of the media was replaced every 2 days, with fresh media containing the compounds at the initial dosing level. Human monocyte derived macrophages (MDMs) were generated by culturing elutriated blood monocytes in RPMI-1640 containing 10% pooled human AB serum and M-CSF. Cells were cultured for 5–7 days before infecting with 105 RT counts of HIV-1 AD8.
RNA unwinding assay
RNA unwinding assay was performed as described previously (9). Briefly pBluescript plasmid was used to generate partially double stranded RNA for the RNA unwinding assay. The pBluescript vector was digested to completion with Kpn I and transcribed with T3 polymerase to generate a 120 base long transcript. The vector was also digested with EcoRI and transcribed with T7 polymerase in the presence of (α32p) UTP. The two complementary transcripts were purified and hybridized at 95° C for 20 minutes. Double stranded RNA was then incubated with purified DDX3 (with or without 4 mM 1 and 2) in ATPase/helicase (20 mm Tris-HCl, pH 8.0, 70 mm KCl, 2 mm MgCl2, 2 mm dithiothreitol, 15 units RNasin, and 2 mm ATP). After incubation for 30 mins at 37 °C, the reactions were stopped by the addition of a solution containing 10 mm EDTA, 40% glycerol, bromphenol blue, xylene cyanol. The reaction was resolved in a 10% polyacrylamide gel for 2 hours. The gel was dried and exposed to a phosphorimaging plate.
Toxicity assays
4 × 106 suspension T cells (MT4) were incubated with or without 1 or 2. 50% of the media was removed every 2 days and replaced with an equivalent amount of complete media containing the inhibitors. At time of each harvest, 100 µl of the cell suspension were measured for viablilty and compared to a similar amount of control (untreated) cells using the Cell Counting kit-8 (Dojindo) according to manufacturer’s instructions. Absorbance was measured at 450 nm using a microplate reader. For in vivo studies 8–10 weeks old BALB/c mice were injected with various concentrations of 1, 2, DMSO, or normal saline twice a week intraperitoneally for 4 weeks and sacrificed for necropsy after week 5. Blood from the mice was collected once per week via the tail vein.
HPLC analysis of mouse serum
Serum samples for HPLC were extracted with 3 volumes of ethyl acetate and lyophilized. Each mouse serum sample was dissolved in a mixture of 100 µl water and 20 µl acetonitrile. The samples were analyzed by reverse-phase HPLC using a C4 Vydac 214TP1010 column (10 × 250 mm). The injection for each run was 20 µl with monitoring at λmax 214 and 254 nm. The column was equilibrated with 75% developer A (water, 0.1%TFA) and 25% developer B (CH3CN, 0.1% TFA), and a gradient was employed to give a final composition of 25% developer A and 75% B over a period of 80 minutes at 3 ml/min. For the HPLC analysis of 1 alone without the mouse serum, the compound was dissolved in a mixture of MeOH:AcCN (1:1).
Acknowledgments
The research was supported in part by a grant (#1 R21 AI071802) (to RSH) from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health, Bethesda, Maryland, and by an unrestricted grant (to RSH) from Nabi Biopharmaceuticals, Rockville, Maryland. Work in KTJ’s laboratory is supported by NIAID intramural research funds and by funds from the Intramural AIDS Targeted Antiviral Program (IATAP) from the office of the Director, NIH.
Footnotes
Abbreviations: REN, Ring expanded nucleoside; MDM, monocyte derived macrophages; DMSO, dimethyl sulfoxide; RT, reverse transcriptase; CCR5, chemokine (C-C motif) receptor 5; CXCR4, chemokine (C-X-C motif) receptor 4; NTPase, nucleoside triphosphatase; TFA, trifluoroacetic acid
Reference List
- 1.Baldwin CE, Berkhout B. HIV-1 drug-resistance and drug-dependence. Retrovirology. 2007;4:78. doi: 10.1186/1742-4690-4-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang WS, Maddukuri A, Teslovich TM, de la FC, Agbottah E, Dadgar S, Kehn K, Hautaniemi S, Pumfery A, Stephan DA, Kashanchi F. Therapeutic targets for HIV-1 infection in the host proteome. Retrovirology. 2005;2:20. doi: 10.1186/1742-4690-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 5.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, Richard S. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 2005;79:124–131. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology. 2006;3:48. doi: 10.1186/1742-4690-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willemsen NM, Hitchen EM, Bodetti TJ, Apolloni A, Warrilow D, Piller SC, Harrich D. Protein methylation is required to maintain optimal HIV-1 infectivity. Retrovirology. 2006;3:92. doi: 10.1186/1742-4690-3-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida T, Hamano A, Koiwa T, Watanabe T. 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology. 2006;3:69. doi: 10.1186/1742-4690-3-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 10.Ishaq M, Hu J, Wu X, Fu Q, Yang Y, Liu Q, Guo D. Knockdown of Cellular RNA Helicase DDX3 by Short Hairpin RNAs Suppresses HIV-1 Viral Replication Without Inducing Apoptosis. Mol. Biotechnol. 2008 doi: 10.1007/s12033-008-9040-0. [DOI] [PubMed] [Google Scholar]
- 11.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 12.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 13.Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 14.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 15.Luking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev. Biochem. Mol. Biol. 1998;33:259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 16.Linder P, Stutz F. mRNA export: travelling with DEAD box proteins. Curr. Biol. 2001;11:R961–R963. doi: 10.1016/s0960-9822(01)00574-7. [DOI] [PubMed] [Google Scholar]
- 17.Marintcheva B, Weller SK. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2001;70:77–118. doi: 10.1016/s0079-6603(01)70014-1. [DOI] [PubMed] [Google Scholar]
- 18.Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, Kibler P, Liuzzi M, Cordingley MG. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 19.Kleymann G, Fischer R, Betz UA, Hendrix M, Bender W, Schneider U, Handke G, Eckenberg P, Hewlett G, Pevzner V, Baumeister J, Weber O, Henninger K, Keldenich J, Jensen A, Kolb J, Bach U, Popp A, Maben J, Frappa I, Haebich D, Lockhoff O, Rubsamen-Waigmann H. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 20.Borowski P, Deinert J, Schalinski S, Bretner M, Ginalski K, Kulikowski T, Shugar D. Halogenated benzimidazoles and benzotriazoles as inhibitors of the NTPase/helicase activities of hepatitis C and related viruses. Eur. J. Biochem. 2003;270:1645–1653. doi: 10.1046/j.1432-1033.2003.03540.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang N, Chen HM, Koch V, Schmitz H, Minczuk M, Stepien P, Fattom AI, Naso RB, Kalicharran K, Borowski P, Hosmane RS. Potent inhibition of NTPase/helicase of the West Nile Virus by ring-expanded ("fat") nucleoside analogues. J. Med. Chem. 2003;46:4776–4789. doi: 10.1021/jm030277k. [DOI] [PubMed] [Google Scholar]
- 22.Zhang N, Chen HM, Koch V, Schmitz H, Liao CL, Bretner M, Bhadti VS, Fattom AI, Naso RB, Hosmane RS, Borowski P. Ring-expanded ("fat") nucleoside and nucleotide analogues exhibit potent in vitro activity against flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J. Med. Chem. 2003;46:4149–4164. doi: 10.1021/jm030842j. [DOI] [PubMed] [Google Scholar]
- 23.Jeang KT, Yedavalli V. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 2006;34:4198–4205. doi: 10.1093/nar/gkl398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chesney M. Adherence to HAART regimens. AIDS Patient. Care STDS. 2003;17:169–177. doi: 10.1089/108729103321619773. [DOI] [PubMed] [Google Scholar]
- 25.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 26.Klase ZA, Van DR, Kashanchi F. Identification of potential drug targets using genomics and proteomics: a systems approach. Adv. Pharmacol. 2008;56:327–368. doi: 10.1016/S1054-3589(07)56011-4. [DOI] [PubMed] [Google Scholar]
- 27.Kibler KV, Miyazato A, Yedavalli VS, Dayton AI, Jacobs BL, Dapolito G, Kim SJ, Jeang KT. Polyarginine inhibits gp160 processing by furin and suppresses productive human immunodeficiency virus type 1 infection. J. Biol. Chem. 2004;279:49055–49063. doi: 10.1074/jbc.M403394200. [DOI] [PubMed] [Google Scholar]
- 28.Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Discov. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cushman DW, Ondetti MA. Design of angiotensin converting enzyme inhibitors. Nat. Med. 1999;5:1110–1113. doi: 10.1038/13423. [DOI] [PubMed] [Google Scholar]
- 30.Menard J, Patchett AA. Angiotensin-converting enzyme inhibitors. Adv. Protein Chem. 2001;56:13–75. doi: 10.1016/s0065-3233(01)56002-7. [DOI] [PubMed] [Google Scholar]
- 31.Miller WR, Anderson TJ, Evans DB, Krause A, Hampton G, Dixon JM. An integrated view of aromatase and its inhibition. J. Steroid Biochem. Mol. Biol. 2003;86:413–421. doi: 10.1016/s0960-0760(03)00352-2. [DOI] [PubMed] [Google Scholar]
- 32.Hook VY, Reisine TD. Cysteine proteases are the major beta-secretase in the regulated secretory pathway that provides most of the beta-amyloid in Alzheimer's disease: role of BACE 1 in the constitutive secretory pathway. J. Neurosci. Res. 2003;74:393–405. doi: 10.1002/jnr.10784. [DOI] [PubMed] [Google Scholar]
- 33.Sarac MS, Cameron A, Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect. Immun. 2002;70:7136–7139. doi: 10.1128/IAI.70.12.7136-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocude C, Truong MJ, Billaut-Mulot O, Delsart V, Darcissac E, Capron A, Mouton Y, Bahr GM. A novel cellular RNA helicase, RH116, differentially regulates cell growth, programmed cell death and human immunodeficiency virus type 1 replication. J. Gen. Virol. 2003;84:3215–3225. doi: 10.1099/vir.0.19300-0. [DOI] [PubMed] [Google Scholar]
- 35.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]