Abstract
Several changes in cell morphology take place during the capping of surface receptors in Entamoeba histolytica. The amoebae develop the uroid, an appendage formed by membrane invaginations, which accumulates ligand–receptor complexes resulting from the capping process. Membrane shedding is particularly active in the uroid region and leads to the elimination of accumulated ligands. This appendage has been postulated to participate in parasitic defense mechanisms against the host immune response, because it eliminates complement and specific antibodies bound to the amoeba surface. The involvement of myosin II in the capping process of surface receptors has been suggested by experiments showing that drugs that affect myosin II heavy-chain phosphorylation prevent this activity. To understand the role of this mechanoenzyme in surface receptor capping, a myosin II dominant negative strain was constructed. This mutant is the first genetically engineered cytoskeleton-deficient strain of E. histolytica. It was obtained by overexpressing the light meromyosin domain, which is essential for myosin II filament formation. E. histolytica overexpressing light meromyosin domain displayed a myosin II null phenotype characterized by abnormal movement, failure to form the uroid, and failure to undergo the capping process after treatment with concanavalin A. In addition, the amoebic cytotoxic capacities of the transfectants on human colon cells was dramatically reduced, indicating a role for cytoskeleton in parasite pathogenicity.
INTRODUCTION
During capping of surface receptors, motile Entamoeba histolytica undergoes striking morphological changes, including highly active membrane folding that leads to the formation of a membrane-accumulated appendage at one pole of the cell called the uroid (Trissl et al., 1977). Ligand–receptor complexes are concentrated in the uroid and eventually are eliminated by membrane shedding into the external medium without damage to the cell. One amoebic protein that undergoes capping is the Gal-GalNAc lectin (Arhets et al., 1995). This lectin participates in the interaction between amoebae and cells upon their direct contact (Ravdin and Guerrant, 1981). Avoidance of lysis by the complement membrane attack complex is also mediated by this protein, which binds to final components of the membrane attack complex and prevents their association with the amoebic plasma membrane (Braga et al., 1992). Another amoebic defensive mechanism is the inhibition of complement activity by the capping of Gal-GalNAc lectin involved in the interaction with complement components and shedding of complement–lectin complexes to the external medium.
Surface receptors are proteins anchored into the plasma membrane that generally contain a cytoplasmic domain that may interact with either regulatory or cytoskeletal proteins. During capping, surface receptors interact with their ligands, leading to activation of signaling pathways and accumulation of receptors in patches. These patches are then translocated in a rearward direction to a pole of the cell and form a cap. The retrograde cell surface flow observed during the capping process is a conserved behavior in migratory cells and has been the basis for models of cell locomotion (Egelhoff and Spudich, 1991). In the case of E. histolytica, capping of surface receptors and formation of the uroid are processes that occur simultaneously. Multiple studies have suggested a role for the actomyosin complex in capping and uroid formation. F-actin and myosin II colocalize with the caps in the uroid (Espinosa-Cantellano and Martinez-Palomo, 1994; Arhets et al., 1995). In addition, comparison of myosin II concentrations in different compartments of amoebae undergoing capping have shown that three times more myosin II is recruited into the uroid region than into any other compartment. Moreover, capping and uroid formation are inhibited by drugs that affect the activity of the actomyosin complex (Arhets et al., 1995).
Myosin II is a hexameric ATPase composed of two heavy chains and two pairs of light chains, as reviewed by Sellers and Goodson (1995). The amino terminus of the heavy chain forms the head region containing the actin and nucleotide binding sites. The carboxyl-terminal portion of the two heavy chains associates to form a coiled coil α-helical rod structure necessary for myosin II filament formation (Warrick and Spudich, 1987). The role of myosin II in the capping of surface receptors has also been studied in several other eukaryotic cell systems. In Dictyostelium discoideum, a nonpathogenic amoeba, a myosin II null mutant is deficient in capping (Pasternak et al., 1989). In other cell types, such as T lymphocytes, the involvement of myosin II and G-proteins mediate pathways that have been proposed as key mechanistic components of the capping process (Campanero et al., 1994). In addition, recruitment of myosin II has also been shown to generate the contractile force necessary for movement and cellular division of eukaryotic cells (De Lozanne and Spudich, 1987; Knecht and Loomis, 1987).
Identification of the mhcA gene, which encodes myosin II in E. histolytica (Raymond-Denise et al., 1993), has opened the way to a genetic analysis of the role of myosin II in pathogenic functions. Recent advances in the manipulation of the E. histolytica genome facilitated the construction of a parasite strain that is defective for myosin II functions. This mutant is the first genetically engineered cytoskeleton-deficient strain of E. histolytica. Analysis of this mutant shows that the inhibition of myosin II activity in E. histolytica leads to the inhibition of capping and uroid formation, motility, and parasite virulence.
MATERIALS AND METHODS
Strains and Culture Conditions
The pathogenic E. histolytica (strain HM1:IMSS) was cultivated axenically in plastic flask containing TYI-S-33 medium (Diamond, 1961) at 35°C for 48 h before each experiment. The human carcinoma Caco-2 cell line was grown to 14 d confluence in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal calf serum at 37°C in a 10% CO2 incubator. The bacterial strain Escherichia coli DH5α:endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 D(lacZYA-argF)U169 F′ F80dlacD(lacZ)M15 was used for plasmid constructions. Ampicillin was used at a concentration of 50 μg/ml.
Construction and Purification of Plasmid DNA
A plasmid expressing a truncated form of E. histolytica myosin II corresponding to the light meromyosin (LMM) was constructed as follows. A DNA fragment of 1041 bp encoding the last 579 amino acids of the myosin II heavy chain (Raymond-Denise et al., 1993) was amplified by PCR using the genomic DNA as a template. The DNA fragment is located between nucleotides 5046 and 6794 of the published myosin II heavy chain gene sequence (Raymond-Denise et al., 1993) and does not contain introns. The following oligonucleotides were used: 5′-GGGGTACCATGTACACCGACATCGAGATGAACCGGCTGGGCAAGAATGCTGATGAAAATGAA-3′ (nucleotides 5046–5063) and 5′-CGGGATCCATGAAATTTAATCAGAATC-3′ (nucleotides 6776–6794) were used. The first oligonucleotide permitted the introduction of an epitope tag at the amino terminus of the light meromyosin (underlined sequence). This tag sequence, encoding a cytoplasmic fragment of the vesicular stomatitis virus glycoprotein (VSV-G) (Kreis, 1986), is preceded by an ATG codon to initiate protein translation. In addition, the first oligonucleotide contains a KpnI site, whereas the second contains a BamHI site. The PCR product was digested by BamHI and KpnI and then cloned into the pExEhNeo plasmid (kindly provided by Dr. E. Tannich, Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany) at the BamHI and KpnI sites. Thus, the DNA encoding LMM is under specific transcription signals from E. histolytica contained in the plasmid vector. This vector also contains the gene conferring geneticin (G418) resistance as a selectable marker The recombinant plasmid was called pExEhNeo/LMM. Both plasmids, pExEhNeo and pExEhNeo/LMM, were used to transform E. coli, and then plasmid DNA was prepared using the QIAGEN (Chatsworth, CA) Maxi kit.
Transfection and Selection of E. histolytica Amoebae
Transfection of both pExEhNeo and pExEhNeo/LMM plasmids into trophozoites was performed by electroporation. Specifically, 107 amoebae were washed twice in cold PBS buffer and once in cold cytomix buffer consisting of 10 mM K2HPO4/KH2PO4 (pH 7.6), 120 mM KCl, 0.15 mM CaCl2, 25 mM HEPES (pH 7.6), 2 mM EGTA, 5 mM MgCl2. The amoebae were then resuspended in 500 μl of cytomix buffer supplemented, immediately before use, with 2 mM ATP, 5 mM glutathione, and 200 μg of plasmid DNA. The amoebae were transferred to an electroporation cuvette (4-mm gap, Bio-Rad, Hercules, CA), and electroporation was performed with the Bio-Rad Gene Pulser (1200 V/cm and 25 μF, with a time constant of 0.4 ms). The electric pulse was applied twice, and electroporated amoebae were transferred into culture medium for 48 h before selecting them in culture medium supplemented with 10 μg/ml G418 (Life Technologies, Gaithersburg, MD). Amoebae expressing G418 resistance were further submitted to increasing concentrations of G418 up to 30 μg/ml.
Preparation of Total Protein Extracts
Transfected amoebae were cultivated in the presence of increasing concentrations of G418, and proteins were prepared as previously described (Burns et al., 1995). Briefly, 5 × 106 trophozoites were washed twice in cold PBS buffer and then lysed in 200 μl of 100 mM 2-[N-morpholino]ethanesulfonic acid (pH 6.8), 1 mM MgCl2, 0.5% Triton-X 100, 2.5 mM EGTA, 2 mM p-hydroxymercuribenzoate, 6 μM leupeptin, 1 mM N-ethylmaleimide for 30 min at 4°C with gentle agitation. Protein concentrations were measured by the Bio-Rad assay with lysozyme as a standard.
Immunodetection of Proteins
Proteins from transfected trophozoites were analyzed by immunoblot. Samples of proteins (10–100 μg) were resolved by SDS-6% PAGE and then electrotransferred onto a nitrocellulose membrane that was saturated with 5% nonfat milk, 1% Tween 20. The membranes were subsequently incubated for 1–2 h at room temperature with antibodies diluted in PBS containing 5% nonfat milk and 1% Tween 20. The following antibodies were used: anti-amoebic myosin II (Rahim et al., 1993), 1:750; anti-VSV-G (hybridoma cell line was a gift from Dr. T. Kreis, University of Geneva Sciences III, Geneva, Switzerland), 1:200; peroxidase-conjugated sheep anti-mouse Ig (Nordic Immunological Laboratories, Tilburg, the Netherlands), 1:10,000; and peroxidase-conjugated goat anti-rabbit Ig (Nordic Immunology), 1:30,000. The last two sera were used as secondary antibodies to detect antigen-antibody complexes. Finally, immunoblots were treated with the ECL Western-blotting detection reagent (Amersham, Buckinghamshire, United Kingdom) and exposed on Kodak (Rochester, NY) X-OMAT x-ray film. To compare the affinity of the anti-myosin II and anti-VSV antibodies, quantification of antibody–protein binding was done. Serial dilutions of amoebic protein extracts, ranging from 5 to 50 μg, were resolved by SDS-6% PAGE and electrotransferred to a membrane as described. The upper part of the blot containing proteins with a molecular mass of >100 kDa was incubated in the presence of anti-myosin II antibody, and the lower part was incubated in the presence of the anti-VSV antibody. The blots were treated as described for ECL Western-blotting detection. Both antibodies gave a clear signal when at least 10 μg of transfected amoebae crude extract were used. The autoradiograms were scanned using a Microtek (Taiwan) E3 Scanmaker, and relative intensities were calculated.
Computer-assisted Analysis of Cellular Movement
The amoebae cell lines generated in this work, as well as the wild-type strain HM1:IMSS, were inoculated in TYI-S-33 medium in a tissue culture flask completely filled with medium. Cells were allowed to grow at 35°C for 24–48 h. The cultures were then observed with an inverted microscope (Olympus Optical, Tokyo, Japan) equipped with a heating plate set at 37°C and a high-resolution Sony (Tokyo, Japan) charge-coupled device video camera. In these experiments 25 wild-type amoebae were analyzed. The culture of amoebae containing the vector alone was grown in the presence of 20 μg/ml G418; from this culture 27 amoebae were analyzed. LMM-transfected amoebae were analyzed from cultures grown in the presence of G418 at 20 μg/ml (11 amoebae analyzed) and 30 μg/ml (18 amoebae analyzed). For image analysis, a dedicated automatic program was developed to detect and track the amoebae as they moved. The program runs on a SPARC station Ultra1 (Sun, Mountain View, CA) to which a Series 151/40 digital image processor (Imaging Technology, Bedford, MA) was connected. Digitization of video sequences was performed using a real-time digitization system perception (Digital Processing Systems, Scarborough, Ontario, Canada). Amoebae were first recognized by a two-step segmentation procedure. First, the gradient image corresponding to the original phase-contrast image was computed. Then, the threshold of the gradient image was delimited at a predefined level that allowed retention of the amoeba outlines; the resulting binary image was processed to fill in the holes, if any, within connected areas. The binary masks representing amoebae were used to compute the following geometrical parameters with a central moment algorithm: centroid, surface, principal axes, and eccentricity. On the basis of these parameters, the algorithm was also able to discriminate between a single amoeba and amoebae aggregates. After all amoebae in the sequence had been characterized, and their coordinates had been stored, a tracking algorithm was used to establish valid trajectories. The algorithm uses a first-order Kalman filtering approach whereby at each frame and on the basis of the previous ones, predictions of the amoeba positions were established and compared with the computed ones. The best matches were selected as trajectory points, and tracks were finally analyzed to compute the data that were used for generating values in Figure 4. For each strain of amoeba, a sequence of ∼2000 images (∼1 min 20 s) was digitized, and every second image was analyzed. Speed values reported here are mean values of the speed of several amoebae and SEs of these group estimates. These values were corrected with the factor introduced by the time lapse recording effect. In addition, we observed one amoeba in 40 pixels; according to the mean size of amoeba (25 μm), an equivalence between 1 pixel and 0.625 μm was established.
Figure 4.
TEM of transfectant amoebae induced for capping with ConA. ExEh30 and LMM30 strains were induced for capping and fixed, and redistribution of ConA–receptor complexes was observed by the labeling of ConA binding sites by peroxidase. The membrane region in which capped complexes are concentrated is stained in black by oxidized diaminobenzidine. Micrograph A (4500× magnification) shows the cellular localization of ConA–receptor complexes in the LMM30 strain (arrows). The membrane appears poorly folded when compared with membrane in the uroid region of the ExEh30 transfectant used as a control (micrograph B, 8000× magnification).
Indirect Immunofluorescence of E. histolytica
For epifluorescence labeling of trophozoites, capping was induced using fluorescein isothiocyanate (FITC)-labeled concanavalin A (ConA, Sigma, St. Louis, MO) as previously described (Arhets et al., 1995). Amoebae were fixed in 3.7% paraformaldehyde for 1 h at 37°C and then permeabilized in PBS and 0.1% Triton for 5 min. For myosin II labeling, amoebae were fixed and permeabilized in methanol at −20°C for 3 min. They were further incubated in 50 mM NH4Cl for 30 min and then blocked in PBS and 1% BSA for 30 min at 37°C. For direct observation of capping, samples of fixed amoebae were mounted on glass slides with PBS and 70% glycerol. For in situ epifluorescence labeling of proteins, trophozoites were incubated for 1 h at 37°C in the following diluted antibodies: amoebic anti-myosin II, 1:10; mouse anti-VSV-G, 1:100; and anti-Gal-GalNAc lectin mAb 8A3, 1:30. After washing with PBS and 1% BSA at 37°C for 30 min, samples were incubated for 30 min in a 1:200 dilution of FITC-labeled goat antirabbit Ig (Sigma) or a 1:250 dilution of rhodamine-labeled goat anti-mouse Ig (Molecular Probes, Eugene, OR) that had been preadsorbed with trophozoites as previously described (Rahim et al., 1993). Labeling of nuclei was performed using propidium iodide (Molecular Probes, Eugene, OR) diluted at 1:200 and included during incubation of samples with the secondary antibodies. Finally, the samples were incubated in PBS and 1% BSA and mounted on glass slide with 10 mg/ml Dabco (Sigma) diluted in 80% glycerol. Results were analyzed using a conventional fluorescence microscope (BH2-RFCA, Olympus Optical).
Confocal Microscopy
Fluorescent samples were examined on a Leica (Heidelberg, Germany) DIAPLAN confocal laser-scanning microscope. Rhodamine-labeled samples excited at 568 nm were visualized with a high-pass RG590 filter. For fluorescein, an interferential filter centered on 535 nm, after excitation at 488 nm, was used. Observations were performed on 15 planes from the bottom to the top of each cell. The distance between scanning planes was 0.5 μm. Photographs were taken on Kodak T-max 400 film with a 35-mm camera mounted on a Polaroid (Cambridge, MA) Freeze Frame video recorder monitor.
Transmission Electron Microscopy
After inducing capping with ConA, 5 × 106 amoebae were washed three times in PBS buffer, and staining of ConA binding sites was performed by incubation of amoebae for 15 min at 37°C in PBS containing 50 μg/ml peroxidase (Boehringer Mannheim, Indianapolis, IN). After four washes in PBS, cells were fixed with 1.5% glutaraldehyde in 0.1 M phosphate buffer (pH 6.3) for 15 min at room temperature. Sedimented amoebae were washed five times in PBS, and peroxidase staining was detected by incubating the samples for 15 min at 4°C with 0.5 mg/ml diaminobenzidine and 0.03% hydrogen in PBS. The enzymatic reaction was stopped by dilution with PBS. After three washes in PBS, cells were postfixed for 1 h at 4°C in PBS containing 2% OsO4. The pellet was rinsed with distilled water, dehydrated in a graded ethanol series, and embedded in Epon resin. Thin sections were prepared on an LKB Nova ultramicrotome (Leica, Vienna, Austria) fitted with a diamond knife and were observed in a JEOL (Tokyo, Japan) 1010 electron microscope operating at 80 kV.
E. histolytica and Caco-2 Cell Interaction
The human Caco-2 cell line was grown for 14 d in 24-well plates. Confluent monolayers were labeled overnight with 2 μCi/ml 51Cr in complete DMEM. Trophozoites grown for 48 h and the Caco-2 cells were washed twice with prewarmed serum-free DMEM. Amoebae were added to the cell monolayer at a ratio of 1 amoeba to 5 cells in DMEM serum-free medium. The coculture was incubated at 37°C in 10% CO2 atmosphere for 0.5–3 h. At designated time intervals, the medium was harvested and centrifuged at 800 × g for five min. The supernatant (fraction A) and the pellet (fraction B) were separated. Any cells remaining in the wells were recovered using 1% SDS in phosphate-NaCl buffer (fraction C). Sampling was done in triplicate. Radioactivity was counted from each fraction, and an average of three samples was determined. The percentage of cell lysis was determined by calculating the counts per minute in fraction B divided by total counts per minute in fractions A–C. Viability of amoebae was determined in parallel by the trypan blue dye exclusion test. Briefly, amoebae were recovered, centrifuged at 800 × g, and resuspended in 0.4% trypan blue in PBS, and then viable amoebae were counted.
RESULTS
Construction of a Myosin II Mutant by Overexpression of LMM
Because gene targeting by homologous recombination is not yet possible in E. histolytica, we took advantage of a recent observation made in D. discoideum in which overexpression of the myosin heavy chain domain required for filament formation was shown to lead to a phenocopy of a null mutation (Burns et al., 1995). We used this approach to create myosin II mutants in E. histolytica. The DNA fragment encoding the end of the tail region of the myosin II heavy chain (light meromyosin, LMM) was amplified by PCR from E. histolytica genomic DNA. Oligonucleotides used in the PCR reaction enabled the introduction of an epitope tag at the amino-terminal fragment of LMM. This epitope tag is recognized by a monoclonal antibody against VSV. The amplified DNA fragment was purified and cloned into the ExEhNeo vector (Figure 1A). The recombinant plasmid expressing LMM as well as the vector alone were introduced into E. histolytica by electroporation. Transfected amoebae were selected by incubation of cells in the presence of G418 at 10 μg/ml. Available vectors for transfection of E. histolytica are maintained in the cell as episomal elements (Hamann et al., 1995). The copy number of these episomal elements varies according to the efficiency of transfection and the concentration of the selective drug in the culture medium. This variability might lead to heterologous phenotypes of transfectant cultures. For this reason, we tried to obtain a clonal population of transfected amoebae. Unfortunately, our efforts to obtain clonal populations were unsuccessful, probably because growth of E. histolytica is highly sensitive to the density of the inoculum (Diamond, 1961). Here, we report the phenotypical analysis of transfected amoebae that may express different levels of recombinant LMM.
Figure 1.
Cloning and expression of light meromyosin in E. histolytica. (A) The 3′ end of the gene encoding the myosin heavy chain was amplified by PCR. Oligonucleotides were designed to incorporate an epitope tag at the amino terminus of the expressed LMM. The generated DNA fragment was cloned into the ExEhNeo vector and introduced into the amoebae by electroporation. (B) Immunoblot of proteins obtained from the transfectant cells revealed by the anti-myosin II and by the anti-VSV-G antibodies. Myosin is at 250 kDa, and VSV-LMM is at 71 kDa. Lane 1, LMM20; lane 2, ExEh20, lane 3, LMM30; lane 4, ExEh30. Analysis shows that the LMM protein is expressed and correctly tagged.
Immunoblot analysis of total cell lysates from two transfectants showed that only the amoebae transfected by the recombinant plasmid expressed truncated myosin II, the LMM fragment (Figure 1B). It was observed that an increase in G418 concentration led to an increase in the expression level of LMM. The complete molecule of myosin II was recognized by a specific antibody against a 100-amino acid sequence on stretch located in the tail region of the myosin II heavy chain, outside of the LMM domain (Rahim et al., 1993). As expected, a prominent band at 250 kDa was visible. LMM was recognized as a 71-kDa band by antibodies against the VSV tag. To estimate the ratio of LMM versus myosin II, scanning of the autoradiogram of immunoblots obtained with serial dilutions of proteins isolated at 30 μg of G418/ml was performed. A ratio of 4 LMM to 1 myosin II was observed in LMM-expressing cells.
Phenotypic Analysis of Cells Overexpressing LMM
To explore the influence of LMM overexpression on the physiology of E. histolytica, we tested the two transfected cell lines for their ability to grow. The growth rate of the transfectants in the presence of diverse concentrations of G418 was examined. Growth rates were determined for wild-type cells as well as for cells containing the vector alone or for cells expressing the gene encoding LMM. Wild-type amoebae and control amoebae containing the vector alone grew roughly at similar mean rates of 10 and 11 h per generation, respectively. In contrast, a modification of the mean generation time was observed in LMM-transfected amoebae. The generation time varied according to the concentration of G418 in the medium. Thus, the generation times for amoebae overexpressing LMM, grown in 20 (LMM20) or 30 (LMM30) μg/ml G418, were 15 and 34 h, respectively.
Qualitative and Quantitative Analysis of Movement in LMM-overexpressing Amoebae
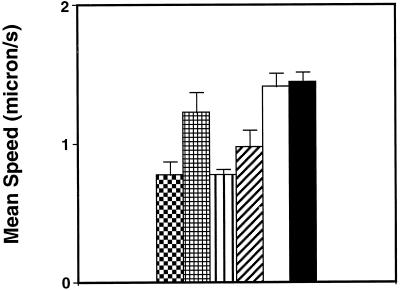
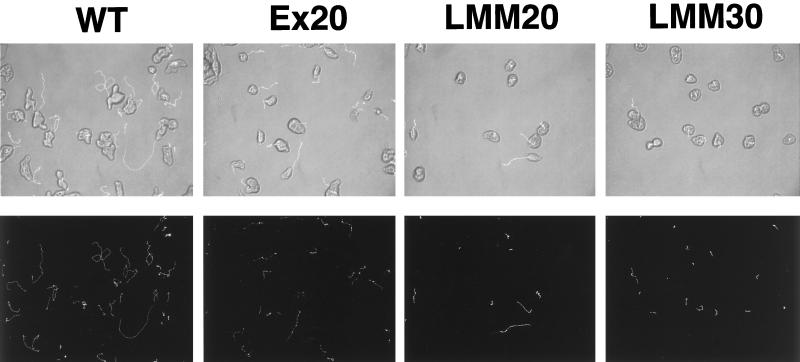
To test whether overexpression of LMM in E. histolytica affected single-cell motility, movement of LMM-overexpressing amoebae in culture medium was compared with that of either the wild-type amoebae or those carrying the vector alone. Trophozoites of each strain were observed by optic phase-contrast microscopy in culture flasks. Movement was video recorded and analyzed as described in MATERIALS AND METHODS. The mean values of average parameters computed for cell motility are presented in Figure 2. Wild-type amoebae moved with a mean velocity of 1.43 μm/s. The vector-transfected amoebae population grown in 20 μg/ml G418 moved with a mean velocity of 1.39 μm/s, a value equivalent to that observed with the wild-type strain. In contrast, LMM-overexpressing amoebae displayed a different behavior. The cell population could be assigned to two categories of movement: slow and very slow (Figure 2). The speed varied according to the concentration of G418 in the growth medium. LMM-overexpressing amoebae grown in 20 μg/ml G418 (LMM20) moved with a rate of between 0.76 and 1.22 μm/s. The rate of movement of LMM30 amoebae was even more reduced, because it ranged from 0.76 to 0.96 μm/s. Trajectories of moving amoebae are summarized in Figure 3. Observation of the trajectories of wild-type cells indicated that they moved randomly on the substrate. Their trajectories were heterogeneous, because some amoebae made straight trajectories, whereas others moved back or circled on a small area. Transfectants containing the vector alone moved according to straight trajectories. In contrast, amoebae expressing LMM wobbled in place around stationary points of contact with the substratum. These results indicate that overexpression of LMM interfered with amoebic movement.
Figure 2.
Analysis of mean speed of wild-type, ExEh20, and LMM transfectant amoebae. The histogram shows the comparison of the means speed of the two subpopulations of LMM20: slower ( ) and LMM20 faster ( ). The same type of representation was chosen for LMM30: slower (▥) and LMM30 faster (▨). Finally, the mean speeds for ExEh20 (□) and wild type (▪) are represented.
Figure 3.
Computer-assisted cellular movement analysis of wild-type, ExEh20, and LMM cells. Images of amoebae were recorded and computer analyzed using conditions described in MATERIALS AND METHODS. Micrographs of analyzed amoebae are shown in the upper panel, and their trajectories are represented in the lower panel. The wild-type strain moves in random directions; some of the amoebae return on their trajectories. ExEh20 cells move at the same rate as the wild-type strain but show more direct trajectories. LMM20 and LMM30 show reduced trajectories. The majority of the amoebae wobble in place around stationary points of contact with the substratum. In addition, LMM transfectants are round, and some of them seem to extend small pseudopods.
Capping of Surface Receptors and Uroid Formation in LMM-expressing amoebae
Modification of the myosin II activity by drugs that inhibit the cytoskeletal activity of E. histolytica leads to inhibition of receptor capping and uroid formation (Arhets et al., 1995). To analyze the influence of LMM expression on receptor capping and uroid formation, transfected amoebae were induced for capping by incubation in the presence of FITC-labeled ConA. The number of amoebae displaying uroids was determined by analyzing the fluorescence of ConA–receptor complexes, which became concentrated in the uroid (Table 1). A correlation was established between an increase in LMM expression and a decrease in the number of cells undergoing capping. Thus, in the LMM30 population only 20% of amoebae formed uroids, whereas among amoebae containing the vector alone, 72% formed uroids. In addition, in some LMM-overexpressing amoebae, fluorescent ConA appeared in large areas of the cell surface, indicating that a modification might occur in the rate of movement of ConA–receptor complexes. To further document this observation, LMM30 amoebae were examined by transmission electron microscopy. The uroid region was visualized by staining the ConA binding sites with peroxidase, which specifically labeled regions in which the ConA–receptor complexes were concentrated (Arhets et al., 1995). In some trophozoites overexpressing LMM, most of the peroxidase-labeled ConA localized in diffuse areas occupying a large surface at one pole of the amoeba, an observation that confirmed the FITC–ConA distribution encountered by confocal analysis. In addition, the membrane in these regions was not folded (Figure 4A), and the cytoplasm appeared less vesiculated when compared with cytoplasmic and uroid regions formed in the amoebae carrying the vector alone (Figure 4B). Thus, among the parasites expressing LMM, the majority of cells were inhibited for uroid formation and capping. Of the amoebae that did undergo capping (20%), a fraction displayed normal uroid formation, whereas the remainder did not form uroids and displayed diffuse caps. These results suggest that overexpression of LMM impairs the capping process. In addition, the endogenous myosin II seems to retain some residual activity but in certain cases is insufficient to assure complete capping or formation of the uroid.
Table 1.
Capping induction in LMM transfectant and control cells
| Amoeba strain | Amoebae with a uroid (%) | Total amoebae counteda |
|---|---|---|
| ExEhNeo 10 | 70.9 | 1031 |
| ExEhNeo 20 | 71.7 | 852 |
| LMM 10 | 68.6 | 868 |
| LMM 20 | 44.7 | 927 |
| LMM 30 | 20.1 | 900 |
Amoebae were counted in two independent experiments. Capping in replicating trophozoites was induced by incubation of the parasites in the presence of FITC-labeled ConA. Cells were fixed, and uroid-containing cells were counted.
Localization of Myosin II, LMM, and Gal-GalNAc Receptor in Resting and Capping Amoebae
Inhibition of myosin heavy-chain phosphorylation leads to the localization of myosin II under the cytoplasmic membrane of capping amoebae (Arhets et al., 1995). To determine the subcellular localization of myosin II, truncated myosin II, and Gal-GalNAc lectin in LMM-overexpressing trophozoites, immunofluorescence labeling of proteins was performed in resting and capping amoebae (Figure 5). The shape of resting cells (80% of the population) appeared essentially round (Figure 5A). In these amoebae, endogenous myosin II was localized under the cytoplasmic membrane at the periphery of the cells, whereas LMM appeared in a diffuse pattern throughout the cytoplasm. In addition, the Gal-GalNAc lectin was concentrated in spots at the membrane level (Figure 5B), indicating that clustering of this receptor still occurred in LMM-transfected amoebae. In the case of cells undergoing capping, both myosins were recruited to the posterior pole of trophozoites and colocalized in the uroid when it was present (Figure 5C). The Gal-GalNAc receptor was similarly recruited (our unpublished data). These results suggested that LMM was recruited to the uroid together with myosin II in response to signaling induced by capping activation.
Figure 5.
Distribution of myosin II, LMM, and Gal-GalNAc lectin in E. histolytica transfected with LMM. Transfectants growing in 30 μg/ml G418 amoebae were fixed and stained for myosin II, VSV-LMM, and Gal-GalNAc lectin. Confocal laser scanning microscopy was performed to detect immunofluorescence. Micrograph A shows rounded cells with myosin II labeling the plasma membrane (panel 1); some discrete myosin II spots are also observed in the cytoplasm. In the same cell, LMM is essentially diffuse throughout the cytoplasm (panel 2). The focal plane shown in this micrograph represents the middle of the cell. Micrograph B shows myosin II distribution in rounded cells (panel 3) and Gal-GalNAc in the same cell (panel 4). Myosin II is located under the plasma membrane as described previously. The Gal-GalNAc lectin is present at the membrane level; some patches of receptor can be seen (arrows). Micrograph C shows transfected amoeba that display a uroid (arrowhead). Myosin II is recruited to and concentrates in the uroid (panel 5); VSV-LMM is also observed in the uroid (panel 6). These results indicate the recruitment of both proteins during capping. Bar, 5 μm.
Effect of LMM Transfectant Amoeba on Caco-2 Cell Viability
To determine the pathogenic consequences of the cytoskeleton modifications observed in LMM transfectants, we assessed monolayer cell damage after contact with amoebae. Monolayers of the human carcinoma cell line Caco-2 were labeled by preincubation with medium containing 51Cr. Amoeba-mediated cytotoxicity was determined by measuring 51Cr release from Caco-2 cells into the incubation medium after amoeba contact (Figure 6). Human cell lysis became evident after 30 min of interaction with wild-type or vector alone–transfected amoebae. After 3 h of interaction, 40% of Caco-2 cells were lysed (wild-type, ±3%; n = 3; and vector transfected, ±1%; n = 3). In contrast, amoeba-mediated cytotoxicity was dramatically reduced when LMM30 transfectant amoebae were used instead of the wild-type or vector transfectant strains. Even after a 3-h incubation, cytolysis was evident only in 5% (±1%; n = 3) of the cell population. The viability of the three amoeba populations was also assessed, and no significant cell death was observed.
Figure 6.
Viability of Caco-2 cells in the presence of LMM transfectants. A 51Cr-labeled, postconfluent monolayer of Caco-2 cells, cultivated in 24-well plates, was infected with growing E. histolytica (1 amoeba for 5 cells) in a 1-ml vol medium and incubated for the indicated times. After incubation, medium was removed and centrifuged. The cytolysis is reflected by the 51Cr molecules liberated in the medium. Percentage of lysis corresponds to the fraction of radioactivity in the supernatant compared with the total radioactivity. Each incubation was condition tested in triplicate, and the experiment was repeated three times. ▴—▴, HM1 wild-type strain (40% ± 3% of lysis; n = 3); ▪—▪, NEO-transfected strain: (40% ± 1% of lysis; n = 3); •—•, LMM transfected strain (5% ± 1% of lysis; n = 3).
DISCUSSION
Myosin II mechanoenzyme assembles into bipolar filaments to exert contractile force on actin filaments. The LMM of the myosin heavy chain is necessary for myosin II assembly, because the deletion of LMM produces a nonfunctional myosin II molecule unable to form filaments (De Lozanne et al., 1987; Fukui et al., 1990). Purified LMM retains the solubility properties of the intact myosin II and polymerizes into paracrystalline-like aggregates under conditions of lower ionic strength. In addition, when the LMM domain is expressed in large quantities in cells, it organizes into α-helical coiled coil molecules and forms cytoplasmic inclusions. These aggregates sequester the native myosin II, resulting in cells that display a myosin II null phenotype (Burns et al., 1995). We used these properties of the LMM domain to impair the functions of myosin II in E. histolytica, a human parasite in which gene targeting is not yet feasible. To construct the dominant negative cell line, the light meromyosin domain from the heavy chain of myosin II was overexpressed. The resultant strain showed phenotypes similar to those of the D. discoideum myosin II null mutant. This is the first genetically engineered parasitic amoebae that is deficient in cytoskeleton activities.
Expression of LMM was obtained by using a multicopy plasmid that can be transfected and expressed in E. histolytica; the copy number of this plasmid is regulated by the concentration of the selecting drug G418 (Nickel and Tannich, 1994). Thus, the quantity of LMM in the transfected population is modulated by gene dosage according to the drug concentration. Mutant phenotypes were more apparent when the concentration of G418 was increased. Transfectant populations, which displayed a ratio of LMM to endogenous myosin II of 4:1, were analyzed in this study. This expression level is lower than that obtained with mutants from D. discoideum, which express 10 times more LMM than endogenous myosin II (Burns et al., 1995). Such a large excess of LMM leads to D. discoideum cells containing large aggregates of tubular structures, a morphological change that does not appear in E. histolytica overexpressing LMM. A deficiency in growth was observed in LMM transfectants, as their growth rates were slower than those of control amoebae. In addition, some of the LMM-overexpressing amoebae became multinucleated. This observation may indicate a modification in the cytokinesis process and/or a deficiency in nutrient uptake. The latter could lead to constant cell death, and, in turn, the apparent generation time would be the net effect of multiplication and decay. These possibilities need to be tested.
LMM transfectant amoebae are characterized by significant changes in their morphology. The amoebae round up and move slowly, demonstrating the need for myosin II in cell morphogenesis and movement. These results suggest, as is the case in D. discoideum, that myosin II participates in cell detachment from the substratum at the rear part of the cell, which is followed by retraction allowing the cell to continue its forward movement (Jay et al., 1995). This function may be also perturbed in E. histolytica overexpressing LMM, because the increase in expression of the truncated myosin inhibits the movement to an extent similar to that observed for other myosin II null mutants (Jay et al., 1995). In rounded, LMM-overexpressing amoebae, the endogenous myosin II is distributed beneath the cell cortex. A model has been suggested to explain morphological changes in amoeboid cells (Shelden and Knecht, 1996). In this model, a round shape may indicate that endogenous myosin II exerts a compressive force throughout the cortex of the cell to balance the hydrostatic force from the extracellular medium. Myosin II could stiffen the cell cortex by binding to actin filaments. Reduction in cortical stiffness in particular areas of the cell, through changes in actin–myosin II interactions, could allow the expansion of cell protrusions (Shelden and Knecht, 1996). In amoebae overexpressing LMM, the modification of actin–myosin II activities may inhibit reduction of stiffness, which in turn accounts for the movement deficiency of these amoebae. Indeed, we have previously shown that movement and morphological changes in E. histolytica require a redistribution of myosin II in which the protein is relocalized to the rear part of amoebae (Arhets et al., 1995). The tail contraction mechanism then allows cytoplasmic streaming to push the cell forward. During movement, amoebae overexpressing LMM are able to extend only very small pseudopods. The reduction in cytoplasmic streaming observed in these cells might account for the reduction of the size of pseudopods; it has been suggested that the intracellular pressure created by cytoplasm streaming is a motive force for cell locomotion (Yanai et al., 1996).
The process of capping of surface receptors is also inhibited in amoebae overexpressing LMM. This phenotype is particularly important with regard to pathogenicity. It has been suggested that capping and uroid formation participate in the adaptation of amoebae to the host immune response. A role for myosin II in receptor-capping activity was previously indicated (Rahim et al., 1993; Arhets et al., 1995) in E. histolytica as well as in D. discoideum (Pasternak et al., 1989) and in T lymphocytes (Campanero et al., 1994). Results reported here are in agreement with these observations. The formation of cap and uroids was dramatically diminished in LMM-overexpressing cells (LMM30). The majority of transfected trophozoites analyzed in this study appeared as resting cells insensitive to ConA activation. In these amoebae, myosin II localized around the cell cortex, and the Gal-GalNAc lectin, an amoebic receptor undergoing capping, formed patches but did not concentrate in a cap. These findings confirmed our previous work in which myosin II activity was suppressed by treatment of amoebae with drugs that inhibit kinases involved in the myosin II heavy-chain phosphorylation and, subsequently, abolish uroid and cap formation (Arhets et al., 1995). The use of amoebae overexpressing LMM allowed direct analysis of the role of myosin II in these cytoskeletal activities. The results obtained in this work reinforce the conclusion that the clustering of receptors in a patch occurs independently of the activity of myosin II, whereas movement of these patches through the uroid and cap formation is dependent on myosin II activity. Analysis of capping demonstrated that the LMM fragment was found associated with endogenous myosin II when amoebae were activated by ConA. It seems likely that, in the minority cell population that undergoes capping, the signaling resulting from Con A stimulation of surface receptors leads to the localization of both myosin II and LMM with the cytoskeleton. The mechanism by which these molecules associate and are stabilized in the cytoskeleton is unknown. One plausible hypothesis may be that myosin II interaction with actin filaments stabilizes heterologous structures formed by myosin II and LMM. For instance, in D. discoideum actin is known to accelerate assembly of myosin filaments (Mahajan et al., 1989). It is also possible that other components of the cytoskeleton bind to and regulate myosin II filament formation.
In conclusion, the phenotypic analysis of an E. histolytica mutant overexpressing LMM has highlighted a critical role for myosin II in capping and uroid formation and in movement. Furthermore, the consequences of the cytoskeletal modifications on E. histolytica pathogenicity were assessed by analysis of amoebic cytotoxicity on Caco-2 cells. Infection of the Caco-2 cell line has been proposed as a model to study initial pathogenic effect of E. histolytica on intestinal epithelial cells (Rigothier et al., 1991; Li et al., 1994). In this study, postconfluent monolayers were used; under such culturing conditions Caco-2 cells are polarized, form a well-defined brush border, and express specific intestinal enzymes (Pinto et al., 1983). Addition of wild-type or vector alone–transfected amoebae to the Caco-2 cells leads to massive monolayer destruction. We observed that 40% of cells were lysed in the first 3 h of interaction with amoebae. The changes in morphology of the Caco-2 cells during the amoebic invasive process essentially correspond to blebbing and cell lysis. In addition, some cells detach without lysis (5% of the population). These morphological observations are indicative of the damage that amoeba exert on the cell integrity and on the functionality of tight junctions. In contrast, analysis of the LMM30 amoebae, in which cytoskeletal functions are impaired, displayed a less toxic phenotype. Only 5% of Caco-2 cells were lysed after contact with LMM30 amoebae for 3 h, and the integrity of the Caco-2 monolayer was similar to that of untreated cells, indicating that LMM30 amoebae do not induce an invasive process. These results indicate that modification of cytoskeletal functions by overexpression of LMM leads to a dramatic decrease in amoeba virulence. Two different mechanisms have been proposed to explain E. histolytica cytotoxicity against tissue culture cells (Li et al., 1994): 1) a cell contact-mediated cytolytic mechanism and 2) secreted virulence factors such as proteases that induce cell detachment without lysis. Results in support of the former mechanism indicate that the introduction of protease inhibitors during the interaction of E. histolytica and Caco-2 cells does not prevent cytotoxicity (Li et al., 1994). Our results suggest that, as a consequence of functional modifications of the amoebic cytoskeleton, LMM30 amoebae and Caco-2 cells do not establish functional cytolytic contacts. This observation may be due to either changes in the efficacy of amoebic adhesion properties and/or inhibition of transport and translocation of amoebic toxins as amoebapores through the cell membrane (Leippe et al., 1991). The myosin II-defective strain described here provides a novel tool to study the influence of the cytoskeletal activities on the pathogenicity of E. histolytica in animal models. According to our previous studies, inhibition of capping may result in parasites that are more susceptible to the host immune system. Thus, this genetically engineered cell line may allow the discovery of new insights of the anti-amoeba human immune response.
ACKNOWLEDGMENTS
We thank E. Tannich for kindly sending the ExEhNEO vector, W. Petri for providing us with Gal-GalNAc antibody, T. Kreis for the anti-VSV-G antibody, F. Briquet-Laugier for help in analyzing the video sequence data, R. Hellio for constant help in confocal analysis, and M. Rathman for critical reading of the manuscript. This work was supported by grants from the French Ministère de l’Education Nationale, de l’Enseignement Supérieur, de la Recherche et de l’Insertion Professionnelle, from the Nord–Sud Institut National de la Santé et de la Recherche Médicale program (grant 2475NS3), and from the Direction de la Recherche et des Techniques du Ministère de la Défense (grant 94/092). P.A. is the recipient of a fellowship from the Fondation Louis Jeantet de Medecine.
REFERENCES
- Arhets P, Gounon P, Sansonetti P, Guillén N. Myosin II is involved in capping and uroid formation in the human pathogen Entamoeba histolytica. Infect Immun. 1995;63:4358–4367. doi: 10.1128/iai.63.11.4358-4367.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga LL, Ninomiya H, McCoy J, Eacker S, Wiedmer T, Pham C, Wood S, Sims PJ, Petri WA. Inhibition of the complement membrane attack complex by the galactose-specific adhesin of Entamoeba histolytica. J Clin Invest. 1992;90:1131–1137. doi: 10.1172/JCI115931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CG, Reedy M, Heuser J, De Lozanne A. Expression of light meromyosin in Dictyosteliumblocks normal myosin II function. J Cell Biol. 1995;130:605–612. doi: 10.1083/jcb.130.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero MR, Sanchez-Mateos P, del Pozo M, Sanchez-Madrid F. ICAM-3 regulates lymphocyte morphology and integrin-mediated T cell interaction with endothelial cell and extracellular matrix ligands. J Cell Biol. 1994;127:867–878. doi: 10.1083/jcb.127.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A, Berlot CH, Leinwand LA, Spudich JA. Expression in Escherichia coli of a functional Dictyosteliummyosin tail fragment. J Cell Biol. 1987;105:2999–3005. doi: 10.1083/jcb.105.6.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyosteliummyosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Diamond LS. Axenic cultivation of Entamoeba histolytica. Science. 1961;134:336–337. doi: 10.1126/science.134.3475.336. [DOI] [PubMed] [Google Scholar]
- Egelhoff TT, Spudich JA. Molecular genetics of cell migration: Dictyosteliumas a model system. Trends Genet. 1991;7:161–166. doi: 10.1016/0168-9525(91)90380-9. [DOI] [PubMed] [Google Scholar]
- Espinosa-Cantellano M, Martinez-Palomo A. Entamoeba histolytica: Mechanism of Surface Receptor Capping. Exp Parasitol. 1994;79:424–435. doi: 10.1006/expr.1994.1104. [DOI] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyosteliummyosin-defective mutant. J Cell Biol. 1990;110:367–387. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann L, Nickel R, Tannich E. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci USA. 1995;92:8975–8979. doi: 10.1073/pnas.92.19.8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay PY, Pham PA, Wong SA, Elson EL. A mechanical function of myosin II in cell motility. J Cell Sci. 1995;108:387–393. doi: 10.1242/jcs.108.1.387. [DOI] [PubMed] [Google Scholar]
- Knecht DA, Loomis WF. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987;236:1081–1086. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leippe M, Ebel S, Schoenberger OL, Horstmann RD, Müller-Eberhard HJ. Pore-forming peptide of pathogenic Entamoeba histolytica. Proc Natl Acad Sci USA. 1991;88:7659–7663. doi: 10.1073/pnas.88.17.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Stenson W, Kunz-Jenkins C, Swanson PE, Duncan R, Stanley S. Entamoeba histolyticainteractions with polarized human intestinal Caco-2 epithelial cells. Infect Immun. 1994;62:5112–5119. doi: 10.1128/iai.62.11.5112-5119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan RK, Vaughn KT, Johns JA, Pardee JD. Actin filaments mediate Dictyosteliummyosin assembly in vitro. Proc Natl Acad Sci USA. 1989;86:6161–6165. doi: 10.1073/pnas.86.16.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel R, Tannich E. Transfection and transient expression of chloramphenicol acetyltransferase gene in the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci USA. 1994;91:7095–7098. doi: 10.1073/pnas.91.15.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak P, Spudich JA, Elson EL. Capping of surface receptors and concomitant cortical tension are generated by conventional myosin. Nature. 1989;341:549–551. doi: 10.1038/341549a0. [DOI] [PubMed] [Google Scholar]
- Pinto M, Robine-Leon S, Appay MD, Kedinger M, Triadou E, Dussaulx B, Lacroix B, Simon-Assman P, Haffen K, Fogh J, Zweibaum A. Enterocyte-like differentiation and polarization of the human carcinoma cell line Caco-2 in culture. Biol Cell. 1983;47:323–330. [Google Scholar]
- Rahim Z, Raymond-Denise A, Sansonetti P, Guillén N. Localization of myosin heavy chain A in the human pathogen Entamoeba histolytica. Infect Immun. 1993;61:1048–1054. doi: 10.1128/iai.61.3.1048-1054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravdin JI, Guerrant RL. Role of adherence in cytopathogenic mechanisms of Entamoeba histolytica.Study with mammalian tissue culture cells and human erythrocytes. J Clin Invest. 1981;68:1305–1313. doi: 10.1172/JCI110377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond-Denise A, Sansonetti P, Guillén N. Identification and characterization of a myosin heavy chain gene (mhcA) from the human parasitic pathogen Entamoeba histolytica. Mol Biochem Parasitol. 1993;59:123–132. doi: 10.1016/0166-6851(93)90013-n. [DOI] [PubMed] [Google Scholar]
- Rigothier MC, Coconnier MH, Servin AL, Gayral P. A new in vitro model of Entamoeba histolyticaadhesion, using the human colon carcinoma cell line Caco-2; scanning electron microscopic study. Infect Immun. 1991;59:4142–4146. doi: 10.1128/iai.59.11.4142-4146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR, Goodson HV. Myosin Prot. Profile. 1995;2:1323–1339. [PubMed] [Google Scholar]
- Shelden E, Knecht DA. Dictyostelium cell shape generation requires myosin II. Cell Motil Cytoskeleton. 1996;35:59–67. doi: 10.1002/(SICI)1097-0169(1996)35:1<59::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Trissl D, Martinez-Palomo A, Argüello C, de la Torre M, de la Hoz R. Surface properties related to concanavalin A-induced agglutination: a comparative study of several Entamoebastrains. J Exp Med. 1977;145:652–665. doi: 10.1084/jem.145.3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick HM, Spudich JA. Myosin structure and function in cell motility. Am Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- Yanai M, Kenyon CM, Butler JP, Macklen PT, Kelly SM. Intracellular pressure is a motive force for cell motion in Amoebae proteus. Cell Motil Cytoskeleton. 1996;33:22–29. doi: 10.1002/(SICI)1097-0169(1996)33:1<22::AID-CM3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]