Abstract
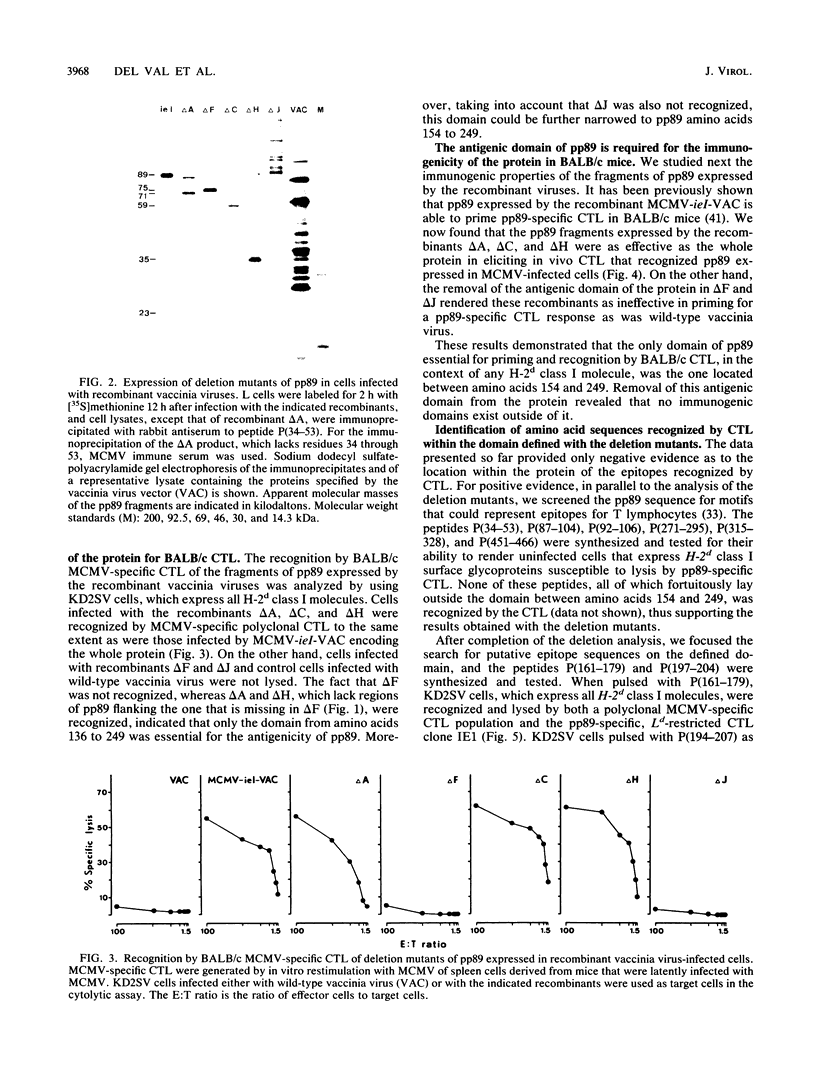
The murine cytomegalovirus protein pp89, which is encoded by gene ieI, is a nonstructural regulatory protein expressed in the immediate-early phase of the viral replication cycle and located mainly in the nucleus of infected cells. Protection of BALB/c (H-2d) mice against a lethal murine cytomegalovirus challenge infection is achieved by vaccination with a recombinant vaccinia virus, MCMV-ieI-VAC, expressing pp89 as the only murine cytomegalovirus gene product. The protection is entirely mediated by T lymphocytes of the CD8+ subset. In the present report, we analyzed the molecular basis of the recognition of pp89 by BALB/c CD8+ cytolytic T lymphocytes. A series of internal and terminal deletion mutants of gene ieI was constructed and cloned in vaccinia virus, and the antigenicity and immunogenicity of the fragments of pp89 expressed by the recombinants were studied. A region of only one-sixth of the protein, from amino acids 154 to 249 and encoded by the fourth exon of gene ieI, was sufficient for both the recognition in vitro of the protein by pp89-specific cytotoxic T lymphocytes and the induction in vivo of pp89-specific cytotoxic T lymphocytes. By using synthetic peptides, the sequence between residues 161 and 179, which is located within the defined domain, was identified as an epitope presented to BALB/C cytotoxic T lymphocytes by the class I major histocompatibility antigen Ld.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avrameas S., Ternynck T. The cross-linking of proteins with glutaraldehyde and its use for the preparation of immunoadsorbents. Immunochemistry. 1969 Jan;6(1):53–66. doi: 10.1016/0019-2791(69)90178-5. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Borysiewicz L. K., Graham S., Hickling J. K., Mason P. D., Sissons J. G. Human cytomegalovirus-specific cytotoxic T cells: their precursor frequency and stage specificity. Eur J Immunol. 1988 Feb;18(2):269–275. doi: 10.1002/eji.1830180214. [DOI] [PubMed] [Google Scholar]
- Briand J. P., Muller S., Van Regenmortel M. H. Synthetic peptides as antigens: pitfalls of conjugation methods. J Immunol Methods. 1985 Apr 8;78(1):59–69. doi: 10.1016/0022-1759(85)90329-1. [DOI] [PubMed] [Google Scholar]
- Cochet M., Kast W. M., Kummer A. M., Transy C., Melief C. J., Kourilsky P. Alternative splicing in the mouse H-2Kd gene is not necessary for the classical Kd antigen function. Immunogenetics. 1986;24(4):267–274. doi: 10.1007/BF00364531. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebeling A., Keil G. M., Knust E., Koszinowski U. H. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J Virol. 1983 Sep;47(3):421–433. doi: 10.1128/jvi.47.3.421-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon G., Shastri N., Cogswell J., Wilbur S., Sadegh-Nasseri S., Krzych U., Miller A., Sercarz E. The choice of T-cell epitopes utilized on a protein antigen depends on multiple factors distant from, as well as at the determinant site. Immunol Rev. 1987 Aug;98:53–73. doi: 10.1111/j.1600-065x.1987.tb00519.x. [DOI] [PubMed] [Google Scholar]
- Guillet J. G., Lai M. Z., Briner T. J., Smith J. A., Gefter M. L. Interaction of peptide antigens and class II major histocompatibility complex antigens. Nature. 1986 Nov 20;324(6094):260–262. doi: 10.1038/324260a0. [DOI] [PubMed] [Google Scholar]
- Jonjić S., del Val M., Keil G. M., Reddehase M. J., Koszinowski U. H. A nonstructural viral protein expressed by a recombinant vaccinia virus protects against lethal cytomegalovirus infection. J Virol. 1988 May;62(5):1653–1658. doi: 10.1128/jvi.62.5.1653-1658.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil G. M., Ebeling-Keil A., Koszinowski U. H. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987 Feb;61(2):526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil G. M., Ebeling-Keil A., Koszinowski U. H. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987 Jun;61(6):1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil G. M., Ebeling-Keil A., Koszinowski U. H. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J Virol. 1984 Jun;50(3):784–795. doi: 10.1128/jvi.50.3.784-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil G. M., Fibi M. R., Koszinowski U. H. Characterization of the major immediate-early polypeptides encoded by murine cytomegalovirus. J Virol. 1985 May;54(2):422–428. doi: 10.1128/jvi.54.2.422-428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszinowski U. H., Keil G. M., Schwarz H., Schickedanz J., Reddehase M. J. A nonstructural polypeptide encoded by immediate-early transcription unit 1 of murine cytomegalovirus is recognized by cytolytic T lymphocytes. J Exp Med. 1987 Jul 1;166(1):289–294. doi: 10.1084/jem.166.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszinowski U. H., Keil G. M., Volkmer H., Fibi M. R., Ebeling-Keil A., Münch K. The 89,000-Mr murine cytomegalovirus immediate-early protein activates gene transcription. J Virol. 1986 Apr;58(1):59–66. doi: 10.1128/jvi.58.1.59-66.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszinowski U. H., Reddehase M. J., Keil G. M., Schickedanz J. Host immune response to cytomegalovirus: products of transfected viral immediate-early genes are recognized by cloned cytolytic T lymphocytes. J Virol. 1987 Jun;61(6):2054–2058. doi: 10.1128/jvi.61.6.2054-2058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. Z., Ross D. T., Guillet J. G., Briner T. J., Gefter M. L., Smith J. A. T lymphocyte response to bacteriophage lambda repressor cI protein. Recognition of the same peptide presented by Ia molecules of different haplotypes. J Immunol. 1987 Dec 15;139(12):3973–3980. [PubMed] [Google Scholar]
- Lemke H., Hämmerling G. J., Hämmerling U. Fine specificity analysis with monoclonal antibodies of antigens controlled by the major histocompatibility complex and by the Qa/TL region in mice. Immunol Rev. 1979;47:175–206. doi: 10.1111/j.1600-065x.1979.tb00293.x. [DOI] [PubMed] [Google Scholar]
- Margalit H., Spouge J. L., Cornette J. L., Cease K. B., Delisi C., Berzofsky J. A. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J Immunol. 1987 Apr 1;138(7):2213–2229. [PubMed] [Google Scholar]
- Meyers J. D. Cytomegalovirus infection following marrow transplantation: risk, treatment, and prevention. Birth Defects Orig Artic Ser. 1984;20(1):101–117. [PubMed] [Google Scholar]
- Meyers J. D., Flournoy N., Thomas E. D. Cytomegalovirus infection and specific cell-mediated immunity after marrow transplant. J Infect Dis. 1980 Dec;142(6):816–824. doi: 10.1093/infdis/142.6.816. [DOI] [PubMed] [Google Scholar]
- Moskowitz L., Hensley G. T., Chan J. C., Adams K. Immediate causes of death in acquired immunodeficiency syndrome. Arch Pathol Lab Med. 1985 Aug;109(8):735–738. [PubMed] [Google Scholar]
- Osborn J. E. Cytomegalovirus: pathogenicity, immunity, and vaccine initiatives. J Infect Dis. 1981 Apr;143(4):618–630. doi: 10.1093/infdis/143.4.618. [DOI] [PubMed] [Google Scholar]
- Quinnan G. V., Jr, Kirmani N., Rook A. H., Manischewitz J. F., Jackson L., Moreschi G., Santos G. W., Saral R., Burns W. H. Cytotoxic t cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N Engl J Med. 1982 Jul 1;307(1):7–13. doi: 10.1056/NEJM198207013070102. [DOI] [PubMed] [Google Scholar]
- Reddehase M. J., Bühring H. J., Koszinowski U. H. Cloned long-term cytolytic T-lymphocyte line with specificity for an immediate-early membrane antigen of murine cytomegalovirus. J Virol. 1986 Jan;57(1):408–412. doi: 10.1128/jvi.57.1.408-412.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase M. J., Keil G. M., Koszinowski U. H. The cytolytic T lymphocyte response to the murine cytomegalovirus. II. Detection of virus replication stage-specific antigens by separate populations of in vivo active cytolytic T lymphocyte precursors. Eur J Immunol. 1984 Jan;14(1):56–61. doi: 10.1002/eji.1830140111. [DOI] [PubMed] [Google Scholar]
- Reddehase M. J., Koszinowski U. H. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature. 1984 Nov 22;312(5992):369–371. doi: 10.1038/312369a0. [DOI] [PubMed] [Google Scholar]
- Reddehase M. J., Mutter W., Münch K., Bühring H. J., Koszinowski U. H. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J Virol. 1987 Oct;61(10):3102–3108. doi: 10.1128/jvi.61.10.3102-3108.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase M. J., Weiland F., Münch K., Jonjic S., Lüske A., Koszinowski U. H. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J Virol. 1985 Aug;55(2):264–273. doi: 10.1128/jvi.55.2.264-273.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase M. J., Zawatzky R., Weiland F., Bühring H. J., Mutter W., Koszinowski U. H. Stable expression of clonal specificity in murine cytomegalovirus-specific large granular lymphoblast lines propagated long-term in recombinant interleukin 2. Immunobiology. 1987 Aug;174(4-5):420–431. doi: 10.1016/s0171-2985(87)80015-3. [DOI] [PubMed] [Google Scholar]
- Rothbard J. B., Lechler R. I., Howland K., Bal V., Eckels D. D., Sekaly R., Long E. O., Taylor W. R., Lamb J. R. Structural model of HLA-DR1 restricted T cell antigen recognition. Cell. 1988 Feb 26;52(4):515–523. doi: 10.1016/0092-8674(88)90464-3. [DOI] [PubMed] [Google Scholar]
- Rothbard J. B., Taylor W. R. A sequence pattern common to T cell epitopes. EMBO J. 1988 Jan;7(1):93–100. doi: 10.1002/j.1460-2075.1988.tb02787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. H. T-lymphocyte recognition of antigen in association with gene products of the major histocompatibility complex. Annu Rev Immunol. 1985;3:237–261. doi: 10.1146/annurev.iy.03.040185.001321. [DOI] [PubMed] [Google Scholar]
- Shastri N., Oki A., Miller A., Sercarz E. E. Distinct recognition phenotypes exist for T cell clones specific for small peptide regions of proteins. Implications for the mechanisms underlying major histocompatibility complex-restricted antigen recognition and clonal deletion models of immune response gene defects. J Exp Med. 1985 Jul 1;162(1):332–345. doi: 10.1084/jem.162.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimonkevitz R., Kappler J., Marrack P., Grey H. Antigen recognition by H-2-restricted T cells. I. Cell-free antigen processing. J Exp Med. 1983 Aug 1;158(2):303–316. doi: 10.1084/jem.158.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Tevethia M. J., Kalderon D., Smith A. E., Tevethia S. S. Clustering of antigenic sites recognized by cytotoxic T lymphocyte clones in the amino terminal half of SV40 T antigen. Virology. 1988 Feb;162(2):427–436. doi: 10.1016/0042-6822(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Taylor P. M., Davey J., Howland K., Rothbard J. B., Askonas B. A. Class I MHC molecules rather than other mouse genes dictate influenza epitope recognition by cytotoxic T cells. Immunogenetics. 1987;26(4-5):267–272. doi: 10.1007/BF00346521. [DOI] [PubMed] [Google Scholar]
- Townsend A. R., Gotch F. M., Davey J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell. 1985 Sep;42(2):457–467. doi: 10.1016/0092-8674(85)90103-5. [DOI] [PubMed] [Google Scholar]
- Townsend A. R., Rothbard J., Gotch F. M., Bahadur G., Wraith D., McMichael A. J. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell. 1986 Mar 28;44(6):959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- Volkmer H., Bertholet C., Jonjić S., Wittek R., Koszinowski U. H. Cytolytic T lymphocyte recognition of the murine cytomegalovirus nonstructural immediate-early protein pp89 expressed by recombinant vaccinia virus. J Exp Med. 1987 Sep 1;166(3):668–677. doi: 10.1084/jem.166.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari E., Dietzschold B., Szokan G., Heber-Katz E. A synthetic peptide induces long-term protection from lethal infection with herpes simplex virus 2. J Exp Med. 1987 Feb 1;165(2):459–470. doi: 10.1084/jem.165.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Gebhard J. R., Lewicki H., Tishon A., Oldstone M. B. Molecular definition of a major cytotoxic T-lymphocyte epitope in the glycoprotein of lymphocytic choriomeningitis virus. J Virol. 1988 Mar;62(3):687–695. doi: 10.1128/jvi.62.3.687-695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]



