Abstract
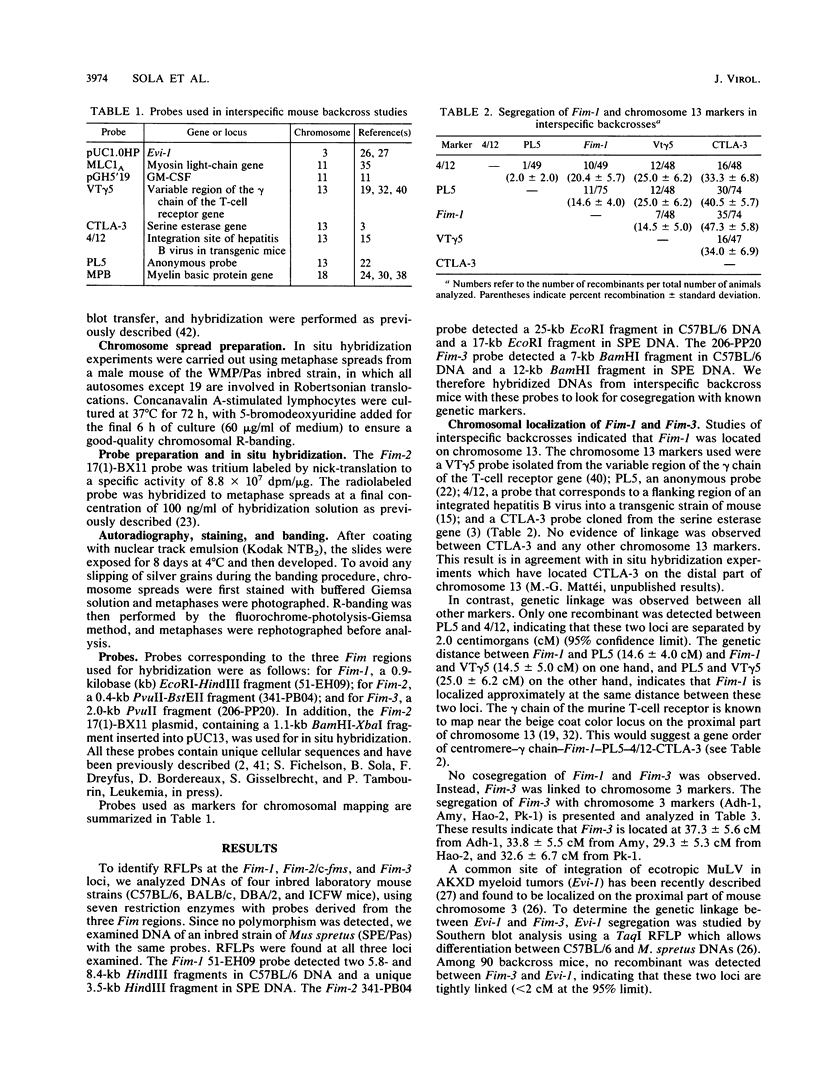
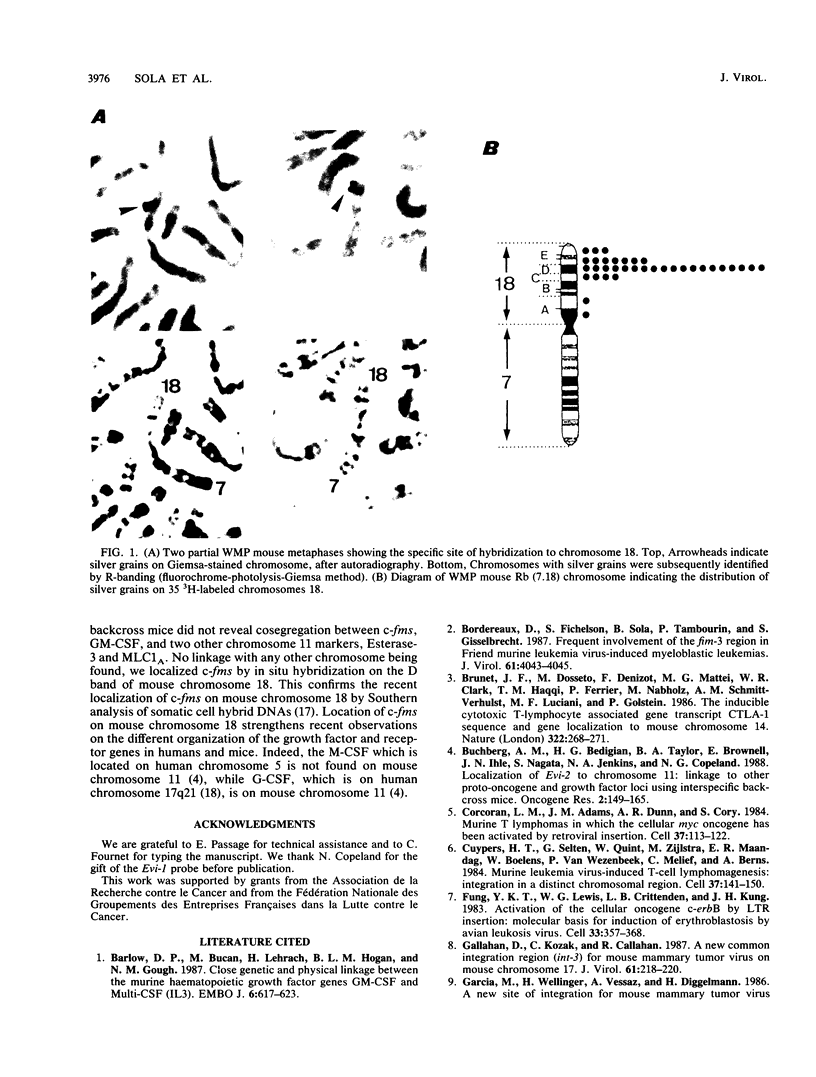
Three common proviral integration sites, Fim-1, Fim-2/c-fms, and Fim-3, have been described in mouse myeloid leukemias induced by the Friend murine leukemia virus. The nature and function of Fim-1 and Fim-3 are still unknown since no transcript from these loci has been detected so far. To identify these two loci, we undertook their chromosomal localization using restriction fragment length polymorphism detected between C57BL/6 mice and the wild-derived inbred strain of Mus spretus. Using interspecific backcross analysis, we mapped Fim-1 to mouse chromosome 13 and Fim-3 to mouse chromosome 3. Interestingly, Fim-3 is tightly linked to Evi-1, another common integration site of ecotropic virus involved in another model of mouse myeloid leukemogenesis. Fim-2 spans the 5' end of the c-fms gene, which encodes for the macrophage-colony-stimulating factor receptor. We located the c-fms gene on the D band of chromosome 18 by in situ hybridization.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barlow D. P., Bućan M., Lehrach H., Hogan B. L., Gough N. M. Close genetic and physical linkage between the murine haemopoietic growth factor genes GM-CSF and Multi-CSF (IL3). EMBO J. 1987 Mar;6(3):617–623. doi: 10.1002/j.1460-2075.1987.tb04799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordereaux D., Fichelson S., Sola B., Tambourin P. E., Gisselbrecht S. Frequent involvement of the fim-3 region in Friend murine leukemia virus-induced mouse myeloblastic leukemias. J Virol. 1987 Dec;61(12):4043–4045. doi: 10.1128/jvi.61.12.4043-4045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet J. F., Dosseto M., Denizot F., Mattei M. G., Clark W. R., Haqqi T. M., Ferrier P., Nabholz M., Schmitt-Verhulst A. M., Luciani M. F. The inducible cytotoxic T-lymphocyte-associated gene transcript CTLA-1 sequence and gene localization to mouse chromosome 14. Nature. 1986 Jul 17;322(6076):268–271. doi: 10.1038/322268a0. [DOI] [PubMed] [Google Scholar]
- Buchberg A. M., Bedigian H. G., Taylor B. A., Brownell E., Ihle J. N., Nagata S., Jenkins N. A., Copeland N. G. Localization of Evi-2 to chromosome 11: linkage to other proto-oncogene and growth factor loci using interspecific backcross mice. Oncogene Res. 1988;2(2):149–165. [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Fung Y. K., Lewis W. G., Crittenden L. B., Kung H. J. Activation of the cellular oncogene c-erbB by LTR insertion: molecular basis for induction of erythroblastosis by avian leukosis virus. Cell. 1983 Jun;33(2):357–368. doi: 10.1016/0092-8674(83)90417-8. [DOI] [PubMed] [Google Scholar]
- Gallahan D., Kozak C., Callahan R. A new common integration region (int-3) for mouse mammary tumor virus on mouse chromosome 17. J Virol. 1987 Jan;61(1):218–220. doi: 10.1128/jvi.61.1.218-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., Wellinger R., Vessaz A., Diggelmann H. A new site of integration for mouse mammary tumor virus proviral DNA common to BALB/cf(C3H) mammary and kidney adenocarcinomas. EMBO J. 1986 Jan;5(1):127–134. doi: 10.1002/j.1460-2075.1986.tb04186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselbrecht S., Fichelson S., Sola B., Bordereaux D., Hampe A., André C., Galibert F., Tambourin P. Frequent c-fms activation by proviral insertion in mouse myeloblastic leukaemias. Nature. 1987 Sep 17;329(6136):259–261. doi: 10.1038/329259a0. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Gough J., Metcalf D., Kelso A., Grail D., Nicola N. A., Burgess A. W., Dunn A. R. Molecular cloning of cDNA encoding a murine haematopoietic growth regulator, granulocyte-macrophage colony stimulating factor. 1984 Jun 28-Jul 4Nature. 309(5971):763–767. doi: 10.1038/309763a0. [DOI] [PubMed] [Google Scholar]
- Graham M., Adams J. M., Cory S. Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. 1985 Apr 25-May 1Nature. 314(6013):740–743. doi: 10.1038/314740a0. [DOI] [PubMed] [Google Scholar]
- Groffen J., Heisterkamp N., Spurr N., Dana S., Wasmuth J. J., Stephenson J. R. Chromosomal localization of the human c-fms oncogene. Nucleic Acids Res. 1983 Sep 24;11(18):6331–6339. doi: 10.1093/nar/11.18.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénet J. L. The contribution of wild derived mouse inbred strains to gene mapping methodology. Curr Top Microbiol Immunol. 1986;127:109–113. doi: 10.1007/978-3-642-71304-0_13. [DOI] [PubMed] [Google Scholar]
- Hadchouel M., Farza H., Simon D., Tiollais P., Pourcel C. Maternal inhibition of hepatitis B surface antigen gene expression in transgenic mice correlates with de novo methylation. Nature. 1987 Oct 1;329(6138):454–456. doi: 10.1038/329454a0. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Hoggan M. D., Halden N. F., Buckler C. E., Kozak C. A. Genetic mapping of the mouse c-fms proto-oncogene to chromosome 18. J Virol. 1988 Mar;62(3):1055–1056. doi: 10.1128/jvi.62.3.1055-1056.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda N., Fukushige S., Murotsu T., Yoshida M. C., Tsuchiya M., Asano S., Kaziro Y., Nagata S. Human gene coding for granulocyte-colony stimulating factor is assigned to the q21-q22 region of chromosome 17. Somat Cell Mol Genet. 1987 Nov;13(6):679–684. doi: 10.1007/BF01534488. [DOI] [PubMed] [Google Scholar]
- Kranz D. M., Saito H., Disteche C. M., Swisshelm K., Pravtcheva D., Ruddle F. H., Eisen H. N., Tonegawa S. Chromosomal locations of the murine T-cell receptor alpha-chain gene and the T-cell gamma gene. Science. 1985 Feb 22;227(4689):941–945. doi: 10.1126/science.3918347. [DOI] [PubMed] [Google Scholar]
- Le Beau M. M., Westbrook C. A., Diaz M. O., Larson R. A., Rowley J. D., Gasson J. C., Golde D. W., Sherr C. J. Evidence for the involvement of GM-CSF and FMS in the deletion (5q) in myeloid disorders. Science. 1986 Feb 28;231(4741):984–987. doi: 10.1126/science.3484837. [DOI] [PubMed] [Google Scholar]
- Lemay G., Jolicoeur P. Rearrangement of a DNA sequence homologous to a cell-virus junction fragment in several Moloney murine leukemia virus-induced rat thymomas. Proc Natl Acad Sci U S A. 1984 Jan;81(1):38–42. doi: 10.1073/pnas.81.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy P., Seboun E., Mattei M. G., Fellous M., Bishop C. E. Testis-specific transcripts detected by a human Y-DNA-derived probe. Development. 1987;101 (Suppl):177–183. doi: 10.1242/dev.101.Supplement.177. [DOI] [PubMed] [Google Scholar]
- Mattei M. G., Philip N., Passage E., Moisan J. P., Mandel J. L., Mattei J. F. DNA probe localization at 18p113 band by in situ hybridization and identification of a small supernumerary chromosome. Hum Genet. 1985;69(3):268–271. doi: 10.1007/BF00293038. [DOI] [PubMed] [Google Scholar]
- Molineaux S. M., Engh H., de Ferra F., Hudson L., Lazzarini R. A. Recombination within the myelin basic protein gene created the dysmyelinating shiverer mouse mutation. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7542–7546. doi: 10.1073/pnas.83.19.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Gachelin F., Tavitian A., Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988 Jan 21;331(6153):277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- Mucenski M. L., Taylor B. A., Copeland N. G., Jenkins N. A. Chromosomal location of Evi-1, a common site of ecotropic viral integration in AKXD murine myeloid tumors. Oncogene Res. 1988 Feb;2(3):219–233. [PubMed] [Google Scholar]
- Mucenski M. L., Taylor B. A., Ihle J. N., Hartley J. W., Morse H. C., 3rd, Jenkins N. A., Copeland N. G. Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol Cell Biol. 1988 Jan;8(1):301–308. doi: 10.1128/mcb.8.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushinski J. F., Potter M., Bauer S. R., Reddy E. P. DNA rearrangement and altered RNA expression of the c-myb oncogene in mouse plasmacytoid lymphosarcomas. Science. 1983 May 20;220(4599):795–798. doi: 10.1126/science.6687762. [DOI] [PubMed] [Google Scholar]
- Neil J. C., Hughes D., McFarlane R., Wilkie N. M., Onions D. E., Lees G., Jarrett O. Transduction and rearrangement of the myc gene by feline leukaemia virus in naturally occurring T-cell leukaemias. 1984 Apr 26-May 2Nature. 308(5962):814–820. doi: 10.1038/308814a0. [DOI] [PubMed] [Google Scholar]
- Newman S., Kitamura K., Campagnoni A. T. Identification of a cDNA coding for a fifth form of myelin basic protein in mouse. Proc Natl Acad Sci U S A. 1987 Feb;84(3):886–890. doi: 10.1073/pnas.84.3.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Owen F. L., Taylor B. A., Zweidler A., Seidman J. G. The murine gamma-chain of the T cell receptor is closely linked to a spermatocyte specific histone gene and the beige coat color locus on chromosome 13. J Immunol. 1986 Aug 1;137(3):1044–1046. [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Pettenati M. J., Le Beau M. M., Lemons R. S., Shima E. A., Kawasaki E. S., Larson R. A., Sherr C. J., Diaz M. O., Rowley J. D. Assignment of CSF-1 to 5q33.1: evidence for clustering of genes regulating hematopoiesis and for their involvement in the deletion of the long arm of chromosome 5 in myeloid disorders. Proc Natl Acad Sci U S A. 1987 May;84(9):2970–2974. doi: 10.1073/pnas.84.9.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert B., Barton P., Minty A., Daubas P., Weydert A., Bonhomme F., Catalan J., Chazottes D., Guénet J. L., Buckingham M. Investigation of genetic linkage between myosin and actin genes using an interspecific mouse back-cross. Nature. 1985 Mar 14;314(6007):181–183. doi: 10.1038/314181a0. [DOI] [PubMed] [Google Scholar]
- Roussel M. F., Sherr C. J., Barker P. E., Ruddle F. H. Molecular cloning of the c-fms locus and its assignment to human chromosome 5. J Virol. 1983 Dec;48(3):770–773. doi: 10.1128/jvi.48.3.770-773.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Ong G. L., Wolff L. Moloney murine leukemia virus-induced myeloid tumors in adult BALB/c mice: requirement of c-myb activation but lack of v-abl involvement. J Virol. 1987 Dec;61(12):3721–3725. doi: 10.1128/jvi.61.12.3721-3725.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman R. L., Conover C. S., Carson J. H. Shiverer gene maps near the distal end of chromosome 18 in the house mouse. Cytogenet Cell Genet. 1985;39(4):241–245. doi: 10.1159/000132151. [DOI] [PubMed] [Google Scholar]
- Silver J., Kozak C. Common proviral integration region on mouse chromosome 7 in lymphomas and myelogenous leukemias induced by Friend murine leukemia virus. J Virol. 1986 Feb;57(2):526–533. doi: 10.1128/jvi.57.2.526-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snodgrass H. R., Dembić Z., Steinmetz M., von Boehmer H. Expression of T-cell antigen receptor genes during fetal development in the thymus. Nature. 1985 May 16;315(6016):232–233. doi: 10.1038/315232a0. [DOI] [PubMed] [Google Scholar]
- Sola B., Fichelson S., Bordereaux D., Tambourin P. E., Gisselbrecht S. fim-1 and fim-2: two new integration regions of Friend murine leukemia virus in myeloblastic leukemias. J Virol. 1986 Nov;60(2):718–725. doi: 10.1128/jvi.60.2.718-725.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Lohse M. A., Szpirer C., Szpirer J., Levan G. Cellular DNA regions involved in the induction of rat thymic lymphomas (Mlvi-1, Mlvi-2, Mlvi-3, and c-myc) represent independent loci as determined by their chromosomal map location in the rat. J Virol. 1985 Dec;56(3):938–942. doi: 10.1128/jvi.56.3.938-942.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Hu L. F. A common region for proviral DNA integration in MoMuLV-induced rat thymic lymphomas. 1983 Mar 31-Apr 6Nature. 302(5907):445–449. doi: 10.1038/302445a0. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Lohse M. A. Concerted DNA rearrangements in Moloney murine leukemia virus-induced thymomas: a potential synergistic relationship in oncogenesis. J Virol. 1985 Oct;56(1):258–267. doi: 10.1128/jvi.56.1.258-267.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Kozak C., Robinson H. L. Dsi-1, a region with frequent proviral insertions in Moloney murine leukemia virus-induced rat thymomas. J Virol. 1987 Apr;61(4):1164–1170. doi: 10.1128/jvi.61.4.1164-1170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydert A., Daubas P., Caravatti M., Minty A., Bugaisky G., Cohen A., Robert B., Buckingham M. Sequential accumulation of mRNAs encoding different myosin heavy chain isoforms during skeletal muscle development in vivo detected with a recombinant plasmid identified as coding for an adult fast myosin heavy chain from mouse skeletal muscle. J Biol Chem. 1983 Nov 25;258(22):13867–13874. [PubMed] [Google Scholar]



