Abstract
Activated GTP-bound Rab proteins are thought to interact with effectors to elicit vesicle targeting and fusion events. Vesicle-associated v-SNARE and target membrane t-SNARE proteins are also involved in vesicular transport. Little is known about the functional relationship between Rabs and SNARE protein complexes. We have constructed an activated allele of VPS21, a yeast Rab protein involved in vacuolar protein sorting, and demonstrated an allele-specific interaction between Vps21p and Vac1p. Vac1p was found to bind the Sec1p homologue Vps45p. Although no association between Vps21p and Vps45p was seen, a genetic interaction between VPS21 and VPS45 was observed. Vac1p contains a zinc-binding FYVE finger that may bind phosphatidylinositol 3-phosphate [PtdIns(3)P]. In other FYVE domain proteins, this motif and PtdIns(3)P are necessary for membrane association. Vac1 proteins with mutant FYVE fingers still associated with membranes but showed vacuolar protein sorting defects and reduced interactions with Vps45p and activated Vps21p. Vac1p membrane association was not dependent on PtdIns(3)P, Pep12p, Vps21p, Vps45p, or the PtdIns 3-kinase, Vps34p. Vac1p FYVE finger mutant missorting phenotypes were suppressed by a defective allele of VPS34. These data indicate that PtdIns(3)P may perform a regulatory role, possibly involved in mediating Vac1p protein–protein interactions. We propose that activated-Vps21p interacts with its effector, Vac1p, which interacts with Vps45p to regulate the Golgi to endosome SNARE complex.
INTRODUCTION
Many vesicle-mediated protein transport events are mediated by functionally conserved sets of proteins that direct the formation of cargo-containing vesicles at donor compartments (Pelham, 1996; Schekman and Orci, 1996; Söllner and Rothman, 1996; Schmid, 1997) or the targeting and fusion of these vesicles with the proper acceptor compartment (Pryer et al., 1992; Bennett and Scheller, 1993; Söllner and Rothman, 1996; Hay and Scheller, 1997). One set of proteins involved in targeting and/or fusion are soluble N-ethylmaleimide-sensitive factor attachment protein receptors or SNARE proteins. It was originally hypothesized that SNARE proteins provided the mechanism of specificity to ensure that specific cargo would reach the proper target organelles (Söllner et al., 1993b). In this model, a specific vesicle-bound SNARE (v-SNARE) binds a specific target membrane-bound SNARE (t-SNARE) to initiate membrane docking and/or fusion (Rothman and Warren, 1994). However, recent studies indicate that SNARE proteins alone cannot direct all the required recognition events (Fischer von Mollard et al., 1997; Hay et al., 1998; Xu et al., 1998), and in fact, v-SNARE and t-SNARE binding may simply be a mechanism of the actual fusion of a docked vesicle, because v- and t-SNARE proteins appear to be all that are required for fusion (Weber et al., 1998). The identity of all the components that participate in targeting specificity and directionality remain to be uncovered.
One class of proteins that may ensure transport specificity is the Ras-like small GTP-binding proteins, the Rabs. More than 40 different Rabs have been identified in mammalian cells (Olkonen and Stenmark, 1997), and 11 have been identified in yeast (Götte and Gallwitz, 1997). Each Rab is thought to act in one particular vesicle-mediated transport step (for review, see Novick and Brennwald, 1993; Pfeffer, 1994). Activated GTP-bound Rab proteins have been shown to interact with unique effector molecules such as Rabphilin 3A, Rabaptin 5, Rim, and p40, and it is thought that these interactions facilitate transport vesicle docking and/or fusion (Shirataki et al., 1993; Stenmark et al., 1995; Díaz et al., 1997; Wang et al., 1997). The functional relationship that Rab proteins or Rab–effector complexes have with the SNARE complex is unknown, although it has been proposed that the function of the Rab is to directly modulate the formation of the SNARE core complex (Sørgaard et al., 1994; Lupashin and Waters, 1997) or to act before and/or independent of SNARE proteins at the initial stage of vesicle docking (Cao et al., 1998).
One potential link between SNARE complexes and Rab proteins are members of the Sec1 protein family. Multiple studies have implicated Sec1 components as t-SNARE-binding proteins that modulate the formation of the SNARE core complex (Hosono et al., 1987; Hata et al., 1993; Garcia et al., 1994; Harrison et al., 1994; Pevsner et al., 1994b; Tellman et al., 1997). However, contradictory evidence exists that implicates the Sec1p-like component as a positive (Banta et al., 1990; Wada et al., 1990; Aalto et al., 1991; Dascher et al., 1991), negative (Hayashi et al., 1994; Pevsner et al., 1994a; Lupashin and Waters, 1997), or both positive and negative (Wu et al., 1998) regulator of SNARE complex formation (for review, see Halachmi and Lev, 1996). Although genetic studies have linked the functions of Rabs and Sec1 proteins (Salminen and Novick, 1987; Dascher et al., 1991), no direct physical interaction between a Sec1p family member and a Rab protein has been reported.
Studies of the transport pathway responsible for delivering proteins to the lysosome-like vacuole of Saccharomyces cerevisiae have led to the identification of a large number of genes (VPS) whose products share sequence and/or functional similarities to proteins found in other vesicle-mediated transport systems (Bennett and Scheller, 1993; Stack et al., 1995). One group of VPS gene products (those affected in the class D vps mutants) (Raymond et al., 1992) facilitate the vesicle-mediated transport of vacuolar proteins from a late Golgi sorting compartment to a prevacuolar endosome (for review, see Horazdovsky et al., 1995). These proteins include the Rab GTPase Vps21p (Horazdovsky et al., 1994; Singer-Krüger et al., 1994), the Vps21p guanine nucleotide exchange factor Vps9p (Hama et al., 1999), the Sec1p homologue Vps45p (Cowles et al., 1994; Piper et al., 1994), the t-SNARE/syntaxin homologue Pep12p (Vps6p) (Becherer et al., 1996), as well as three other proteins, Vac1p (Pep7p or Vps19p) (Weisman and Wickner, 1992; Burd et al., 1997; Webb et al., 1997b), Vps8p (Horazdovsky et al., 1996), and Vps3p (Raymond et al., 1990). The v-SNARE homologue Vti1p, which has a role in cis-Golgi transport, also serves as the putative v-SNARE partner for the t-SNARE Pep12p (Fischer von Mollard et al., 1997). A number of studies have uncovered multiple genetic and physical interactions among many of these genes and gene products (Horazdovsky et al., 1996; Burd et al., 1997; Fischer von Mollard et al., 1997; Webb et al., 1997a; Hama et al., 1999).
The role of Vac1p in Golgi-to-endosome vesicle-mediated protein sorting is largely unknown. An analysis of its primary structure revealed that Vac1p is a 515-amino acid protein that contains a N-terminal classical TFIIIA-like zinc finger, two putative zinc-binding RING fingers, and a C-terminal coiled coil region (Burd et al., 1997; Webb et al., 1997b). The more C-terminal Vac1p RING finger belongs to a unique class of RING fingers (later termed FYVE fingers) that are found in other proteins implicated in vesicular transport (Mu et al., 1995; Piper et al., 1995; Stenmark et al., 1996). The FYVE finger–containing protein EEA1 is involved in mammalian endosomal protein trafficking and has been shown to colocalize to endosomes with the mammalian Vps21p sequence homologue Rab5 (Stenmark et al., 1996). EEA1 interacts directly with the Q79L-Rab5 mutant protein (Simonsen et al., 1998), and its endosomal localization is sensitive to the phos-phoinositide inhibitor wortmannin (Patki et al., 1997). Recent studies have provided in vitro biochemical confirmation that FYVE fingers bind PtdIns(3)P (Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998), but the functional role of PtdIns(3)P binding remains unknown.
This study demonstrates that Vac1p specifically interacts with the GTPase-defective form of Vps21p (Q66L) and the Sec1p homologue Vps45p. The C-terminal RING (FYVE) finger of Vac1p is shown to be required for Vac1p function and is responsible for the ability of Vac1p to efficiently interact with Vps21p and Vps45p. Unlike the mammalian FYVE domain–containing protein EEA1, a functional Vac1p FYVE domain and PtdIns(3)P are not required for Vac1p membrane association. A genetic interaction between VPS21 and VPS45 is also uncovered. We propose that Vac1p-like proteins represent a class of molecules that act as the molecular link between activated Rab proteins and the SNARE complex machinery via a Sec1p-like component.
MATERIALS AND METHODS
Reagents
Bacterial strains were grown in Luria–Bertani medium containing ampicillin (50 μg/ml) (Miller, 1972). Yeast strains were grown in medium containing 2% peptone, 1% yeast extract, and 2% glucose (YPD) or in synthetic medium supplemented with the appropriate amino acids as required (Sherman et al., 1979). Restriction and modifying enzymes were purchased from Boehringer Mannheim (Indianapolis, IN), Life Technologies (Gaithersburg, MD) or New England Biolabs (Beverly, MA). PCRs were carried out with Vent polymerase from New England Biolabs. [35S]ProMix, [γ-32P]GTP, peroxidase-conjugated anti-rabbit immunoglobulin G (IgG), and peroxidase-conjugated anti-mouse IgG were purchased from Amersham (Arlington Heights, IL). Protein A-Sepharose and protein G-Sepharose were purchased from Pharmacia Biotech (Piscataway, NJ) and Sigma (St. Louis, MO), respectively. Bacterial and yeast strains used in this study are listed in Table 1.
Table 1.
Strains and plasmids
| Strains and plasmids | Description | Source |
|---|---|---|
| Yeast strains | ||
| L40 | MATa trp1 leu2 his3 LYS2::(lexAop)4-HIS3 URA3::(lexAop)4-lacZ | Vojtek et al., 1993 |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1Δ901 lys2-801 suc2-Δ9 | Robinson et al., 1988 |
| BHY10 | SEY6210; leu2-3, 112::pBHY11(CPY-Inv LEU2) | Horazdovsky et al., 1994 |
| CCY120 | SEY6210; vps45Δ2::HIS3 | Cowles et al., 1994 |
| PHY102 | SEY6210; vps34Δ1::TRP1 | Herman and Emr, 1990 |
| CBY31 | SEY6210; Δpep12::HIS3 | Burd et al., 1997 |
| DDY3477 | SEY6210; vps34ts | This study |
| GTY102 | SEY6210; vac1Δ2::URA3 | This study |
| GTY104 | SEY6210; vac1Δ3::NEO | This study |
| GTY105 | BHY10; vac1Δ3::NEO | This study |
| GTY106 | SEY6210; vsp21::HIS3 vac1Δ3::NEO | This study |
| GTY107 | SEY6210; vps45Δ2::HIS3 vac1Δ3::NEO | This study |
| GTY108 | SEY6210; Δpep12::HIS3 vac1Δ3::NEO | This study |
| GTY109 | SEY6210; vps34Δ1::TRP1 vac1Δ3::NEO | This study |
| GTY110 | SEY6210; vsp34ts vac1Δ3::NEO | This study |
| GTY111 | SEY6210; Δpep12::HIS3 vac1Δ3::NEO vps34Δ1::TRP1 | This study |
| GTY112 | BHY10; vac1Δ2::URA3 vps21Δ3::NEO | This study |
| E. coli strains | ||
| XL1Blue | (recA1 endA1 gyrA96 thi-1 hsdR17 supE44) | Stratagene |
| M15 | (NalS StrS rifS lac− ara− gal− mtl− F− recA+ uvr+) | Qiagen |
| Plasmids | ||
| pVJL11 | Bait Vector | Jullien-Flores et al., 1995 |
| pGADGH | Prey Vector | Hannon et al., 1993 |
| pSM491 | Triple HA-tag with NotI linkers in pBS | Obtained from Pamela A. Marshall and Joel M. Goodman |
| pRS series Vectors | yeast expression vectors | Christianson et al., 1992 |
| pBS | pBluescript II | Stratagene |
| pQE31 | Histidine-tagged E. coli expression vector | Qiagen |
| pGT21-1 | Wild-type Vps21 bait in pVJL11 | Hama et al., 1999 |
| pGT21-2 | S21N-Vps21p bait in pVJL11 | Hama et al., 1999 |
| pGT21-3 | Q66L-Vps21p bait in pVJL11 | This study |
| pGT21-4 | Q66L-vps21 in pQE31 | This study |
| pBHY21-10 | VPS21 in pRS414 | Horazdovsky et al., 1994 |
| pBHY21-13 | Q66L-VPS21 in pRS414 | This study |
| pBHY21-23 | Q66L-VPS21 in pRS424 | This study |
| pBHY21-49 | VPS21-49 allele in pRS416 | This study |
| pBHY21-79 | Wild-type VPS21 in pQE31 | This study |
| pLW102 | vac1Δ2::URA3 in pRS316 | Weisman and Wickner, 1992 |
| pGT102 | vac1Δ2::URA3 in pBS | This study |
| pGT104 | vac1Δ3::NEO in pBS | This study |
| pGT1-1 | Wild-type VAC1 in pRS415 | This study |
| pVAC1-425 | Wild-type VAC1 in pRS425 | Weisman and Wickner, 1992 |
| pGT1-2 | Wild-type VAC1 in pRS413 | This study |
| pGT1-3 | Wild-type VAC1 in pRS423 | This study |
| pGT1-4 | C221S-vac1 in pRS413 | This study |
| pGT1-5 | C237S-vac1 in pRS415 | This study |
| pGT1-6 | C292S-vac1 in pRS413 | This study |
| pGT1-7 | C221/292S-vac1 in pRS413 | This study |
| pGT1-8 | Wild-type Vac1 bait in pVJL11 | This study |
| pGT1-9 | Wild-type Vac1 prey in pGADGH | This study |
| pGT1-10 | C221S-Vac1p prey in pGADGH | This study |
| pGT1-11 | C292S-Vac1p prey in pGADGH | This study |
| pGT1-12 | C221/292S-Vac1p prey in pGADGH | This study |
| pGT1-16 | VAC1 w/C-terminal HA-tag in pRS413 | This study |
| pGT1-17 | VAC1 w/C-terminal HA-tag in pRS415 | This study |
| pGT1-18 | C221S-VAC1 w/C-terminal HA-tag in pRS415 | This study |
| pGT1-19 | C292S-VAC1 w/C-terminal HA-tag in pRS415 | This study |
| pGT1-20 | C221/292S-VAC1 w/C-terminal HA-tag in pRS415 | This study |
| pGT1-21 | VAC1 w/C-terminal HA-tag in pRS423 | This study |
| pGT45-1 | Vps45p bait in pVJL11 | This study |
| pGT45-2 | Vps45p prey in pGADGH | This study |
| pVPS45-1 | original VPS45 genomic clone | Cowles et al., 1994 |
| pBHY45-CC2 | VPS45 in pRS414 | This study |
| pBHY45-3 | VPS45 in pRS416 | This study |
| pBHY45-4 | VPS45 in pRS426 | This study |
| pBHY45-7 | vps45 ts mutant in pRS414 | This study |
Plasmid and Strain Constructions
An oligonucleotide-directed mutagenesis procedure was previously described for constructing VPS21 point mutant alleles (Horazdovsky et al., 1994). This procedure was used to create plasmids pBHY21-13 and pBHY21-23, which carry the Q66L-vps21 point mutant allele in pRS414 and pRS424, respectively (Christianson et al., 1992). To create His6-tagged Vps21p and Q66L-Vps21p fusion proteins, the VPS21 and Q66L-vps21 genes were amplified from pBHY21-10 (Horazdovsky et al., 1994) and pBHY21-23, respectively, using the primers Vps21-15 (5′-GCGGGATCCGATGAACACATCAGTCACTTCC-3′) and Vps21-17 (5′-GCGCGCGCGTCGACTC-TAACAACTGCAAGCACTGTT-3′). These PCR products were digested with BamHI and SalI and cloned into the same sites in pQE31 (Qiagen, Hilden, Germany) to create the His6-tagged Vps21p and His6-tagged Q66L-Vps21p expression constructs pBHY21-79 and pGT21-4.
The PvuII–PvuII fragment from a pRS425 plasmid containing a HindII–HindII VAC1 genomic fragment (Weisman and Wickner, 1992) was inserted into the PvuII–PvuII sites of pRS415, pRS413, and pRS423 to create the wild-type VAC1 expression constructs pGT1-1, pGT1-2, and pGT1-3. To create the C-terminally triple hemagglutinin (HA)-tagged Vac1p expression constructs pGT1-16 (VAC1HA-pRS413) and pGT1-17 (VAC1HA-pRS415), the VAC1 gene was amplified from pGT1-1 with the T7 primer and the primer Vps19-10 (5′-TTAGCGGCCGCCATTAAACCCATGGTCACCCAGCTT-3′).This PCR product was cut with XhoI and ligated into the XhoI and Klenow-blunted NotI sites of pRS413 and pRS415 to create VAC1 intermediate constructs that had a C-terminal NotI site and stop codon introduced by the Vps19-10 primer. The triple HA epitope was amplified from pSM491 with the primers HA-5′ (5′-GGGAGATCTGCGGCCGCATCTTTACCCATACG-3′) and HA-3′-2 (5′-TAGTTCTACCGCGGCCTCACTGAGCAGCG-3′) (obtained from Pamela A. Marshall and Joel M. Goodman). This HA fragment was digested with SacII and NotI and ligated into the SacII–NotI sites of the intermediate VAC1 constructs. The vac1 point mutant alleles C221S (pGT1-4), C237S (pGT1-5) and C292S (pGT1-6) were constructed by PCR site-directed mutagenesis (Stack et al., 1995). The PCR products were then used to replace the analogous wild-type fragment of DNA within VAC1 in pGT1-1 or pGT1-2. The same procedure was used to create the C221S/C292S double point mutant (pGT1-7), except that the C221S mutagenic oligonucleotide was used with pGT1-6 as PCR template. To create C-terminal HA-tagged VAC1 point mutant constructs, the procedure was performed in the same manner as for the construction of pGT1-16 and pGT1-17, except that pGT1-4, pGT1-6, and pGT1-7 were used as PCR templates.
To create a strain deleted for VAC1, a plasmid (pGT102) containing the VAC1 gene disrupted with a URA3 cassette was made by inserting the KpnI–SpeI fragment of pLW102 (Weisman and Wickner, 1992) into the KpnI–SpeI sites of pBluescript II (Stratagene, La Jolla, CA). GTY102 (vac1Δ2::URA3) was made by transforming SEY6210 with a KpnI–SpeI digest of pGT102. Disruption of the VAC1 locus was confirmed by prototrophy of these yeast to uracil, PCR, and scoring of a Vps− phenotype.
Another VAC1 disruption construct (pGT104) containing the G418 resistance marker (NEO) within the VAC1 coding sequence was created by excising the URA3 cassette out of pGT102 with BglII. The BglII ends were blunted with Klenow polymerase, and the EcoRV kanMX4 module from pBS-NEO (Hama et al., 1999) was inserted. vac1Δ3::NEO yeast were created by transforming the appropriate strain with a KpnI–SpeI digest of pGT104 (Table 1). Disruption of the VAC1 locus was confirmed by resistance of these yeast to G418, PCR, and/or scoring of a Vps− phenotype.
The VPS45 SacI–ClaI fragment from the VPS45 genomic clone pBHY45-1 (Cowles et al., 1994) was cloned into those sites in pRS414, pRS416, and pRS426 (Christianson et al., 1992) to create the VPS45 expression plasmids pBHY45-CC2, pBHY45-3, and pBHY45-4, respectively.
A wild-type Vps21 bait plasmid, pGT21-1, and an S21N-Vps21 bait plasmid, pGT21-2, were previously constructed (Hama et al., 1999). In this study, a Q66L-Vps21 bait plasmid, pGT21-3, was constructed in the same manner as pGT21-1 and pGT21-2, except that pBHY21-23 was used as the PCR template. Full-length wild-type and point mutant Vac1 baits and preys were constructed by amplifying the VAC1 gene from the corresponding wild-type or mutant VAC1 plasmids with the PCR primers Vps19-1 (5′-CTTGGATCCTATGGATCTTGAAAATGTTTC-3′) and Vps19-2 (5′-CATTAACTGTCGACATCCTTTAAC-3′). The PCR products were then digested with BamHI and SalI and ligated into the BamHI and SalI sites of pVJL11 (bait) (Jullien-Flores et al., 1995) or pGADGH (preys) (Hannon et al., 1993) to create the wild-type Vac1 bait (pGT1-7), the wild-type Vac1 prey (pGT1-8), the C221S Vac1 prey (pGT1-9), the C292S Vac1 prey (pGT1-10), and the C221/C292S Vac1 prey (pGT1-11). The Vps45 bait and prey were constructed by amplifying the VPS45 gene from pVPS45-4 with the primers Vps45-3 (5′-GCGGGATCCGATGAACCTTTTTGATGTGGCT-3′) and Vps45-4 (5′-GCGCGCGCGTCGACTTTATTTTGCAGATCTAATAGAATCC-3′) (Cowles et al., 1994). The PCR product was then digested with BamHI and SalI and ligated into the BamHI and SalI sites of pVJL11 (Jullien-Flores et al., 1995) and pGADGH (Hannon et al., 1993) to create the Vps45 bait pGT45-1 and the Vps45 prey pGT45-2. VPS21 dominant mutants and vps45 temperature-sensitive for function mutants (ts) were constructed using a mutagenic PCR-based technique (Muhlrad et al., 1992). Plasmid pBHY21-49, which carries the dominant VPS21-49 allele, was isolated using this procedure and was found to exhibit a partial Vps− sorting defect when expressed in a wild-type (BHY10) background. Sequencing analysis of pBHY21-49 revealed that its DNA sequence had been altered to encode a Vps21 protein with a serine-to-leucine substitution at amino acid position 21. The vps45ts mutant allele, vps45-7, was also constructed using the same PCR mutagenesis technique that was used to construct the VPS21-49 allele. CCY120 (vps45Δ2) transformed with pBHY45-7, which carries the vps45-7 ts allele, was found to exhibit a Vps− phenotype at 38°C and a wild-type Vps+ phenotype at 25°C. The vps34 temperature-conditional allele was constructed using a PCR mutagenesis procedure similar to that described above using the plasmid vector and oligonucleotides described by Stack et al. (1995). Linearized mutant vps34ts DNA together with pRS316 were used to transform PHY102 (vps34Δ1::TRP1) selecting for Ura+ transformants. Trp− and Ura− transformants were identified and tested for temperature-conditional carboxypeptidase Y (CPY) sorting. The presence of the integrated vps34ts allele was confirmed by PCR.
Vps21 Protein Purification
M15 Escherichia coli transformed with pBHY21-79 or pGT21-4 were grown to an OD600 of 0.7–0.9. Isopropyl-β-d-thiogalactopyranoside was then added to 2 mM, and the cultures were grown an additional 5 h. The cells were suspended in 5-vol equivalents of sonication buffer (50 mM NaH2PO4, pH 8.0 and 300 mM NaCl). Lysozyme was added to 1.0 mg/ml, and the cell suspensions were incubated on ice for 30 min. The cells were then sonicated with a tip sonicator for 30 s. RNase was added to 10 μg/ml, and the lysate was passed in and out of an 18-gauge needle 5 times. The cellular debris was removed using a 10,000 × g, 20-min centrifugation. Four milliliters of 50% Ni-nitriloacetic acid resin (equilibrated with sonication buffer) were added, and the mixture was incubated on a roller at 4°C for 30 min. The mixture was then loaded into a 1-cm-diameter column and washed four times with 10 ml of wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, and 10% glycerol). Twelve milliliters of wash buffer containing 20 mM imidazole were then passed through the column. His-tagged Vps21p was eluted off the column by running 12 ml of wash buffer containing 500 mM imidazole. The eluted Vps21p was then concentrated in a Centri-prep concentrator and dialyzed against wash buffer containing no imidazole overnight at 4°C.
GTP Binding and GTPase Assays
Recombinant purified His6-Vps21p and His6-Q66L-Vps21p (2 μg) were incubated in 100-μl GTP binding reactions containing 1 mM DTT, 2 mM EDTA, 0.5 mg/ml BSA, 20 mM Tris-HCl, pH 7.5, and 10 mM MgCl2, with 0.25 μl of [γ-32P]GTP (6.5 × 104 cpm/μl). The reactions were incubated for 15 min at 30°C, and a 10-μl aliquot was removed and added to 1.0 ml of cold buffer B (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 50 mM NaCl). This solution was then passed through a 0.45-μm nitrocellulose filter and subsequently washed with 3.0 ml of buffer B. The filters were dried and subjected to scintillation counting. Counts that remained on the filters correspond to the amount of [γ-32P]GTP that was associated with Vps21p.
To assess the GTPase activities of Vps21p and Q66L-Vps21p, His6-Vps21p and His6-Q66L-Vps21p (3.29 pmol each) were incubated in 50-μl GTP loading reactions containing 0.025 μM [γ-32P]GTP in loading buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1 mM DTT, 0.5 μg/ml BSA, and 0.5 μM GTP) for 15 min at 30°C. Vps21p bound [γ-32P]GTP was stabilized by the addition of 2.75 μl of 100 mM MgCl2 and 47.25 μl of buffer A (50 mM Tris-HCl, pH 8.0, 1 mM DTT, and 5 mM MgCl2) and kept at 4°C. To initiate a GTPase reaction, 90 μl of the stabilized loading reaction was brought up to 450 μl in buffer A at 30°C, with the final concentrations of ATP and GTP being adjusted to 1 mM. At 0, 5, 15, 30, 60, and 120 min after the start of the reaction, a 50-μl aliquot was removed and added to 1.0 ml of charcoal suspension (0.1 M HCl, 10% ethanol, and 50 mM KH2PO4) and was briefly vortexed. The charcoal mixture was centrifuged at 13,000 × g for 2 min, and the amount of soluble [32P]orthophosphate was determined by liquid scintillation counting.
Yeast Two-Hybrid Assays
Two-hybrid filter assays were performed as previously described (Hama et al., 1999). Quantitative two-hybrid assays were performed by growing L40 yeast expressing the appropriate baits and preys to an OD600 of ∼1.0 at 30°C in YNB media lacking tryptophan and leucine; 1.5 ml of these cultures were then washed in 1.0 ml of Lac-Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4, pH 7.0) and resuspended in 300 μl of Lac-Z buffer. Three 100-μl aliquots of the cell suspensions were then frozen in liquid nitrogen and subsequently thawed at 37°C. Six hundred eighty-one microliters of Lac-Z buffer and 19 μl of β-mercaptoethanol were then added to each 100-μl aliquot of yeast. At time 0, 160 μl of 4 mg/ml o-nitrophenyl-β-d-galactoside in Lac-Z buffer was added to each reaction, and the reaction mixtures were incubated at 30°C. After the desired incubation time, the reactions were quenched with 400 μl of 1 M Na2CO3. The reaction mixtures were centrifuged at 15,000 × g for 5 min, and the A420 was recorded for the reaction supernatants. β-Galactosidase units were calculated as follows: β-galactosidase units = 10,000 × A420/(500 μl × OD600).
Protein Cross-Linking
For the Vps45p/Vac1p cross-linking experiments, GTY107 (CCY120; vac1Δ3::NEO) yeast expressing (pGT1-1) Vac1HAp from a CEN plasmid and/or (pVPS45-4) Vps45p from a 2μ plasmid were grown in YNB media at 30°C supplemented with appropriate amino acids as well as 2% caseamino acids. Spheroplasts were made from 5 OD600 units of each culture. The spheroplasts were washed once in YNB and 1 M sorbitol and subsequently lysed in 1.0 ml of 0.02 M KH2PO4, pH 7.5, containing 10 μg of PMSF and Boehringer Mannheim protease inhibitor mixture tablets at the manufacturer’s recommended concentration. Lysates were then incubated with either 60 μl of a 20 mg/ml (in DMSO) solution of dithiobis(succinimidylpropionate) (DSP) cross-linker or with 60 μl of DMSO alone for 30 min at room temperature. The cross-linking reactions were quenched by the addition of 20 μl of 1 M hydroxylamine, and proteins were precipitated by the addition of trichloroacetic acid (TCA) to 10%. The precipitates were resuspended in 1.0 ml of acetone by sonication and subsequently washed with 1 ml of acetone. The precipitates were dried, suspended in 100 μl of boiling buffer (50 mM Tris, pH 7.5, and 1% SDS) by sonication, and incubated at 100°C for 5 min. One milliliter of Tween buffer (0.5% Tween 20, 50 mM Tris, pH 7.5, 150 mM NaCl, and 0.1 mM EDTA) containing 1 mg of BSA was added to the protein suspensions, and insoluble material was removed by a 13,000 × g centrifugation. Five microliters of Vps45p antiserum were added to each 13,000 × g supernatant and gently shaken overnight at 4°C. Protein A-Sepharose was then added and incubated for 1.5 h. The Sepharose–antibody–antigen complexes were precipitated at 13,000 × g and washed twice with Tween buffer and once with Tris-buffered saline (TBS). Antigens and cross-linked proteins were removed from the Sepharose by incubation with 50 μl of sample buffer (6% SDS, 10% 2-mercaptoethanol, 20% glycerol, 125 mM Tris, pH 6.8, and 1% bromophenol blue) at 100°C. The immunoprecipitates were resolved by SDS-PAGE and transferred to nitrocellulose, and the blots were probed with a 1:500 dilution of monoclonal HA antibodies (Babco, Richmond, Ca) in TBSTM (TBS, 0.05% Tween 20, 5% powdered milk, and 10 mM NaN3) overnight at 4°C. The blot was washed with TBSTM and incubated with a 1:1000 dilution of anti-mouse IgG conjugated to HRP for 1.5 h at room temperature. The blot was then washed with TBS, and Vac1HAp was detected using Blaze (Pierce, Rockford, IL) chemilluminescent detection reagents.
Native Immunoprecipitations
Spheroplasts were generated from 25 OD600 equivalents of GTY112 (BHY10; vac1Δ2::URA3, vps21Δ3::NEO) expressing Q66L-Vps21p (pBHY21-23), Vps21p (pBHY21-19), and Vac1HAp (pGT1-21) or Q66L-Vps21p and Vac1HAp. The spheroplasts were lysed in 1.0 ml of native lysis buffer (50 mM NaH2PO4, pH 8.0, 150 mM potassium acetate, 5 mM DTT, and 5 mM MgCl2) using a Dounce homogenizer. The lysates were centrifuged at 16,000 × g for 10 min, and 10 μl of HA antisera were added to the supernatants. After incubation at 4°C for 30 min, protein G-Sepharose was added, and the incubations were continued for 30 min. Sepharose–antibody–antigen complexes were pelleted at 2000 × g and washed five times with native lysis buffer. Proteins were eluted from the beads in sample buffer, resolved by SDS-PAGE, and subjected to Western analysis using Vps21p antisera. Prenylated (22 kDa) and unprenylated Vps21p* (23 kDa) proteins were detected using Blaze chemilluminescent detection reagents.
Subcellular Fractionation of Vac1HAp
GTY104 (SEY6210 vac1Δ3::NEO) was transformed with pGT1-16 (VAC1HA) or pGT1-20 (C221/292S-VAC1HA). GTY106 (SEY6210; vps21::HIS3, vac1Δ3::NEO), GTY107 (SEY6210; vps45Δ2, vac1Δ3::NEO), GTY108 (SEY6120; Δpep12::HIS3, vac1Δ3::NEO), GTY109 (SEY6210; vps34Δ1::TRP1, vac1Δ3::NEO), and GTY111 (SEY6210; vps34Δ1::TRP1, vac1Δ3::NEO, Δpep12::HIS3) were transformed with pGT1-16. These strains were grown in YNB-glucose with appropriate amino acids plus 2% casamino acids, and spheroplasts were generated from 20 OD600 units of these cells. The spheroplasts were resuspended in 2.0 ml of lysis buffer (50 mM Tris, pH 7.5, 200 mM Sorbitol, and 1 mM EDTA) containing 10 μg of PMSF and Boehringer Mannheim protease inhibitor mixture tablets at the manufacturer’s recommended concentration and lysed with five strokes of a Dounce homogenizer. Unbroken cells were removed by centrifugation at 500 × g. The supernatant (5 OD600 equivalents) was fractionated into a 13,000 × g pellet (P13) and supernatant (S13). An equivalent fraction of the S13 was further fractionated into a 100,000 × g pellet (P100) and supernatant (S100). Proteins in the S13 and S100 were precipitated by the addition of 50 μl of TCA. The TCA pellets were washed twice in 1.0 ml of acetone and dried, and proteins were suspended by sonication in 75 μl of urea sample buffer (sample buffer with 6 M urea). The P13 and P100 samples were resuspended directly in 75 μl of urea sample buffer. All samples were incubated at 65°C for 5 min, and 25 μl of each were resolved by SDS-PAGE. HA-tagged Vac1p was detected by Western analysis as described above.
Labeling and HPLC Analysis of Phosphoinositides
Yeast strains were grown for 16 h at 25 or 30°C in minimal media containing [3H]myo-inositol (23 Ci/mmol; New England Nuclear, Boston, MA) as previously described (Stack et al., 1995). YPD media was added at a concentration of 1/10 total volume, and the incubation was continued for 2.5 h at 25 or 30°C. One 30°C culture was shifted to 38.5°C for the last 30 min of incubation. TCA was added to a final concentration of 5%, and samples were stored on ice for 1 h. Cells were harvested and washed twice with H2O and suspended in 500 μl H2O, 750 μl of ethanol/diethyl ether/pyridine (15:5:1) were added, and the samples were incubated at 57°C for 30 min. Cell debris was removed by centrifugation, and the lipid extracts were dried under nitrogen. The extracted lipids were deacylated (Serunian et al., 1991) and separated by HPLC using a Whatman Partisil 10 SAX column as previously described (Stack et al., 1995), except a 5–700 mM ammonium phosphate, pH 3.8 gradient was used. Glycerophosphoinositol 3-phosphate [gPI(3)P] and glycerophosphoinositol 4-phosphate [gPI(4)P] standards were generated using partially purified yeast PtdIns and crude cell extracts from a Δvps34 yeast strain as enzyme sources.
RESULTS
Characterization of the Q66L-VPS21 Point Mutant
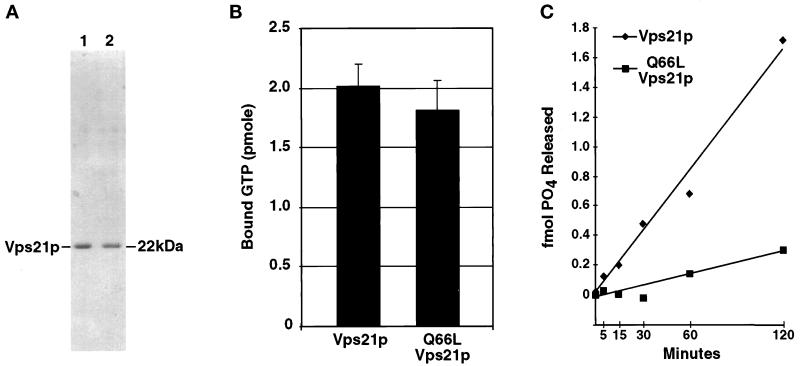
A number of studies describe point mutations in small GTPases that render these proteins constitutively active (Der et al., 1986; Stenmark et al., 1994; Martinez et al., 1997). One of these involves mutation of a conserved glutamine in GTP binding motif II. In the case of Ras, the Q61L point mutation leads to the production of an oncogenic (active) protein that exists predominantly in a GTP-bound form (Der et al., 1986). The Q79L point mutation in Rab5 results in a protein that stimulates mammalian endosomal membrane fusion events (Stenmark et al., 1994). To uncover factors that specifically associate with an activated form of the small GTPase involved in yeast Golgi-to-endosome membrane trafficking, the analogous mutation was made in VPS21 (Q66L) and used in this study. Initial characterization of Q66L-Vps21p demonstrated that the mutant protein was functional and able to facilitate the proper localization of CPY to the vacuole (our unpublished data). To better characterize the nucleotide binding capacity of Q66L-Vps21p, N-terminally hexahistidine (His6)-tagged recombinant forms of the mutant and wild-type Vps21 proteins were expressed in E. coli and purified using Ni-agarose affinity resin (see MATERIALS AND METHODS). This purification protocol yielded wild-type and mutant Vps21 proteins at >90% purity (Figure 1A). Both proteins were then assayed for their abilities to bind GTP. His6-Vps21p and His6-Q66L-Vps21p were incubated in a binding reaction containing [γ-32P]GTP, and protein–GTP complexes were isolated on nitrocellulose filters. As seen in Figure 1B, both Vps21p and Q66L-Vps21p bound GTP to very similar extents. When a large excess of unlabeled GTP was added to either binding reaction, no significant [γ-32P]GTP binding was observed (our unpublished data).
Figure 1.
Purification and characterization of GTP binding and GTPase activities of Vps21p and Q66L-Vps21p. (A) His6-Vps21p and His6-Q66L-Vps21p were expressed in E. coli and purified using Ni-agarose as described in MATERIALS AND METHODS. One microgram of purified Vps21p (lane 1) and purified Q66L-Vps21p (lane 2) were resolved by SDS-PAGE and stained with Coomassie brilliant blue to visualize proteins. (B) His6-Vps21p and His6-Q66L-Vps21p (5.2 pmol each) were incubated in a GTP-binding reaction containing 0.025 mM [γ-32P]GTP (5000 Ci/ml) and 50 mM GTP for 15 min at 30°C. Aliquots from the binding reactions were passed through nitrocellulose filters, washed, dried, and the amount of protein-associated [γ-32P]GTP was determined by scintillation counting. (C) His6-Vps21p and His6-Q66L-Vps21p (2.96 pmol each) were prebound with [γ-32P]GTP and then incubated at 30°C in GTPase reactions as described in MATERIALS AND METHODS. At the specific time points indicated, an aliquot was removed from each reaction (equivalent to 329 fmol of Vps21p) and quenched by the addition of charcoal suspension. The charcoal and bound Vps21p, Vps21p-GTP, and free GTP were pelleted, and the amount of hydrolyzed γ-32P that remained in the supernatant was determined by liquid scintillation counting. No GTPase activity was seen using a BSA control (our unpublished data).
The Q61L mutation in Ras results in a mutant protein with very low intrinsic GTPase activity compared with wild type (Der et al., 1986). To determine whether the Q66L mutant form of Vps21p also shared this property, the intrinsic GTPase activities of His6-Vps21p and His6-Q66L-Vps21p were determined (Figure 1C). His6-Vps21p and His6-Q66L-Vps21p were preloaded with [γ-32P]GTP. The GTPase reaction was initiated by the addition of Mg2+, and the amount of inorganic phosphate released was monitored as a function of time. The Q66L mutant form of Vps21p exhibited a 5.3-fold lower intrinsic rate of GTP hydrolysis than that of wild-type Vps21p (Figure 1C). This result indicated that like the Ras Q61L mutant, the GTPase activity of Q66L-Vps21p was greatly impaired, resulting in a protein that is likely to exist predominantly in the activated GTP-bound state.
Q66L-Vps21p and Vac1p Interact in the Yeast Two-Hybrid System
The class D vps mutants (including vps21) share many similar phenotypes, including enlarged vacuolar morphologies, defects in mother to bud vacuole segregation, and vacuolar protein missorting phenotypes (Raymond et al., 1992; Burd et al., 1997). Most of the gene products affected in these mutants appear to act at a similar point in the vacuolar protein sorting pathway, vesicle-mediated transport of vacuolar proteins from the late-Golgi to the prevacuolar endosome (Cowles et al., 1994; Horazdovsky et al., 1994, 1995, 1996; Burd et al., 1997; Bryant et al., 1998; Hama et al., 1999). These proteins are likely to be some of the best candidates for interaction partners that function as effectors and/or modulators of Vps21p. In our earlier work, we demonstrated that S21N mutant Vps21p, which is primarily found in the GDP-bound state, interacts with a member of this class of proteins, Vps9p, in the yeast two-hybrid system and in vivo (Hama et al., 1999). This observation led to the identification of Vps9p as the guanine nucleotide exchange factor for Vps21p.
Because the Q66L-vps21 point mutant encodes a protein that represents the active, GTP-bound form of Vps21p, we reasoned that it could be a useful reagent for uncovering downstream effectors of activated Vps21p. To investigate protein–protein interactions that occur between Q66L-Vps21p and its potential effectors, we used the yeast two-hybrid system. In this set of experiments, a plasmid encoding a LexA DNA binding domain–Q66L-Vps21 fusion protein bait (pGT21-3) was constructed. Plasmids encoding GAL4 activation domains (GAL4AD) fused to the wild-type genes that complement the class D vps mutants (class D preys) were also constructed (VPS8, VPS9, PEP12, VPS45, and VAC1). The Q66L-Vps21p, wild-type Vps21p, and S21N-Vps21p bait constructs were independently cotransformed with each of the class D prey constructs into the yeast strain L40. This yeast reporter strain contains lacZ and HIS3 reporter genes that are activated when the LexA-Vps21p and the Gal4AD-Vps protein hybrids interact.
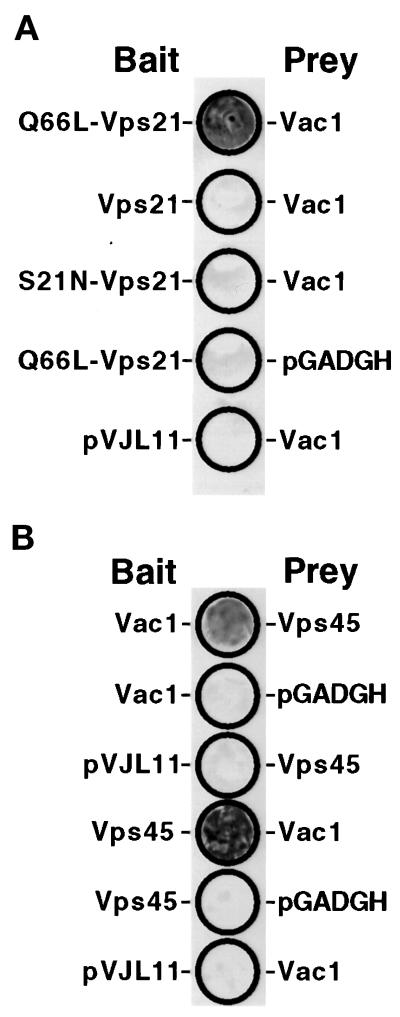
L40 yeast expressing the various combinations of the class D baits and preys were patched onto a complete synthetic medium to test for activation of the L40 lacZ reporter using a β-galactosidase filter assay (see MATERIALS AND METHODS). Coexpression of the Q66L-Vps21p bait and the Vac1p prey resulted in detectable β-galactosidase activity (Figure 2A, patch 1). Expression of the other combinations of Vps21p baits and the Vac1p prey did not result in any detectable activity (Figure 2A: Vps21p bait/Vac1p prey, patch 2; S21N-Vps21p bait/Vac1p prey, patch 3; Q66L-Vps21p bait/pGADGH, patch 4; and pVJL11/Vac1p prey, patch 5). These results indicated that Vac1p may be an effector of Vps21p in that it specifically interacts with the GTP-bound form of Vps21p.
Figure 2.
Vac1p interacts with Q66L-Vps21p and Vps45p in the yeast two-hybrid system. (A) The following L40 yeast cotransformants were patched onto a YNB-glucose plate lacking tryptophan and leucine: pGT21-3 (Q66L-Vps21 bait) and pGT1-9 (Vac1 prey), pGT21-1 (Vps21 bait) and pGT1-9, pGT21-2 (S21N-Vps21 bait) and pGT1-9, pGT21-3 and pGADGH, and pVJL11 and pGT1-9. After 3 d of growth at 30°C, these yeast were subjected to the β-galactosidase filter activity assay (Hama et al., 1999). Detectable β-galactosidase activity indicates a positive interaction between bait and prey proteins. (B) The following L40 yeast transformants were subjected to the β-galactosidase filter activity assay in the same manner as in A: pGT1-8 (Vac1 bait) and pGT45-2 (Vps45 prey), pGT1-8 and pGADGH, pVJL11 and pGT45-2, pGT45-1 (Vps45 bait) and pGT1-9 (Vac1 prey), pGT45-1 and pGADGH, and pVJL11 and pGT1-9.
Vac1p Interacts with Vps45p in the Yeast Two-Hybrid System
Previous studies have uncovered genetic interactions among VAC1, VPS45, and PEP12 (Burd et al., 1997; and Webb et al., 1997a). Vac1p and Vps45p both appear to physically interact with Pep12p in vitro (Burd et al., 1997). In this study, Vac1p baits and preys were used to uncover additional physical interactions among proteins encoded by the wild-type genes affected in class D vps mutants. Vps45p bait (pGT45-1) and prey (pGT45-2) constructs were made and tested in the yeast two-hybrid system with the wild-type Vac1p bait (pGT1-8) and prey (pGT1-9). The Vac1p prey/Vps45p bait and Vps45p bait/Vac1p prey L40 cotransformants were able to activate transcription of the L40 lexAop::lacZ reporter, whereas the Vac1p bait/empty prey vector, empty bait vector/Vps45p prey, Vps45p bait/empty prey vector, and empty bait vector/Vac1p prey cotransformants did not (Figure 2B). Greater β-galactosidase activity was reproducibly exhibited by the Vps45p bait/Vac1p prey cotransformant when compared with the Vac1p bait/Vps45p prey cotransformant. We attributed this to the fact that in the two-hybrid system, hybrid proteins possessing either the LexA DNA binding domain or Gal4 activation domain may result in fusion proteins with slightly different biochemical properties. These results provide evidence that Vac1p and Vps45p physically interact. To exclude the possibility that the Vac1p/Vps45p yeast two-hybrid interaction was indirect and possibly being bridged by Vps21p, the identical experiments were performed in L40 yeast possessing a deleted VPS21 gene. It was found that the Vac1p/Vps45p interactions were still detectable in the absence of Vps21p (our unpublished data). This result indicated that the association of Vps45p with Vac1p is not dependent on Vps21p.
Vac1p Interacts with Vps45p and Vps21p In Vivo
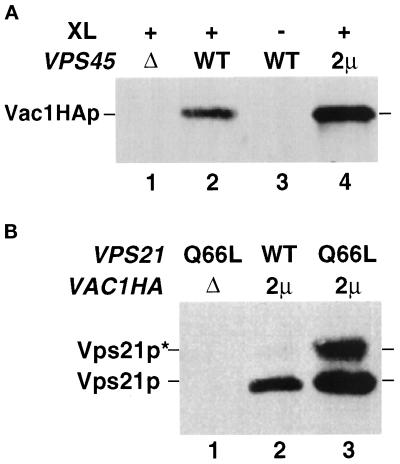
To confirm that the two-hybrid interaction seen between Vps45p and Vac1p represented an authentic in vivo interaction, yeast lysates were prepared from strains deleted for the chromosomal copy of VAC1 (GTY104) or VAC1 and VPS45 (GTY107) but expressing Vac1HAp from a CEN-based and/or Vps45p from a 2μ-based plasmid. These lysates were cross-linked with the reducible homobifunctional cross-linking agent DSP and subjected to immunoprecipitation with Vps45p antiserum. To detect Vac1HAp in these immunoprecipitates, the precipitated proteins were resolved by SDS-PAGE and transferred to nitrocellulose filters, and the blots were probed with HA antibodies. As shown in Figure 3, Vac1HAp was present in the Vps45p immunoprecipitate from the strain expressing endogenous Vps45p and Vac1HAp from a CEN-based plasmid (Figure 3, lane 2). The presence of Vac1HAp in this immunoprecipitate was dependent on the presence of Vps45p and cross-linking agent (Figure 3A, lanes 1 and 3). Additionally, when Vps45p was overexpressed from a 2μ-based plasmid, significantly more Vac1HAp cross-linked with Vps45p (Figure 3A, lane 4). These results confirm that Vps45p and Vac1p interact in vivo.
Figure 3.
Vac1p interacts with Vps45p and Vps21p in vivo. (A) Spheroplasts (5 OD600 equivalents) of the following strains were prepared: GTY107 (Δvps45Δvac1) expressing Vac1HAp (lane 1), GTY104 (Δvac1) expressing Vac1HAp (lanes 2 and 3), and GTY104 expressing Vac1HAp and overexpressing Vps45p (lane 4). The spheroplasts were lysed, and the lysates were treated with (+) or without (−) the reducible cross-linker DSP (XL) as described. Vps45p and cross-linked proteins were immunoprecipitated with Vps45p antibodies. The immunoprecipitates were resolved by SDS-PAGE and transferred to nitrocellulose, and the blots were probed with HA antiserum. Vac1HAp was visualized using Blaze chemilluminescent detection reagents. (B) Spheroplasts of GTY112 (Δvac1Δvps21) (25 OD600 equivalents) overexpressing Q66L-Vps21p (lane 1), Vps21p and Vac1HAp (lane 2), or Q66L-Vps21p and Vac1HAp (lane 3) were lysed and centrifuged at 16,000 × g. HA antisera (10 μl) was added to the cleared supernatants and incubated for 30 min at 4°C. Protein G-Sepharose was added, and the reactions were incubated an additional 30 min at 4°C. Sepharose–antibody–antigen complexes were pelleted at 2000 × g and washed five times with native lysis buffer. Proteins were eluted with sample buffer, resolved by SDS-PAGE, and subjected to Western analysis using Vps21p antisera. Prenylated (22 kDa, Vps21p) and unprenylated (23 kDa, Vps21p*) Vps21p were detected using Blaze chemilluminescent detection reagents.
Our two-hybrid results also indicate that Vac1p specifically interacts with GTPase-defective Vps21p. To confirm this interaction through biochemical means, yeast lysates expressing Vac1HAp, Vac1HAp and Vps21p, or Vac1HAp and Q66L-Vps21p were immunoprecipitated with HA antibodies under native conditions. The immunoprecipitants were resolved by SDS-PAGE, and the presence or Vps21p in the Vac1HAp immunoprecipitants was determined by Western blot analysis. As shown in Figure 3B, when Vac1HAp is expressed, both Q66L-Vps21p and wild-type Vps21p are found in the Vac1HAp immunoprecipitants (Figure 3B, lanes 3 and 2, respectively). The Q66L-Vps21p point mutation in Vps21p results in the production of a protein that is a poorer substrate for in vivo prenylation when compared with wild-type Vps21p (our unpublished observations). Accordingly, unprenylated Q66L-Vps21p immunoprecipitated in the procedure (Figure 3B, lane 3, Vps21*). This indicates that prenylation of Vps21p is not required for the interaction between Vac1p and Vps21p. Including both the prenylated and unprenylated forms, ∼10-fold more Q66L-Vps21p coimmunoprecipitated with Vac1p than wild-type Vps21p (Figure 3B, compare lanes 2 and 3). This result confirms the two-hybrid result in which Vac1p preferentially interacts with the GTP-bound conformation of Vps21p.
VPS21 and VPS45 Genetically Interact
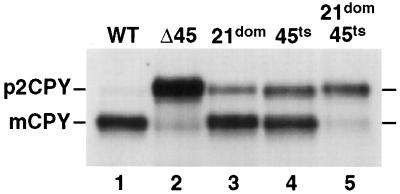
Several genetic interactions between genes encoding small GTPases of the Rab family and Sec1-like proteins have been reported (Salminen and Novick, 1987; Dascher et al., 1991). We were interested in determining whether VPS21 and the SEC1 sequence homologue VPS45 (Cowles et al., 1994) exhibited a similar interaction. To uncover any potential genetic interactions, the compartmental processing of CPY in several vps21 and vps45 double mutants was examined. A dominant mutant of VPS21 was constructed (see MATERIALS AND METHODS) and combined with different temperature-conditional vps45 mutants to construct double mutant strains. The ability of these double mutants to properly localize CPY to the vacuole was examined under a number of temperature conditions. As shown in Figure 4, when the dominant VPS21-49 allele (S21L, 21dom) was expressed in wild-type cells at 28.5°C, ∼20% of CPY was found in its Golgi-modified p2 precursor form as a result of improper vacuolar protein sorting (Figure 4, lane 3). A temperature-conditional allele of VPS45 (vps45-7, 45ts) accumulated ∼30% of the unsorted p2 precursor at 28.5°C (Figure 4, lane 4). In wild-type cells, >95% of CPY was found in its fully mature form, indicating delivery to the vacuole (Figure 4, lane 1), and in cells that lack Vps45p, the vast majority of CPY was found in its unsorted precursor form (Figure 4, lane 2). Interestingly, when VPS21-49 and vps45-7 were combined to generate double mutant cells, the vast majority (>90%) of the CPY was found in the Golgi-modified p2 form (Figure 4, lane 5). The synthetic phenotype displayed by the double mutant provides evidence that VPS21 and VPS45 functionally interact. Considering the physical interaction data presented above, this functional interaction may be mediated through the action of Vac1p.
Figure 4.
vps45ts and VPS21dom display a synthetic CPY missorting phenotype. SEY6210 (WT, lane 1), CCY45-2 (Δ45, lane 2), SEY6210 expressing pBHY21-49 (21dom, lane 3), CCY45Δ2 expressing pBHY45-7 (45ts, lane 4), and CCY45Δ2 expressing pBHY21-49 and pBHY45-7 (21dom/45ts, lane 5) were pulse labeled with [35S]ProMix for 10 min at 28.5°C in YNB-glucose containing 25 mM KH2PO4, pH 5.4. Chase solution with unlabeled methionine and cysteine was added, and the cells were incubated for an additional 30 min before being quenched with TCA. CPY was then immunoprecipitated from the combined intracellular and extracellular fractions of these cells and resolved by SDS-PAGE. The positions of mature vacuolar (m) and the Golgi-modified forms (p2) of CPY are denoted.
The Vac1p FYVE Zinc Finger Is Required for Efficient Vac1p Interaction with Vps21p and Vps45p
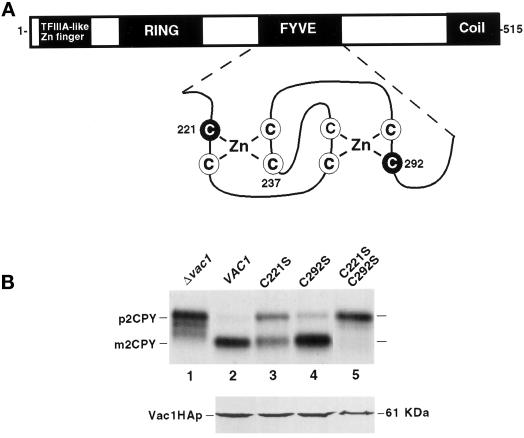
Vac1p contains a FYVE finger sequence motif that is composed of two sets of four conserved cysteine residues and is thought to be responsible for binding two zinc ions (Stenmark et al., 1996; Burd et al., 1997). A FYVE finger domain in the mammalian protein EEA1 has in fact been shown to bind two molecules of zinc (Stenmark et al., 1996). Mutational analysis also revealed that an intact EEA1 FYVE finger appears to be required for functional membrane association (Stenmark et al., 1996). Previous genetic studies have suggested that the Vac1p FYVE finger may be important for Vac1p function in that mutations in several conserved residues in the Vac1p FYVE finger result in partial vacuolar protein sorting defects (Burd et al., 1997; Webb et al., 1997a).
To determine the functional role that the Vac1p FYVE finger may have in mediating Vac1p association with Vps21p and Vps45p, cysteine residues from the first (C221) and second (C292) putative zinc binding pockets of the Vac1p FYVE finger were mutated to encode serine residues individually and in combination (Figure 5A). Each of these point mutant vac1 genes was expressed from CEN-based plasmids in a strain deleted for VAC1, and these strains were tested for their abilities to sort CPY to the vacuole. The single point mutant C221S-vac1 missorted ∼50% of newly synthesized CPY (Figure 5B, lane 3; Burd et al., 1997). The C292S-vac1 mutant, however, exhibited a very slight CPY sorting defect (Figure 5B, lane 4) when compared with the wild-type (Figure 5B, lane 2) and Δvac1 (Figure 5B, lane 1) cells. Interestingly, the double vac1 mutant C221/292S-vac1, in which cysteines from both putative zinc binding pockets are mutated, showed a severe CPY missorting defect (Figure 5B, lane 5) comparable with the null vac1 CPY sorting defect (Figure 5B, lane 1). To confirm that the missorting phenotypes seen in the C221S-, C292S-, and C221/292S-vac1 mutants were not the result of Vac1 mutant protein instabilities, plasmids expressing C-terminal HA-tagged versions of wild-type Vac1p (pGT1-16), C221S-Vac1p (pGT1-18), C292S-Vac1p (pGT1-19), and C221/292S-Vac1p (pGT1-20) were constructed and expressed in GTY104 (SEY6210; vac1Δ3::NEO). The CPY sorting phenotypes of these HA-tagged vac1 alleles revealed that the tags did not interfere with Vac1p function, because these alleles exhibited phenotypes identical to those of their untagged counterparts (our unpublished data). All of these proteins were present at very similar steady-state levels as determined by Western blot analysis (Figure 5B, lower panel). This indicated that the sorting defects seen in the vac1 FYVE finger point mutants were most likely a result of dysfunctional proteins and were not a result of mutant Vac1 protein instability. The data presented in Figure 5, demonstrate that the Vac1p FYVE finger is required for Vac1p function and that both zinc binding pockets of the FYVE finger contribute to the function of Vac1p.
Figure 5.
Characterization of vac1 FYVE finger mutations. (A) Vac1p consists of the following previously identified domains: a C2H2 TFIIIA-like zinc finger located proximal to the N terminus, an H6 variant zinc-binding RING finger, a zinc-binding FYVE RING finger, and a region predicted to form coiled coil structures located at the C terminus (Burd et al., 1997). Shown below the FYVE finger is an enlargement of the predicted structure of this domain along with the highlighted cysteine residues that have been mutated to serines individually or in combination (C221S, C237S, and C292S). (B) The abilities of yeast strains expressing vac1 alleles encoding Vac1 proteins with mutant FYVE domains to sort CPY were assessed. GTY104 (SEY6210; vac1Δ3:: NEO; lane 1) was transformed with plasmids pGT1-1 (WT VAC1) (lane 2), pGT1-4 (C221S-vac1) (lane 3), pGT1-6 (C292S-vac1) (lane 4), and pGT1-7 (C221/292S-vac1) (lane 5). CPY was immunoprecipitated from the labeled lysates that were generated from these strains as described in MATERIALS AND METHODS and in Figure 4 but at 30°C (upper panel). The expression levels of C-terminal HA-tagged Vac1p FYVE finger point mutants were also examined under steady-state conditions. GTY104 (1.0 OD600 equivalent) expressing the following HA-tagged vac1 alleles were lysed in urea sample buffer: pGT1-17 (WT Vac1HA) (lane 2), pGT1-18 (C221S-Vac1HA) (lane 3), pGT1-19 (C292S-Vac1HA) (lane 4), and pGT1-20 (C221SC292S-Vac1HA) (lane 5). The lysates were resolved by SDS-PAGE, and the Vac1HA proteins were identified by Western blot analysis using HA antibodies. Vac1HAp was detected using the Blaze chemilluminescent detection system (lower panel).
The two-hybrid system was used to examine whether the sorting defects observed in the vac1 point mutants were a manifestation of the mutated Vac1 protein(s) inability to interact with either Vps21p and/or Vps45p. Wild-type and vac1 mutant preys were constructed and coexpressed in L40 yeast with the Q66L-Vps21p bait or the Vps45p bait. Yeast lysates were generated from these cotransformants, and the amount of β-galactosidase activity in each extract was determined (Table 2). Increased β-galactosidase activity generally correlates to the strength of interaction between a bait and prey in the two-hybrid system. The C221S-Vac1p prey interacted with the Q66L-Vps21p bait with approximately half the affinity of that of the wild-type Vac1p prey, whereas the C292S- and C221/292S Vac1p prey/Q66L-Vps21p interactions exhibited a very low affinity (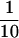 of the wild-type Vac1p prey/Q66L-Vps21p bait interaction). Interestingly, the degrees of decreased interaction strengths seen between the Q66L-Vps21 bait and the mutant Vac1 preys did not directly correlate with the pattern of CPY sorting defects observed in the mutant vac1 strains. In the CPY sorting analyses, the C292S-vac1 mutant exhibited phenotypes similar to those of wild-type VAC1, whereas the C221S-vac1 and C221/292S-vac1 mutants both exhibited CPY sorting defects (compare Figure 5 and Table 2).
of the wild-type Vac1p prey/Q66L-Vps21p bait interaction). Interestingly, the degrees of decreased interaction strengths seen between the Q66L-Vps21 bait and the mutant Vac1 preys did not directly correlate with the pattern of CPY sorting defects observed in the mutant vac1 strains. In the CPY sorting analyses, the C292S-vac1 mutant exhibited phenotypes similar to those of wild-type VAC1, whereas the C221S-vac1 and C221/292S-vac1 mutants both exhibited CPY sorting defects (compare Figure 5 and Table 2).
Table 2.
β-Galactosidase activity
| WT Vps21p bait | Q66L Vps21p bait | Vps45 bait | |
|---|---|---|---|
| Vac1 prey | 2.4 | 287.9 | 378.1 |
| C221S-Vac1p prey | 2.1 | 152.6 | 90.1 |
| C292S-Vac1p prey | 2.7 | 18.5 | 29.6 |
| C221/292S Vac1p prey | 5.0 | 25.4 | 1.9 |
β-Galactosidase activity is expressed as units per yeast cell density using the procedure described in MATERIALS AND METHODS.
The mutant Vac1p preys/Vps45p bait extracts all displayed decreased activity when compared with the wild-type Vac1p prey/Vps45p bait extract (Table 2). Interestingly, the C221S-Vac1p prey/Vps45p bait extract had significantly more β-galactosidase activity than the C292S-Vac1p prey/Vps45p bait extract (Table 2). This is similar to the result that was observed for the Vac1p prey/Q66L-Vps21p interactions. The most striking difference, however, was the interaction strength that was observed between the double C221/292S-Vac1p prey and either the Q66L-Vps21p bait or the Vps45p bait. The double point mutant Vac1p prey appears to still have the ability to weakly interact with the Q66L-Vps21p bait, but it does not appear to be able to interact with the Vps45p bait to any significant extent.
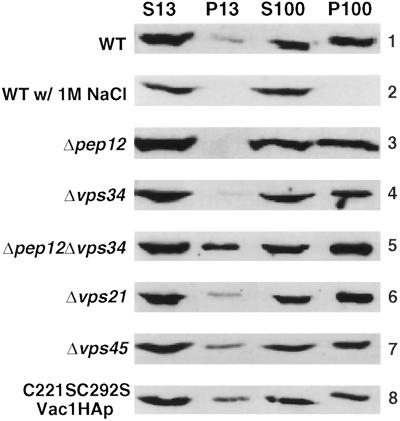
The Membrane Association of Vac1p Is Not Dependent on Pep12p, Vps21p, Vps34p, Vps45p, or the Vac1p FYVE Finger
Much of the understanding of FYVE finger function has come from an analysis of the mammalian endosomal transport factor EEA1. The EEA1 FYVE finger has been shown to be responsible for the membrane association of EEA1 (Stenmark et al., 1996), presumably by binding PtdIns(3)P (Patki et al., 1997; Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998). We were interested in determining whether the Vac1p FYVE finger and/or PtdIns(3)P was responsible for the membrane association of Vac1p. To this end, subcellular fractionation experiments of Vac1HAp were carried out in a number of different genetic backgrounds. C-terminal HA-tagged Vac1p was expressed from a CEN-based plasmid in a strain deleted for the chromosomal copy of VAC1. Figure 6, panel 1, shows the results of the subcellular fractionation pattern of Vac1HAp at steady state in this strain. A portion of Vac1HAp was found in both the soluble (S) and membrane (P) fractions. The majority of membrane-bound Vac1HAp cosedimented with membranes associated with the pellet of a 100,000 × g centrifugation (P100). A small, poorly reproducible population of Vac1HAp was also found to be associated with larger membranes found in a 13,000 × g membrane pellet fraction (P13). These results coincide with the previously published results of Webb et al. (1997b), who found that ∼77% of the newly synthesized membrane pool of Vac1HAp was in a P100 membrane fraction with the remainder in a P13 membrane fraction. It has also been established that newly synthesized Vac1HAp was solubilized from the membrane fractions when 1 M NaCl was used during the fractionation procedure (Webb et al., 1997b), and our results with steady-state Vac1HAp are consistent with this finding (Figure 6, panel 2). The fractionation patterns of Vac1HAp were also determined in strains lacking Pep12p (Figure 6, panel 3), Vps34p (Figure 6, panel 4), Vps21p (Figure 6, panel 6), and Vps45p (Figure 6, panel 7). The membrane pool of Vac1HAp was not significantly altered in these strains when compared with the wild-type background (Figure 6, panel 1). Interestingly, the fractionation pattern of C221/292S-Vac1HAp did not significantly differ from that of wild-type Vac1HAp (Figure 6, panel 8).
Figure 6.
The membrane association of Vac1HAp is not dependent on the functions of Pep12p, Vps34p, Vps21p, Vps45p, or the Vac1p FYVE finger. The subcellular fractionation patterns of Vac1HAp were assessed in the following strains that expressed Vac1HAp from a CEN plasmid; GTY104 (WT, panels 1 and 2), GTY108 (Δpep12, panel 3), GTY109 (Δvps34, panel 4), GTY111 (Δpep12Δvps34, panel 5), GTY106, (Δvps21, panel 6), and GTY107 (Δvps45, panel 7) and that expressed C221/292S-Vac1HAp from a CEN plasmid, GTY104 (C221/292S-Vac1HAp, panel 8). Spheroplasts (20 OD600 equivalents) of these strains were lysed, and unbroken cells were cleared by centrifugation at 500 × g. Five OD600 equivalents of these supernatants were then centrifuged at 13,000 × g to generate a P13 pellet and an S13 supernatant. Five OD600 equivalents of the S13 were further fractionated by spinning at 100,000 × g to generate a P100 pellet and an S100 supernatant. Supernatants were precipitated with TCA and resuspended in urea sample buffer as described. Pellets were directly resuspended in urea sample buffer. All fractions were resolved by SDS-PAGE, and Vac1HA proteins were identified by Western blot analysis using HA antibodies. Vac1HAp and C221/292S-Vac1HAp were visualized using Blaze chemilluminescent detection reagents.
To test the possibility that the syntaxin homologue Pep12p and the PtdIns 3-kinase Vps34p produce two independent means by which Vac1p could associate with cellular membranes, Vac1HAp membrane association in a Δpep12Δvps34 strain was examined. As was seen in single mutant strains (Figure 6, lanes 3 and 4), Vac1HAp membrane association was unaltered in the double mutant strain (Figure 6, Panel 5). These results indicate that Pep12p, Vps21p, Vps34p, and Vps45p do not mediate Vac1HAp membrane association. Moreover, the FYVE finger of Vac1HAp and the Vps34p enzymatic product PtdIns(3)P are also not required for Vac1p membrane association. These results differ dramatically from those obtained for EEA1 and raise the possibility that some as yet unknown component of the vacuolar protein sorting machinery is involved in mediating Vac1p membrane association.
CPY Sorting Defects of vac1 Point Mutants Are Suppressed in Cells That Have Decreased PtdIns(3)P Levels
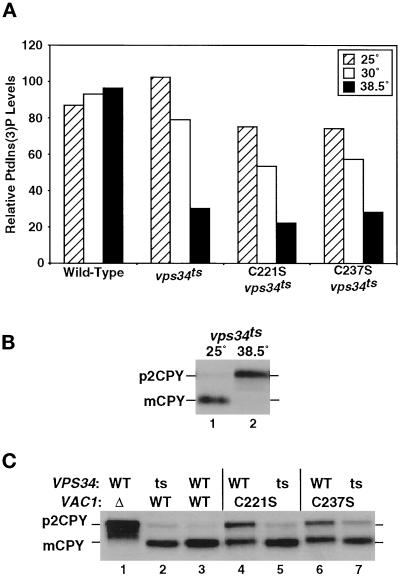
FYVE domain-containing proteins have been shown to bind PtdIns(3)P (Patki et al., 1997; Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998). To investigate the functional significance of Vac1p PtdIns(3)P binding in vivo, the effects of combined mutations in VAC1 and in the gene encoding the PtdIns 3-kinase VPS34 (Schu et al., 1993) on vacuolar protein sorting were determined. A temperature-sensitive for function allele of VPS34 was constructed using error-prone PCR mutagenesis, and the mutant vps34ts allele was integrated at the VPS34 chromosomal locus as described in MATERIALS AND METHODS. The abilities of strains expressing this allele to produce PtdIns(3)P were examined at permissive (25°C), semipermissive (30°C), and nonpermissive (38.5°C) temperatures (Figure 7). Strains expressing the vps34ts allele alone or in combination with the C221S-vac1 or C237S-vac1 mutants as well as wild-type yeast were labeled to steady-state levels with [3H]myo-inositol. After labeling, the cultures were maintained at either 25 or 30°C or shifted to 38.5°C for 30 min as described in MATERIALS AND METHODS. As shown in Figure 7A, the levels of PtdIns(3)P remained relatively constant in wild-type cells at 25, 30, or 38.5°C. In contrast, cells containing the vps34ts allele showed decreases in PtdIns(3)P levels at the semipermissive (30°C) and nonpermissive (38.5°C) temperatures. These temperature-dependent decreases were seen in vps34ts cells expressing the wild-type or mutant forms of Vac1p (although the vps34ts/vac1 double mutants generated slightly lower levels of PtdIns(3)P at all temperatures) (Figure 7A). The levels of PtdIns(3)P correlated well with the ability of the vps34ts cells to sort vacuolar proteins. At 25°C, the vps34ts strain properly sorted CPY as indicated by the presence of mature, vacuolar localized CPY (Figure 7B, lane 1). At the nonpermissive temperature (38.5°C), CPY was completely mislocalized; only the Golgi modified p2 precursor form of the protein was seen (Figure 7B, lane 2). Consistent with the moderate decrease in PtdIns(3)P levels at 30°C, only a small amount of CPY was mislocalized at this temperature (Figure 7C, lane 2), whereas wild-type cells properly sorted CPY (Figure 7C, lane 3).
Figure 7.
A vps34ts mutant suppresses the partial missorting phenotypes of vac1 FYVE finger mutants. (A) DDY3477 (vps34ts) displays temperature-sensitive PtdIns(3) kinase activity. Wild-type yeast and DDY3477 yeast expressing wild-type VAC1, C221S-vac1, or C237S-vac1 were grown for 16 h at 25 or 30°C in minimal media containing [3H]myo-inositol. A 0.1 vol of YPD was added, and incubations were continued for 2.5 h. One 30°C culture was shifted to 38.5°C for the last 30 min of the 2.5-h incubation. Deacylated lipid extracts were prepared from lysates of these cultures and separated by HPLC. This procedure allowed the complete separation of the deacylated glycerophosphoinositols gPI(3)P and gPI(4)P. For each strain, the amount of gPI(3)P that was produced is expressed relative to the amount of gPI(4)P, whose level remained largely unchanged at 25, 30, and 38.5°C. (B) DDY3477 (vps34ts) displayed a temperature-sensitive vacuolar protein sorting defect. The ability of the vps34ts strain DDY3477 to sort CPY was determined as described in Figure 4 and MATERIALS AND METHODS, except that cultures were incubated at 25 or 38.5°C for 10 min before labeling at those temperatures. (C) The abilities of vac1 FYVE finger mutants to sort CPY to the vacuole were compared in wild-type and vps34ts backgrounds at the semipermissive temperature of 30°C. GTY104 (Δvac1) expressing no Vac1p (lane 1), VAC1 (lane 3), C221S-vac1 (lane 4), and C237S-vac1 (lane 6) and GTY110 (Δvac1vps34ts) expressing VAC1 (lane 2), C221S-vac1 (lane 5), and C237S-vac1 (lane 7) were pulse labeled with [35S]ProMix for 10 min and chased with unlabeled amino acids for 30 min at 30°C. CPY was immunoprecipitated from the combined intracellular and extracellular fractions of lysates generated from these cells as described in Figure 4 and MATERIALS AND METHODS.
We next examined the severity of the CPY sorting defects in a number of vac1 FYVE finger point mutants when these vac1 alleles were combined with the vps34ts allele at the semipermissive temperature of 30°C. The C221S and C237S vac1 point mutants were found to exhibit partial CPY sorting defects at 30°C (Figures 5B and 7C, lanes 4 and 6). Surprisingly, when these vac1 alleles were combined with the vps34ts allele, the vac1 mutant sorting defects were largely suppressed (Figure 7C, lanes 5 and 7). This suppression was not seen in strains that carried other vac1 point mutations or in a strain that carried the C221/292S-vac1 double point mutant (our unpublished data). In addition, the sorting defects of the C221- and C237-vac1 alleles shown at 30°C were also tested at 25°C. The partial sorting defects of these vac1 alleles were found to be significantly less severe at 25°C, and importantly, these defects were not suppressed by the vps34ts allele (our unpublished data). These data indicate that Vac1p and Vps34p or the Vps34p product, PtdIns(3)P functionally interact in vivo.
DISCUSSION
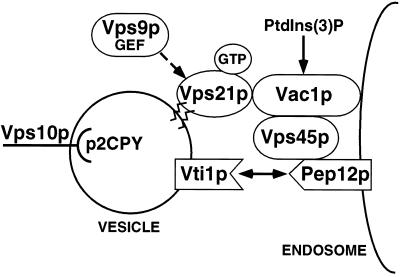
In this study we demonstrate two novel protein–protein interactions; one between an activated-Rab GTPase, Vps21p, and a FYVE finger–containing protein, Vac1p, and another between Vac1p and the Sec1p homologue Vps45p. We demonstrate that the Vac1p zinc-binding FYVE finger is required for Vac1p function and show that this motif is necessary for efficient interaction with Vps21p and Vps45p. Unlike other FYVE finger proteins such as EEA1, we conclude that the Vac1p FYVE finger is not involved in mediating Vac1p association with membranes, through its interaction(s) with PtdIns(3)P. Genetic suppression analyses indicate that PtdIns(3)P may serve to regulate Vac1p function. Together these data indicate that Vac1p is an effector of Vps21p, and Vps45p is an effector of Vac1p function. This series of interactions may represent a Rab signaling cascade that regulates Golgi-to-endosome SNARE pairing and vesicle fusion (Figure 8).
Figure 8.
Model of yeast Golgi-to-endosome vesicle-mediated protein transport. Golgi-modified p2 CPY is bound by the sorting receptor Vps10p and packaged into transport vesicles that bud from the Golgi. The v-SNARE Vti1p and the GTPase Vps21p that has been activated by the Vps21p exchange factor Vps9p are present on these vesicles as they move through the cytoplasm to the prevacuolar endosomal compartment. Vac1p then interacts with activated GTP-bound Vps21p on the vesicle and also with Vps45p that is peripherally associated with the endosome via the t-SNARE homologue Pep12p. These interactions may be regulated by PtdIns(3)P. The signal transduced from activated Vps21p through Vac1p and Vps45p enables the t-SNARE homologue Pep12p to interact with Vti1p to promote vesicle fusion.
Rab GTPase effectors such as Rabphilin 3A (Shirataki et al., 1993), Rabaptin 5 (Stenmark et al., 1995), Rim (Wang et al., 1997), and p40 (Díaz et al., 1997) have been defined by two criteria: 1) they physically interact with the activated form of their respective Rabs, and 2) they are either required for or stimulate vesicular transport. By these criteria, Vac1p is an effector of the Rab GTPase Vps21p, in that both Vps21p (Horazdovsky et al., 1994) and Vac1p (Weisman and Wickner, 1992) have been shown to be required for vacuolar protein sorting in yeast, and Vac1p associates with the activated GTP-bound form of Vps21p. Vac1p is a member of a group of gene products involved in regulating the vesicle-mediated transport of soluble vacuolar hydrolases between the yeast Golgi and a prevacuolar endosome. Other members of this group include Vps21p, Vps45p, and the t-SNARE Pep12p. vps21, vac1, vps45, and pep12 mutants display common class D vacuolar protein sorting–defective phenotypes (Vps) (Raymond et al., 1992), including a block in transport of soluble CPY to the vacuole, altered vacuolar morphologies, as well as a phenotype unique to this group of class D vps mutants, the accumulation of 40- to 60-nm vesicles (Weisman and Wickner, 1992; Cowles et al., 1994; Horazdovsky et al., 1994; Becherer et al., 1996). These shared phenotypes and the observation that both Vps21p (our unpublished observations) and Vps45p (Bryant et al., 1998) are not required for the internalization and vacuolar localization of the styryl dye FM4-64, indicate that the two proteins facilitate Golgi-to-endosome vesicle–mediated transport.
In addition to mutant phenotypes, genetic interactions have also been uncovered that that link the functions of VPS21, VAC1, VPS45, and PEP12. VPS45 and PEP12 were identified as high-copy suppressors of the vac1 C252W mutant (pep7–20) (Webb et al., 1997a). vac1ts/vps45ts, and vac1ts/pep12ts double mutants display synthetic Vps missorting phenotypes at permissive temperature (Burd et al., 1997). In this study, we observed a genetic interaction between VPS21 and VPS45. Coexpression of a weak VPS21 dominant mutant with a vps45ts allele at semipermissive temperature resulted in a synthetic Vps missorting phenotype. A similar result was obtained for the essential yeast Rab GTPase involved in the late stages of yeast secretory pathway Sec4p. Salminen and Novick (1987) observed that coexpression of the temperature-sensitive sec4-8 and sec1-1 alleles resulted in a synthetic lethal phenotype at a permissive temperature. In addition, the SEC1 homologue SLY1 and the Rab homologue YPT1 encode yeast gene products involved in mediating endoplasmic reticulum-to-Golgi transport and have also been shown to interact genetically (Ossig et al., 1991). The genetic interactions between these three Rabs and their cognate SEC1 homologues strongly indicate that the action of these two types of proteins are interrelated. These data also raise the possibility that Rab and Sec1 proteins may be members of the same protein complex. However, no physical interactions between Rab and Sec1-like proteins have been reported, and our two-hybrid and cross-linking results indicate that Vps45p does not interact directly with Vps21p, Q66L-Vps21p, or S21N-Vps21p (our unpublished data). Rab GTPases may therefore interact with Sec1 proteins through an unknown third component.
This study provides evidence that in yeast Golgi-to-endosome transport, this third component is Vac1p. Despite the very conserved nature of proteins that function in vesicle transport systems, Vac1p does not share significant overall sequence identity with other known proteins. However, Vac1p does contain a consensus zinc-binding FYVE finger domain that is shared by proteins involved in regulating many different protein trafficking pathways (Piper et al., 1995; Stenmark et al., 1996; Burd et al., 1997). For several of these proteins, the FYVE finger has been shown to bind PtdIns(3)P (Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998). Although the exact role of PtdIns(3)P binding is unknown, the presence of PtdIns(3)P is required for endosomal transport in mammalian cells (Jones and Clague, 1995; Li et al., 1995; Spiro et al., 1996) and for vacuolar protein sorting in yeast (Schu et al., 1993). One possibility is that the FYVE domain facilitates PtdIns(3)P-mediated membrane association. Mutations to conserved zinc coordinating cysteine residues in the FYVE finger of the Rab5 effector EEA1 abolish its ability to bind PtdIns(3)P and, in so doing, abolish its ability to bind membranes (Stenmark et al., 1996). Treatment of cells with the potent phosphoinositide kinase inhibitor wortmannin (Arcaro and Wymann, 1993) results in the solubilization of EEA1 (Patki et al., 1997) and blocks endosome fusion events (Jones and Clague, 1995; Li, et al., 1995; Spiro et al., 1996). These data strongly indicate a role for PtdIns(3)P binding in membrane association. However, we found that the membrane association of Vac1p was not altered in a strain that did not produce PtdIns(3)P (Δvps34) and was also not altered when the FYVE domain was mutated (C221/292S-vac1 mutant) (Figure 6). In contrast to the role PtdIns(3)P seems to play in mediating EEA1 membrane association in mammalian endosomal transport and despite the fact that Vac1p does bind PtdIns(3)P (Burd and Emr, 1998), PtdIns(3)P appears to play no role in Vac1p membrane association. However, an intact Vac1p FYVE domain is important for protein–protein interactions. Mutations in the FYVE finger of Vac1p result in mutant Vac1 proteins that no longer efficiently interact with Vps21p or Vps45p (Table 2). The FYVE domain of Vac1p may therefore perform a regulatory role in the yeast vacuolar protein sorting pathway by facilitating interactions with different proteins in the PtdIns(3)P-bound versus unbound states.
Further evidence that PtdIns(3)P may perform a regulatory role in Vac1p function comes from the observation that mutations in the Vac1p FYVE finger can be suppressed in strains that have decreased PtdIns 3- kinase (Vps34p) activity (Figure 7). In EEA1, mutations that affect one of the two FYVE finger zinc binding pockets result in EEA1 proteins that are still capable of binding one molecule of zinc (Stenmark et al., 1996). Similar mutations in Vac1p result in Vac1 proteins that are partially functional (Figure 5; Burd et al., 1997; Webb et al., 1997b). One possibility is that when one of the zinc binding domains of the FYVE finger is defective, PtdIns(3)P is still capable of binding but does so largely in a nonproductive manner, leading to the partial sorting defects displayed by these vac1 mutants. Suppression of the Vac1p FYVE domain mutations occurs when the levels of PtdIns(3)P are reduced, thereby decreasing the amount of nonproductive binding of PtdIns(3)P to mutant Vac1p. Consistent with this model, when the single vac1 FYVE mutants were overexpressed in cells with normal levels of PtdIns(3)P, these vac1 mutants were found to fully complement the null vac1 missorting phenotypes (our unpublished observations), indicating that higher levels of the mutant proteins may be able to overcome the nonproductive effects of the wild-type levels of PtdIns(3)P. An equally probable explanation for how decreased PtdIns(3)P levels may suppress these vac1 mutants is that general flux through the vacuolar protein sorting pathway may be decreased under these conditions, enabling partially defective Vac1 mutant proteins to function adequately. Whether PtdIns(3)P plays a direct or indirect role in regulating Vac1p function needs to be elucidated, but it is clear that fluctuations in the cellular levels of PtdIns(3)P can negatively and/or positively influence vacuolar protein sorting through Vac1p. To adequately assess the role PtdIns(3)P plays in mediating Vac1p function, direct correlation between the cellular levels of PtdIns(3)P with the abilities of mutant Vac1 proteins to bind PtdIns(3)P and facilitate vacuolar protein transport will need to be determined.
Activated GTP-bound Rab proteins have long been postulated to influence the function of the SNARE complex (for review, see Pfeffer, 1994). In a recent model, Schimmöller et al. (1998) predicted that vesicle-bound activated Rab proteins recruit factors (effectors) that cause the displacement of Sec1-like proteins from t-SNAREs. t-SNAREs that have been relieved of the Sec1p block are now able to pair with v-SNAREs to promote vesicle fusion (Pevsner et al., 1994a; Schimmöller et al., 1998). Vac1p may serve as this displacement factor in Golgi-to-endosome vesicle transport in the vacuolar protein sorting pathway. In the model presented here (Figure 8), activated GTP-bound Vps21p binds its effector, Vac1p, which in turn binds directly to Vps45p. The Vac1p-Vps45p interaction may result in 1) the displacement of Vps45p from the t-SNARE Pep12p or 2) the activation of Pep12p, which allows subsequent SNARE pairing and vesicle fusion. This regulated series of interactions serves to elicit the effect of activated Rab GTPases. Further studies will be required to understand the precise mechanistic roles used by Vps21p, Vac1p, PtdIns(3)P, and Vps45p in regulating Vti1p/Pep12p SNARE complex formation and function.
Note.
Several of the Vac1p interactions uncovered in this study are also described by Peterson et al. (1999).
ACKNOWLEDGMENTS
We thank members of the Horazdovsky laboratory for many helpful discussions. This work was supported by National Institutes of Health grant GM-55301 (to B.F.H.), American Cancer Society grant RPG-97-017-01-CB (to B.F.H.), and National Science Foundation grant MCB-9630108 (to D.B.D.). G.G.T. is a member of the Biochemistry and Molecular Biology Graduate Program, Division of Cellular and Molecular Biology.
REFERENCES
- Aalto MK, Ruohonen L, Hosono K, Keranen S. Cloning and sequencing of the yeast Saccharomyces cerevisiae SEC1 gene localized on chromosome IV. Yeast. 1991;7:643–650. doi: 10.1002/yea.320070613. [DOI] [PubMed] [Google Scholar]
- Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil reponses. Biochem J. 1993;296:297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta LM, Vida TA, Herman PK, Emr SD. Characterization of the yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol Cell Biol. 1990;10:4638–4649. doi: 10.1128/mcb.10.9.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Scheller RH. The molecular machinery for secretion is conserved from yeast to neurons. Proc Natl Acad Sci USA. 1993;90:2559–2563. doi: 10.1073/pnas.90.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Piper RC, Gerrard SR, Stevens TH. Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur J Cell Biol. 1998;76:43–52. doi: 10.1016/S0171-9335(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Burd CG, Peterson M, Cowles CR, Emr SD. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Emr SD, Horazdovsky BF. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J Cell Sci. 1994;107:3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- Dascher C, Rainer O, Gallwitz D, Dieter Schmitt H. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human RasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Díaz E, Schimmöller F, Pfeffer SR. A novel Rab9 effector required for endosome-to-TGN transport. J Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNARES Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EP, Gatti E, Butler M, Burton J, De Camilli P. A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci USA. 1994;91:2003–2007. doi: 10.1073/pnas.91.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- Götte LT, Gallwitz D. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- Halachmi N, Lev Z. The Sec1 family: a novel family of proteins involved in synaptic transmission and general secretion. J Neurochem. 1996;66:889–897. doi: 10.1046/j.1471-4159.1996.66030889.x. [DOI] [PubMed] [Google Scholar]
- Hama, H., Tall, G.G., and Horazdovsky, B.F. (1999). Vps9p Is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. (in press). [DOI] [PubMed]
- Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes & Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- Harrison SD, Broadie K, van de Goor J, Rubin GM. Mutations in the Drosophila Rop gene suggest a function in general secretion and synaptic transmission. Neuron. 1994;13:555–566. doi: 10.1016/0896-6273(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Hay JC, Klumperman J, Oorschot V, Steegmaier M, Kuo CS, Scheller RH. Localization, dynamics, and protein interactions reveal distinct roles for ER and Golgi SNAREs. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sydhof TC, Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins and assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Busch GR, Emr SD. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Cowles CR, Mustol P, Holmes M, Emr SD. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J Biol Chem. 1996;271:33607–33615. doi: 10.1074/jbc.271.52.33607. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, DeWald DB, Emr SD. Protein transport to the yeast vacuole. Curr Opin Cell Biol. 1995;7:544–551. doi: 10.1016/0955-0674(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Hosono R, Sassa T, Kuno S. Mutations affecting acetylcholine levels in the nematode Caenorhabditis elegans. J Neurochem. 1987;49:1820–1823. doi: 10.1111/j.1471-4159.1987.tb02442.x. [DOI] [PubMed] [Google Scholar]
- Jones AT, Clague MJ. Phosphatidylinositol 3-kinase activity is required for early endosome fusion. Biochem J. 1995;311:31–34. doi: 10.1042/bj3110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- Li G, D’Souza-Schorey C, Barberi MA, Roberts RL, Klippel A, Williams LT, Stahl PD. Evidence for phosphatidylinositol 3-kinase as a regulator of endocytosis via activation of Rab5. Proc Natl Acad Sci USA. 1995;92:10207–10211. doi: 10.1073/pnas.92.22.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin VV, Waters MG. t-SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- Martinez O, Antony C, Pehau-Arnaudet G, Berger EG, Salamero J, Goud B. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc Natl Acad Sci USA. 1997;94:1828–1833. doi: 10.1073/pnas.94.5.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mu FT, et al. EEA1, an early endosome-associated protein, EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers”and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Novick P, Brennwald P. Friends and family: the role of the Rab GTPases in vesicular traffic. Cell. 1993;75:597–601. doi: 10.1016/0092-8674(93)90478-9. [DOI] [PubMed] [Google Scholar]
- Olkonen VM, Stenmark H. Role of Rab GTPases in membrane traffic. Int Rev Cytol. 1997;176:1–85. doi: 10.1016/s0074-7696(08)61608-3. [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional PtdIns(3)P-binding motif. Nature. 1998;394:433–434. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- Patki V, Virbasius J, Lane WS, Toh BH, Shpetner HS, Corvera S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 1997;94:7326–7330. doi: 10.1073/pnas.94.14.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR. The dynamic organization of the secretory pathway. Cell Struct Funct. 1996;21:413–419. doi: 10.1247/csf.21.413. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Burd CG, Emr SE. Vac1p coordinated Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–162. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakos N, Ting AE, Bennett MK, Scheller RH. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994a;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Scheller RH. n-Sec1: a neural specific syntaxin-binding protein. Proc Natl Acad Sci USA. 1994b;91:1445–1449. doi: 10.1073/pnas.91.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol. 1994;6:522–526. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post Golgi transport vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- Pryer NK, Wuestehube LJ, Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevendon I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, O’Hara PJ, Eichinger G, Rothman JH, Stevens TH. Molecular analysis of the yeast VPS3 gene and the role of its product in vacuolar protein sorting and vacuolar segregation during the cell cycle. J Cell Biol. 1990;111:877–892. doi: 10.1083/jcb.111.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a postGolgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schimmöller F, Simon I, Pfeffer SR. Rab GTPases, directors of vesicle docking. J Biol Chem. 1998;273:22161–22164. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Serunian LA, Auger KR, Cantley LC. Identification and quantitation of polyphosphoinositides produced in response to platelet-derived growth factor stimulation. Methods Enzymol. 1991;198:78–87. doi: 10.1016/0076-6879(91)98010-4. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence LW. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M, Takai Y. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. J Biol Chem. 1993;265:2333–2337. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger B, Stenmark H, Dusterhoft A, Philippsen P, Soo J-S, Gallwitz D, Zerial M. Role of three rab5-like GTPases. Ypt51p, Ypt52p, Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993b;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Söllner TH, Rothman JE. Molecular machinery mediating vesicle budding, docking and fusion. Cell Struct Funct. 1996;21:407–412. doi: 10.1247/csf.21.407. [DOI] [PubMed] [Google Scholar]
- Sørgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchausen T, Rothman JE, Sollner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Spiro DJ, Boll W, Kirchausen T, Wessling-Resnick M. Wortmannin alters the transferrin receptor endocytic pathway in vivo and in vitro. Mol Biol Cell. 1996;7:355–367. doi: 10.1091/mbc.7.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport:regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh BH, D’Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- Tellman JT, Macaulay SL, McIntosh S, Hewish DR, Ward CW, James DE. Characterization of Munc-18c and syntaxin-4 in 3T3–L1 adipocytes. Putative role in insulin-dependent movement of GLUT-4. J Biol Chem. 1997;272:6179–6186. doi: 10.1074/jbc.272.10.6179. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wada Y, Kitamoto K, Kanbe T, Tanaka K, Anraku Y. The SLP1 gene of Saccharomyces cerevisiae is essential for vacuolar morphogenesis and function. Mol Cell Biol. 1990;10:2214–2223. doi: 10.1128/mcb.10.5.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Sudhof TC. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- Webb GC, Hoedt M, Poole LJ, Jones EW. Genetic interactions between a pep7 mutation and the PEP12 and VPS45 genes: evidence for a novel SNARE component in transport between the Saccharomyces Golgi complex and endosome. Genetics. 1997a;147:467–478. doi: 10.1093/genetics/147.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb GC, Zhang J, Garlow SJ, Wesp A, Riezman H, Jones EW. Pep7p provides a novel protein that functions in vesicle-mediated transport between the yeast Golgi and endosome. Mol Biol Cell. 1997b;8:871–895. doi: 10.1091/mbc.8.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachi M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weisman LS, Wickner W. Molecular characterization of VAC1, a gene required for vacuolar inheritance and vacuole protein sorting. J Biol Chem. 1992;267:618–623. [PubMed] [Google Scholar]
- Wu MN, Littleton JT, Bhat MA, Prokop A, Bellen HJ. ROP, the Drosophila Sec1 homolog, interacts with syntaxin and regulates neurotransmitter release in a dosage-dependent manner. EMBO J. 1998;17:127–139. doi: 10.1093/emboj/17.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Sato K, Wickner W. LMA1 binds to vacuoles at Sec18p (NSF), transfers upon ATP hydrolysis to a t-SNARE (Vam3p) complex, and is released during fusion. Cell. 1998;93:1125–1134. doi: 10.1016/s0092-8674(00)81457-9. [DOI] [PubMed] [Google Scholar]