Abstract
The cyclic peptide AF17121 (VDECWRIIASHTWFCAEE) is a library-derived antagonist for human Interleukin-5 receptor α (IL5Rα). We have previously demonstrated that AF17121 mimics Interleukin-5 (IL5) by binding in a region of IL5Rα that overlaps the IL5 binding epitope. In the present study, in order to explore the functional importance of the amino acid residues of AF17121 required for effective binding to, and antagonism of, IL5Rα, each charged residue was subjected to site-directed mutagenesis, and examined for IL5Rα interaction by using a surface plasmon resonance biosensor. One residue, Arg6, was found to be essential for receptor antagonism; its replacement with either alanine or lysine completely abolished the interaction between AF17121 and IL5Rα. Other charged residues play modulatory roles. One class consists of the N-terminal acidic cluster (Asp2 and Glu3) for which alanine replacement decreased the association rate. A second class consists of His11 and the C-terminal acidic cluster (Glu17 and Glu18) for which alanine replacement increased the dissociation rate. Binding model analysis of the mutants of the latter class of residues indicated the existence of conformational rearrangement during the interaction. Based on these results, we propose a model in which Arg6 and N-terminal acidic residues drive the encounter complex while Arg6, His11 and C-terminal acidic residues are involved in stabilizing the final complex. These data argue that the charged residues of AF17121 are utilized asymmetrically in the pathway of inhibitor-receptor complex formation to deactivate the receptor function. The results also help focus emerging models for the mechanism by which IL5 activates the IL5Rα-βc receptor system.
Interleukin 5 (IL5)1 is a T cell-derived cytokine that plays a central role in proliferation and maturation of eosinophils 1 and has been implicated in the pathogenesis of eosinophil-associated allergic inflammations such as asthma 2 3. Eosinophil activation by IL5 is a dynamic process on the cell surface encompassing specific interaction of IL5 with IL5 receptor α (IL5Rα), formation of oligomeric receptor complexes with common receptor β (βc), and initiation of cytoplasmic phosphorylation events 4. These steps culminate in various cellular responses. Hence, the lineage-specific effects of IL5 on eosinophils have generated intense interest to characterize the IL5-receptor interaction and to discover receptor antagonists for alternative asthma treatment 5.
Extensive studies on the IL5-IL5Rα interaction have been carried out over the past decade and have unveiled several important findings on epitope usage. It was shown by in vitro experiments that IL5 binds to IL5Rα with 1:1 stoichiometry 6, even though IL5 is a symmetric homodimer folded into two four-helix bundles 7 each of which could be expected to support receptor recognition. The ability to form active monomers by recombinant reconstruction 8 9 confirmed that only one binding site in the IL5 molecule is required for receptor activation. Regions in human IL5 important for this binding interaction with IL5Rα were found to be dominated by charged residues in helix B (His38, Lys39, His41), CD turn (Glu88, Glu89, Arg90, and Arg91) and helix D (Glu110) 10 11 12. The IL5 binding residues in IL5Rα also are charged. These are located in domain 1 (Asp55, Asp56, Glu58), domain 2 (Lys186, Arg188) and domain 3 (Arg297) of the three fibronectin-type III domains of the extracellular region of IL5Rα 13. The homology-deduced IL5Rα structure suggests that the binding interface of receptor α comprises two charged clusters, one a cluster of negatively-charged residues from D1 domain and the second a cluster of positively-charged residues from D2 and D3 domains. These clusters may form charge-complementary sites for epitopes in IL5. Thermodynamic and kinetic analyses suggest that conformational changes occur from IL5-receptor binding, and high resolution structural considerations suggest that it is the receptor which is likely to undergo the predominant rearrangement upon interactions of the two charge-complementary sites on IL5Rα 6. Such charge-complementary interactions and ensuing conformational isomerization are likely steps in receptor activation. Molecules which interfere with binding of IL5 at the two receptor sites could block this activation process and hence would be leads for antagonism of IL5 function.
Important advances have been made to discover several specific inhibitors for the IL5-IL5Rα interaction from random recombinant peptide libraries. A potent disulfide-cyclized 18-mer peptide, AF17121, inhibits the binding of IL5 to IL5Rα and blocks IL5 dependent eosinophil activation with an IC50 of 50 nM 14. We have evaluated the antagonist activities of AF17121 and mutational analogues showing that alanine substitution of Arg6 had completely reduced competition activity, while alanine substitution of individual acidic residues retained substantial competition activity, with multiple replacements in these residues leading to fractional loss of potency 15. Using recombinant DNA techniques, we have recently developed a thioredoxin-fused AF17121, which facilitated direct interaction analysis with IL5Rα mutational variants by using a surface plasmon resonance biosensor 16. We have found that the binding epitopes of IL5Rα for AF17121 are residues Asp55, Arg188 and Arg297, which represents a significant subset of the IL5Rα residues for IL5 binding. The fact that AF17121 mimics the binding properties of IL5 but antagonizes the receptor prompts the question of how complex formation of AF17121-receptor leads to an alternative recruitment mode that prevents receptor activation.
Real-time interaction analysis can elucidate kinetic parameters of association and dissociation rate constants which control the formation and breakdown of protein complexes. The interaction pathway can be viewed as a stepwise process, in which the protein components initially encounter by multiple interaction mode, followed by rearrangements that evolve the final stabilized complex, while the dissociation pathway can be viewed as the destabilization of the final complex. Therefore, these kinetic constants are important information for better understanding the dynamics of molecular recognition and subsequent antagonist design. The purpose of the present study is to elucidate the dynamic mechanism of AF17121 required for antagonism of IL5Rα. Here, we replaced each charged amino acid of the peptide with alanine, and used surface plasmon resonance technology to directly measure the binding kinetics of the mutational variants with IL5Rα. We found that all of the charged amino acid residues play a substantive role in binding of AF17121 to IL5Rα. Replacement of Arg6 with alanine or lysine completely abolished the interaction between AF17121 and IL5Rα, demonstrating that the guanidino group of Arg6 is a key player for receptor antagonism. We further demonstrated that other charged residues play different roles in the kinetic pathway of the AF17121-IL5Rα interaction. In this article, we have developed a model in which Arg6 and the N-terminal acidic residues are involved in stabilizing the encounter complex formation, while Arg6, His11 and the C-terminal acidic residues are involved in stabilizing the final matured complex. The consequent mechanism proposed for receptor recognition by AF17121 provides an improved rationale for the deactivation mechanism of IL5Rα and low molecular weight antagonist design.
Materials and Methods
Materials
All the oligo DNA primers, Drosophila Schneider 2 (S2) cells, cell culture media and L-glutamine solution (200 mM) were purchased from Invitrogen Inc. (Carlsbad, CA). The anti HIV gp120 monoclonal antibody, 17b, was prepared as described previously 17 and used as a negative control in this project. For surface plasmon resonance (SPR) measurements, the sensor chip CM5, surfactant P20, N-ethyl-N-(3-dimethylaminopropyl)carbodiimide (EDC), N-hydroxysuccinimide (NHS), and 1 M ethanolamine (pH 8.5) were purchased from Biacore Inc. (Piscataway, NJ).
Insect Cell Expression and Purification of Soluble IL5Rα
The soluble form of human IL5Rα (sIL5Rα) was produced in a Drosophila cell culture system (Invitrogen). The Drosophila expression vector pMT-IL5Rα-V5-His 13 and the blasticidin resistant vector pCoBlast (Invitrogen) were co-transfected into S2 cells using Cellfectin reagent (Invitrogen). Blasticidin-resistant cells were selected as a stable polyclonal population and grown in Schneider's Drosophila media with 10% FCS (Hyclone) and 25 μg/mL blasticidin S (Invitrogen). This cell line was adapted and expanded to Drosophila serum-free medium supplemented with 20 mM L-glutamine, and protein expression was induced by addition of 600 μM copper sulfate.
Sterile-filtered, conditioned Drosophila media were dialyzed against buffer A (50 mM Tris-HC1, pH 7.4, 50 mM NaCl) and loaded onto a HiTrap Chelating HP column (Amersham Biosciences) pre-equilibrated with buffer A. Non-specific binding proteins were washed away with buffer B (50 mM Tris-HC1, pH 7.4, 25 mM Imidazole, 300 mM NaCl), and the sIL5Rα protein was eluted with a linear gradient from buffer B to buffer C (50 mM Tris-HC1, pH 7.4, 300 mM Imidazole). The pooled fraction was loaded onto Sephacryl-200 (26 × 600 mm) and eluted with phosphate buffer saline (PBS) buffer (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4). The resulting protein solution was concentrated and stored below -80°C. The purity was confirmed by SDS-PAGE (4-20% linear gradient gel, Bio-Rad Laboratories), and the binding activity for IL5 was verified with an SPR biosensor as described previously 13.
Production and Characterization of Thioredoxin-fused AF17121 Mutants
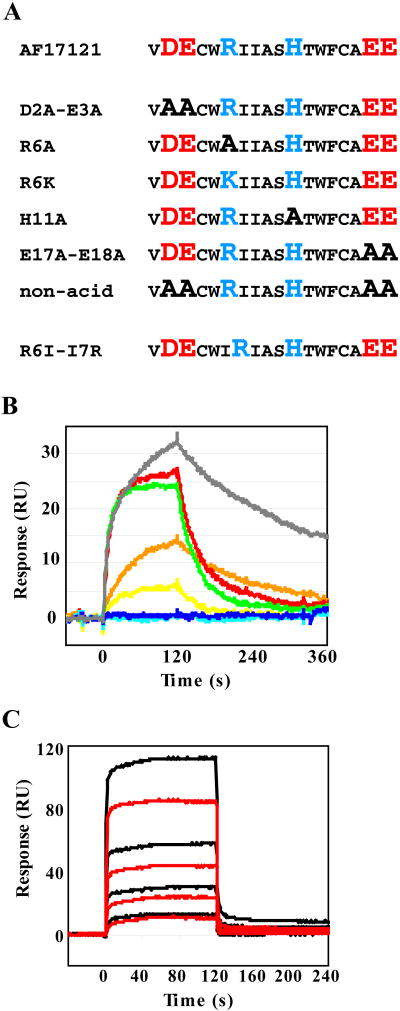
For the thioredoxin-fused AF17121 (trx-AF17121) mutational variants at positions 2, 3, 6, 11, 17 and 18 (numbering within the AF17121 sequence), each point-mutation was introduced into the bacterial expression vector pTRXaf17 16 with a QuikChange site-directed mutagenesis kit (Stratagene). The names of the mutational variants and their amino acid sequences are shown in Figure 1A. The oligonucleotide forward primers used for this study are as follows:
Figure 1.
Amino acid sequence of AF17121 mutational variants and comparison of kinetic characteristics. A. All the charged amino acid residues were singly or doubly substituted by alanine. Arginine residue 6 was substituted by lysine and also the arginine was swapped with Leu6. Acidic and basic residues are shown as red and blue, respectively. B. The sIL5Rα-coupled surface was challenged with 1 μM of each mutational variant of trx-AF17121. In this assay, rapid increase in the association phase (0-120 s) indicates fast association of the kinetic interaction, while rapid decrease in the dissociation phase (120-360 s) indicates fast dissociation. Sensorgrams of trx-AF17121, D2A-E3A (D2A and E3A), R6A, R6K, H11A, E17A-E18A (E17A and E18A), and non-acid (D2A, E3A, E17A and E18A) variants are shown as grey, orange, blue, cyan, red, green and yellow lines, respectively. C. The swapped mutant R6I-I7R was injected at concentrations of 4, 8, 16, 32 and 64 nM. Binding to the IL5Rα-immobilized surface (black lines) as well as the control surface (red lines) was detected. The non-specific effects were subtracted from the experimental binding data, leading to net sensorgrams reflecting a weak binding affinity of approximately 4 mM.
D2A-E3A, 5′-CGACAAGGCCATGGTTGCCGCATGCTGGCGTATCATC-3′;
H11A, 5′-GCTGGCGTATCATCGCTTCCGCCACCTGGTTCTGCGCTGAAG-3′;
R6A, 5′-GCCATGGTTGACGAATGCTGGGCTATCATCGCTTCCCACACCTGG-3′;
R6K, 5′-GCCATGGTTGACGAATGCTGGAAAATCATCGCTTCCCACACCTGG-3′;
E17A-E18A, 5′-CCTGGTTCTGCGCTGCAGCATGAAAGCTTGCGGCC-3′,
R6I-I7R, 5′-CCATGGTTGACGAATGCTGGATTCGCATCGCTTCCCACACCTGG-3′.
For production of trx-AF17121 mutational variants, the plasmids were transformed into the host strain BL21-CodonPlus (DE3)-RIL (Stratagene). The proteins were produced and purified as described previously 16. Oxidative cyclization of AF17121 mutants was confirmed with Ellman's reagent and purity of the fusion peptides was evaluated by analytical reverse phase HPLC. The integrity of the fusion peptides was certified by MALDI MS (Wistar Institute, Philadelphia, PA). Protein quantitation was achieved by measuring the absorbance at 280 nm and calculating the concentration using the molar extinction coefficient (ε280 = 25,230 M-1cm-1) according to Pace et al 18.
Kinetic Interaction Analysis
The kinetic interaction assay was carried out using an SPR biosensor, Biacore 3000 (Biacore, Uppsala, Sweden). The SPR experiments were conducted at 25°C in PBS buffer (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) with 0.005% P20, unless otherwise noted.
Immobilization and binding assay
Immobilization of sIL5Rα on a CM5 sensor chip was conducted by the amine coupling method (BIAapplication handbook; Biacore). Briefly, 10 μM protein solution was diluted 20 times in 10 mM acetate (pH 4.5) and injected onto a biosensor surface which had been pre-activated with a 20 μL injection of 1:1 mixture of 200 mM EDC and 50 mM NHS, followed by the injection of 1 M ethanolamine-HCl (pH 8.5). The amounts of immobilization were around 1,500 resonance units (RU). Anti HIV gp120 monoclonal antibody 17b was immobilized and used as a reference surface. The real-time interaction was measured by injecting purified trx-AF17121 and its mutational variants over these surfaces. All the procedures were automated to create repetitive cycles of injection of 0-10 μM of the fusion peptides (flow rate 50 μL/min). The sensor surfaces were regenerated by running PBS buffer for 5 min (flow rate 100 μL/min) after the dissociation phase. We observed no dependency of the measured association rates on the flow rates, confirming that there is no mass transfer effect in this system.
pH titration
To examine the effect of pH on the binding of trx-AF17121 and the H11A variant, the instrument was equilibrated with buffers of different pH. The fusion peptides were diluted in the same buffer, and the kinetic binding assay was performed as described above. The buffers used were: pH 5.0, 20 mM sodium acetate; pH 6.0; 20 mM sodium phosphate; pH 7.0, 20 mM sodium phosphate; pH 8.0, 20 mM Tris-HCl; pH 9.0, 20 mM Tris-HCl; and pH10.0, 20 mM glycine-NaOH. In addition, every buffer contained 1M NaCl and 0.005% P20.
Data analysis
The sensorgrams were analyzed using the BIAevaluation software (Biacore) and CLAMP software 19 which employ numerical integration and global fitting algorithms 20. Prior to the calculation, the binding data were corrected for non-specific interaction by subtracting the reference surface data from the reaction surface data, and further corrected for buffer effect by subtracting the signal from buffer injections from those of protein sample injections (double referencing; ref. 21). The data thus obtained were globally fit using a simple 1:1 binding (or Langmuir interaction) model:
| (1) |
In this model, A and B are free proteins and AB is a final complex. The equilibrium dissociation constant (Kd) was calculated from the association and dissociation rate constants (kon and koff) as Kd = koff/kon.
In some cases, the interaction curves were also analyzed using a two-step reaction (or conformational change) model:
| (2) |
In this model, A binds to B to form an intermediate (or encounter) complex (A:B) similar to simple 1:1 binding, but the complex can undergo a conformational change to form a final complex (AB). We use the symbols k1 and k-1 to denote the association rate and dissociation rate constants for the transition from free proteins A and B to A:B, while k2 and k-2 denote the forward and reverse rate constants for the transition from A:B to AB. The equilibrium dissociation constant of the two-step model was calculated as Kd = k-1k-2/ k1(k2 + k-2). Details of the rate equations for both models are described elsewhere 20. Individual kinetic parameters were obtained from at least three separate experiments.
Molecular Dynamics Simulation
We performed molecular dynamics (MD) simulations of AF17121 peptide (cyclic) at 300 K using the GROMOS96 software package 22 and the GROMOS96 43A1 force field parameters 22. The starting cyclic structure was first built by energy-minimizing an extended conformation (φ = ψ = 180°) of linear AF17121 peptide in vacuum using the SYBYL 6.8 software (Tripos). A disulfide bond constraint was introduced between Cys4 and Cys15 when the distance of the sulfur atoms was reduced to 5 Å. The peptide was modified by the addition of an acetyl group at the N-terminus and an amide group at the C-terminus. For MD simulation, the cyclic peptide was solvated in a cubic box with the simple point charge water model 23. The clearance between the peptide molecule and the edge of the box was at least 14 Å. The resulting system was composed of 202 peptide atoms and 14,373 water atoms. All chemical bonds were constrained using SHAKE 24, allowing a time step of 2 fs for the integration of the equation of motion. During the MD run, the temperature and pressure were controlled to ensure isothermal and isobaric conditions. The equilibration protocol for all simulations consisted of 1,000 steps of steepest-descent energy minimization applied to the solvent molecules with the solute atom positions restrained by a force constant of 250 kJ mol-1 nm-2. This was followed by steepest-descent energy minimization of the entire system, without positional restraints, to eliminate any residual strain. The minimization was terminated when the energy change converged to less than 0.1 kJ mol-1. The MD simulation was then continued for 20 ns with initial velocities assigned from a Maxwellian distribution at 300 K. Trajectory coordinates and energies were recorded at every 0.5 ps (250 steps) for subsequent analysis. We also performed molecular dynamic simulations of the linear AF17121 (cysteine-to-alanine mutant) in the same manner as the cyclic AF17121, except for the initial energy-minimizing process.
The entire analysis of trajectories was performed using the GROMOS96 software package. Atom-positional RMSD (root mean square deviation) similarity criterion cutoff of 1.2 Å was used to cluster the structures. The stereochemistry of the cluster structures was evaluated using the PROCHECK software 25.
Results
Design and Production of Thioredoxin-fused AF17121 Mutational Variants
We previously constructed thioredoxin-fused AF17121 peptide (trx-AF17121) for better solubility and sensitivity in the surface plasmon resonance (SPR) biosensor assay, and successfully defined the epitope residues of IL5Rα for trx-AF17121 binding. In the current work, we performed a kinetic interaction analysis of the mutational variants of trx-AF17121, in order to identify the functional impact of each charged amino acid on the kinetic interaction with IL5Rα. The fusion method facilitates production and characterization of mutational variants because trx-AF17121 can be over-expressed in E.coli and purified easily by affinity column chromatography. Site-directed mutagenesis on the AF17121 sequence was focused on charged amino acid residues such as aspartic acid, glutamic acid, arginine, and histidine (Fig. 1A). This is because the binding residues of IL5Rα for AF17121 are charged 16, and these amino acid residues can be surmised to interact with oppositely charged residues of AF17121 through salt bridge or hydrogen bond interactions. Our previous results have demonstrated that Arg6 of AF17121 is crucial for the inhibitory effect, and the two-residue successive acidic sequences on the N- or C-terminal regions also contribute to the inhibitory effect 15. For the purpose of this analysis, we treated the pair of Asp2 and Glu3 of trx-AF17121 as a functionally-indivisible unit. Thus we replaced these residues doubly with alanine (D2A-E3A). Likewise, Glu17 and Glu18 of trx-AF17121 were doubly replaced with alanine (E17A-E18A). We also replaced all these acidic residues with alanine (non-acid). His11 of trx-AF17121 was singly replaced with alanine (H11A). To examine whether positive charge on position 6 is important or arginine itself is important, Arg6 of trx-AF17121 was replaced with lysine (R6K) as well as alanine (R6A), and finally Arg6 was swapped with a neighbor residue Ile7 (R6I-I7R).
SPR Kinetic Interaction Analysis
In order to investigate the kinetics of receptor-ligand interactions, we have recently employed a sandwich SPR-based binding assay, in which receptor is captured by its antibody providing an oriented surface for the subsequent quantitative ligand binding measurement. This biosensor assay has enabled us to perform medium-throughput kinetic assays of receptor mutagenesis using on-chip receptor purifications 10 13. Despite the tight interaction between antigen and antibody, the effect of captured receptor dissociating from the antibody surface cannot be neglected for accurate kinetic analysis in some cases 26. This decaying surface would be more problematic when comparing the kinetic parameters determined from different experimental conditions such as pH and ionic strength, because these conditions may affect the dissociation of the captured receptor as well as receptor-ligand interaction and thus complicate data analysis of the binding responses. To eliminate the possibility of this complexity, we used a covalent immobilization method in the present study. The soluble form of IL5Rα (sIL5Rα) was immobilized on a carboxymethylated dextran matrix by the standard amine coupling method, which was used before for characterization of the IL5-IL5Rα interaction in our group 27. Even though 1,500 RU of sIL5Rα was immobilized, we found that it could only bind 50 RU of trx-AF17121. This indicates that a large portion of sIL5Rα on the surface was either inactivated or inaccessible. Nonetheless, the Kd value of trx-AF17121 binding to the sIL5Rα-coupled surface was determined to be 23 nM, with a kon value of 1.3 × 105 M-1 s-1 and a koff value of 2.9 × 10-3 s-1 (Table 1) which were almost the same as previously observed using the oriented receptor surface 16. This suggests that the kinetic interaction is not affected by a randomly-immobilized receptor surface. The covalent immobilization method provided a stable surface, which was readily regenerated by passing the running buffer for 5 min at a high flow rate. This enabled us to perform in excess of 100 repetitive binding assays while maintaining the same surface activity. The kinetic assays were performed at a flow rate of 50 μL/min, and no mass transfer effect was observed. It was previously shown that the stoichiometry of the AF17121-IL5Rα interaction is 1:1 14 16. Therefore, in order to determine the kinetic constants of association and dissociation in the current study, sensorgrams recorded at different concentrations of trx-AF17121 were fitted globally to a simple 1:1 binding model.
Table 1.
Kinetic parameters and biological effects of trx-AF17121 and its mutational variants
|
kon × 10-4 (M-1 s-1) |
koff × 103 (s-1) |
Kd × 108 (M) |
Chi2 | # IC50 | |
|---|---|---|---|---|---|
| AF17121 | 13 ± 1 | 2.9 ± 0.2 | 2.3 ± 0.2 | 0.11 ± 0.03 | 1 |
| D2A-E3A | 1.8 ± 0.4 | 5.6 ± 0.5 | 31 ± 3 | 0.10 ± 0.02 | 4 |
| R6A | N.D. | N.D. | |||
| R6K | N.D. | N.D.1 | |||
| H11A | 7.7 ± 0.2 | 17 ± 1 | 22 ± 2 | 0.33 ± 0.05 | 8.7 1 |
| E17A-E18A | 5.9 ± 0.2 | 10 ± 1 | 17 ± 1 | 0.50 ± 0.15 | 1.8 |
| non-acid | 0.76 ± 0.04 | 12 ± 1 | 160 ± 12 | 0.89 ± 0.36 | N.D.1 |
N.D., not detected.
The relative IC50 values are the concentrations to inhibit half-maximal proliferation of TF-1 cell. Data are taken from R. Piotr et al. (2004).
Unpublished data (M. Bhattacharya et al).
Mutational Effect of Charged Amino Acid Residues
The above experimental condition was applied to measure the kinetics of receptor binding for the mutational variants of trx-AF17121, namely D2A-E3A, R6A, R6K, H11A, E17A-E18A and non-acid. Figure 1B shows representative sensorgrams recorded when each variant of 1 μM concentration was injected over the same amount of immobilized IL5Rα. Both arginine mutants, R6A and R6K, showed impaired binding to IL5Rα even when the concentrations of the mutants were increased up to 10 μM. This result was unanticipated as arginine and lysine are often interchangeable without much change in binding activity, suggesting that the specific structural elements of the guanidino group are the major factor rather than just the positive charge of Arg6. To confirm the positional importance of this arginine, the position-swapping mutant R6I-I7R, was examined for receptor binding. This latter mutant showed non-specific binding to the control surface as well as to the IL5Rα-coupled surface at the concentration of 4 μM or higher (Fig 1C), though such non-specific binding was not observed in case of R6A and R6K variants. Little specific binding of the position-swapped mutant was observed. This finding suggests that the position of arginine is important for the specific, high-affinity interaction between AF17121 and IL5Rα. The finding that Arg6 of AF17121 was crucial for the direct interaction with IL5Rα is consistent with our previous data showing the importance of this arginine for its inhibitory effect in a cell proliferation assay 15. All other mutations resulted in a marked reduction in the binding affinity, that is, an increase in the equilibrium dissociation constant, the Kd value (Table 1). Double mutation of the N-terminal acidic cluster (D2A-E3A) and the C-terminal acidic cluster (E17A-E18A) caused 15- and 7-fold decreases in binding affinity, respectively. Of note, the same tendency has been observed in the biological assay showing that the N-terminal acidic cluster had two times more impact on the IC50 value than the C-terminal acidic cluster (Table 1). Depletion of all four acidic side chains (non-acid) still allowed the peptide to interact with IL5Rα although it caused a 70-fold decrease in the binding affinity. These data confirm that Arg6 is the key player for the peptide-receptor interaction. We demonstrated that His11 is also involved in the interaction between AF17121 and IL5Rα. Alanine replacement of His11 resulted in a 10-fold reduction of binding activity for IL5Rα, and the inhibition assay showed an 8.7-fold increase in the IC50 value upon this mutation (Table 1).
Kinetic parameter analysis demonstrated that these mutations of trx-AF17121 caused increases in the Kd value for the interactions due to the combination of decreases in the kon value and increases in the koff value (Table 1). Interestingly, which effect is more dominant, the association rate (on-rate) or dissociation rate (off-rate), was different depending on the residues varied. Replacement of the N-terminal acidic cluster was found to more greatly affect the on-rate, showing a 7.2-fold reduction in the kon value and a 1.9-fold increase in the koff value. In contrast, H11A and E17A-E18A variants were found to more greatly affect the off-rate, showing 1.7- and 2.2-fold reductions in the kon values and 5.9- and 3.4-fold increases in the koff values, respectively. The non-acid variant caused a decrease in the kon value and increase in the koff value by factors of 17 and 4, respectively. The mutational effect of all four acidic residues appears to be additive versus the mutational effects of the N- and C-terminal acidic clusters separately.
We realized that data fitting for such mutations as H11A, E17A-E18A and non-acid to a simple 1:1 binding model yielded higher chi-square values than fitting for trx-AF17121 and D2A-E3A (Table 1). The unsatisfied fitting for the former cases can be seen visually as reflected by residuals with non-random deviations about zero (Fig. 2A). Non-random deviation from the simple 1:1 model can be explained either by a more complex interaction model or by a non-ideal experimental condition. The latter reasons include mass transfer limitations and immobilization problems like crowding, steric hindrance and heterogeneity of coupled species. Mass transfer limitations can be a major artifact in an SPR biosensor assay 28. As previously found, however, the binding signal was not dependent on the level of immobilization or the flow-rate in the AF17121-IL5Rα interaction system 16, and we thus ruled out the possibility of mass transfer limitation. Indeed, no improved fit was observed when using a binding model for the 1:1 binding with explicit mass transfer limitation. Immobilization problems can be ruled out as a cause of the latter because all the mutational variants were injected over the same sIL5Rα-coupled surface and only H11A, E17A-E18A and non-acid variants failed to fit the simple 1:1 model. Furthermore, we observed non-random deviations in fitting to the simple 1:1 model for H11A, E17A-E18A and non-acid variants using the antibody-captured IL5Rα surface which is more homogeneous than the randomly-immobilized surface of sIL5Rα (data not shown).
Figure 2.
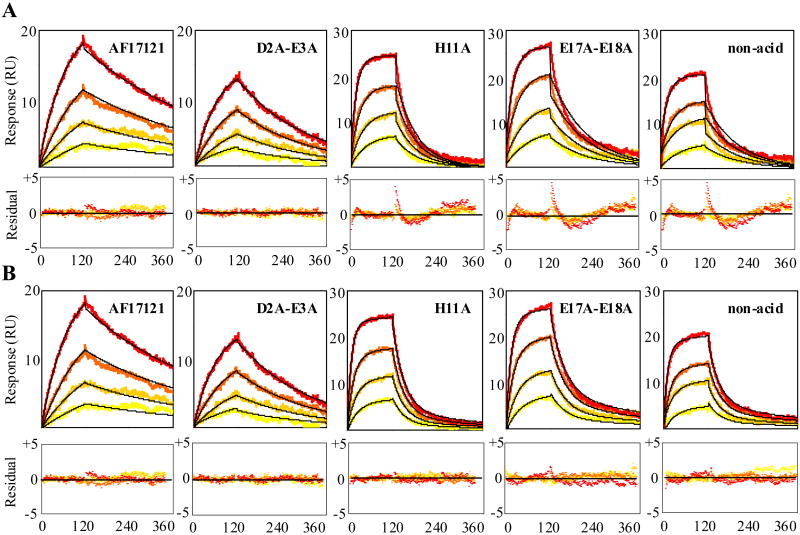
Global fitting analyses of the interactions of trx-AF17121 mutational variants with IL5Rα. Various concentrations of trx-AF17121 and its alanine-mutants, D2A-E3A, H11A, E17A-E18A and non-acid, were injected over the sIL5Rα-coupled surface. For trx-AF17121, sensorgrams of sequential injections of 10, 20, 40 and 80 nM (yellow to orange lines) are shown. For D2A-E3A, H11A and E17A-E18A variants, sequential injections of 80, 160, 320 and 640 nM (yellow to orange lines) are shown. For non-acid variant, sensorgrams of sequential injections of 640, 1,280, 2,560 and 5,120 nM (yellow to orange lines) are shown. Nonlinear least-squares analyses and numerical integration methods were used to fit the data to equations corresponding to a simple 1:1 binding model (A) and a two-step reaction model (B). Black lines show calculated sensorgrams. Residuals from the fits are shown in lower panels.
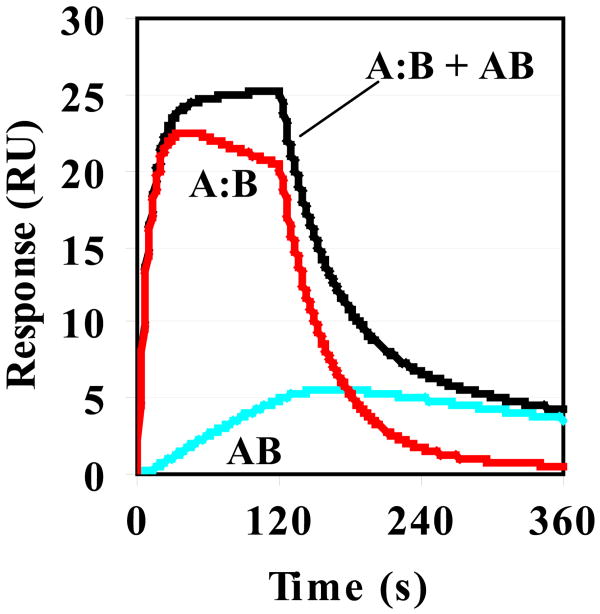
Based on the above, we applied complex models such as a two-step reaction model and a bivalent binding model to fit the data of H11A, E17A-E18A and non-acid variants. The data fit better to both the two-step reaction and bivalent binding models than the simple 1:1 model. Because AF17121 binds to IL5Rα with 1 to 1 stoichiometry 16 and does not induce dimerization of receptor 14, the bivalent binding model is inappropriate for the AF17121-IL5Rα interaction system. Thus, the interaction of these mutational variants with IL5Rα is most likely explained by the two-step reaction model. The improved quality of fits using the two-step reaction model was judged quantitatively as lower chi-square values (Table 2) and visually as the residuals with random deviation about zero (Fig. 2B). For these mutational variants, the association rate and dissociation rate constants of the first step (k1 and k-1) and the forward and reverse rate constants (k2 and k-2) of the second step are shown in Table 2. The Kd values calculated from the kinetic parameters of the two-step model (Table 2) were almost the same as those of the simple 1:1 binding model (Table 1). Based on these kinetic parameters, the time dependences of complex formation of the first and second step reactions were simulated. Figure 3 shows the simulated sensorgrams of the E17A-E18A mutational variant.
Table 2.
Kinetic parameters for H11A, E17A-E18A and non-acid variants using a two-step reaction model
|
k1 × 10-4 (M-1 s-1) |
k-1 × 103 (s-1) |
k2 × 103 (s-1) |
k-2 × 103 (s-1) |
Kd × 108 (M) |
Chi2 | |
|---|---|---|---|---|---|---|
| H11A | 9.7 ± 0.6 | 31 ± 2 | 1.9 ± 0.5 | 5.5 ± 1.2 | 24 ± 4 | 0.10 ± 0.01 |
| E17A-E18A | 8.0 ± 0.3 | 22 ± 1 | 2.4 ± 0.1 | 3.1 ± 0.4 | 16 ± 1 | 0.18 ± 0.02 |
| non-acid | 1.1 ± 0.1 | 29 ± 2 | 2.3 ± 0.4 | 3.7 ± 0.7 | 163 ± 15 | 0.24 ± 0.03 |
Figure 3.
Simulated sensorgrams generated by numerical integration of the rate equation for a two-step model. The simulation was carried out by using CLAMP software 19. The rate constants of the E17A-E18A variant were used as input values which are listed in Table 3. The concentration of injected protein was set at 1 μM and the maximum response at 30 RU. The output response (black line) is the sum of the response of the encounter complex (A:B, red line) and the final complex (AB, blue line).
Effects of Ionic State and Temperature
We demonstrated that His11 of AF17121 is involved in the interaction with IL5Rα (see above). The pKa value for imidazole is approximately 6, which can be slightly varied depending on its environment in proteins. Therefore, the histidine side-chain is either protonated or deprotonated at physiological pH. In order to determine which ionic state of the imidazole side-chain of His11 is most used in the interaction with IL5Rα, the effect of pH on the binding of trx-AF17121 to IL5Rα was studied in the pH range 5.0 to 10.0. To avoid non-specific interaction between the fusion-peptides and carboxylate groups of the dextran surface at lower pH, we added 1M NaCl to the running buffers. Note that such high salt concentration did not affect the binding affinity or the kinetic parameters of the AF17121-IL5Rα interaction at physiological pH (Table 1 and 3). The pH titration experiments showed that the Kd value of the AF17121-IL5Rα interaction was unaffected in the acidic conditions (pH 5-7), but it increased about 10-fold when the pH was increased to 8.0 (Table 3). This substantial change of the Kd value around the neutral pH strongly suggests that protonated imidazole groups of histidine are involved in the AF17121-IL5Rα interaction. This effect is more likely to be due to the histidine from AF17121 rather than that from IL5Rα, because previous pH titration experiments on the IL5-IL5Rα interaction have shown that the Kd value remains constant for the range of pH 6.0 to 9.5 27. To confirm this, we examined the effect of pH on the H11A-IL5Rα interaction (Table 3). The Kd value was little affected up to pH 8.0 and was around 500 nM, which was similar to the Kd value of the AF17121-IL5Rα interaction at pH 8.0. Thus, the mutational effect of H11A was comparable to the deprotonation effect of AF17121 at pH 8.0, supporting the idea that the cationic form of His11 of AF17121 plays a stabilizing role in the interaction between AF17121 and IL5Rα. Both AF17121 and the H11A variant showed almost the same increased Kd values both at pH 9.0 and 10.0. The pH effect on the Kd value for AF17121 was attributed to both a decrease in the kon and an increase in the koff, while the H11A variant showed an increase in the koff without affecting the kon in the pH range 5.0-10.0.
Table 3.
pH and temperature dependency of kinetics parameters
|
kon × 10-4 (M-1 s-1) |
koff × 103 (s-1) |
Kd × 108 (M) |
|
|---|---|---|---|
| pH 5.0 | 14 ± 2 | 2.9 ± 0.4 | 2.3 ± 0.4 |
| pH 6.0 | 13 ± 1 | 1.9 ± 0.2 | 1.5 ± 0.6 |
| pH 7.0 | 8.3 ± 0.3 | 2.6 ± 0.3 | 3.2 ± 0.3 |
| pH 8.0 | 2.2 ± 0.3 | 8.4 ± 0.2 | 40 ± 7 |
| pH 9.0 | 2.0 ± 0.3 | 16 ± 2 | 79 ± 7 |
| pH 10.0 | 2.6 ± 0.1 | 51 ± 1 | 200 ± 8 |
|
| |||
| H11A, pH 5.0 | 1.2 ± 0.1 | 9.5 ± 0.1 | 79 ± 6 |
| H11A, pH 6.0 | 3.5 ± 0.3 | 16 ± 2 | 45 ± 3 |
| H11A, pH 7.0 | 3.4 ± 0.1 | 17 ± 1 | 51 ± 2 |
| H11A, pH 8.0 | 3.4 ± 0.1 | 18 ± 0.9 | 54 ± 3 |
| H11A, pH 9.0 | 2.9 ± 0.1 | 25 ± 1 | 89 ± 7 |
| H11A, pH 10.0 | 2.6 ± 0.4 | 47 ± 2 | 190 ± 33 |
|
| |||
| 5 °C | 0.62 ± 0.4 | 0.63 ± 0.4 | 0.97 ± 0.2 |
| 15 °C | 8.3 ± 0.5 | 1.3 ± 0.2 | 1.6 ± 0.3 |
| 25 °C | 12 ± 0.7 | 3.4 ± 0.5 | 2.9 ± 0.4 |
| 35 °C | 12 ± 0.7 | 9.4 ± 1.0 | 7.7 ± 1.5 |
1M NaCl was included to avoid non-specific interaction for pH titration experiments.
The effect of temperature on the binding affinity and rate constants of the AF17121-IL5Rα interaction was also examined at 5, 15, 25 and 35°C. The results are shown in Table 3. The Kd value increased with increasing temperature. This correlates most strongly with a progressive increase in the koff value. In contrast, the change in kon value was found to be less continuous over the temperature range examined, although the association rate was found to be one-order lower at 5 °C. In any case, that the Kd value of the AF17121-IL5Rα interaction increased with increasing temperature (Table 3) is consistent with the view that the interaction is dominated by charged and other hydrophilic structural elements.
Discussion
Asymmetric Usage of the Charged Residues in the Kinetic Pathway of AF17121-IL5Rα Interaction
Analyzing the kinetic pathway of a protein-protein interaction is meaningful for understanding the molecular recognition mechanism in complex biological systems 29. A protein-protein interaction reaction can be viewed as a process with multiple steps. For instance, Gideon Schreiber has proposed a four-state (three-step) model of the association-dissociation pathway in which two free proteins form an encounter complex by diffusion, followed by conformational rearrangement and desolvation leading to an intermediate complex and then formation of the final stable complex 30. In this model, the encounter complex redissociates faster than its transition into the intermediate complex, and therefore the formation of intermediate complex is the rate-limiting step. Depending on the stability of the encounter and intermediate complexes, the association may appear to follow a three-state (two-step) or two-state (one-step) model for some interactions.
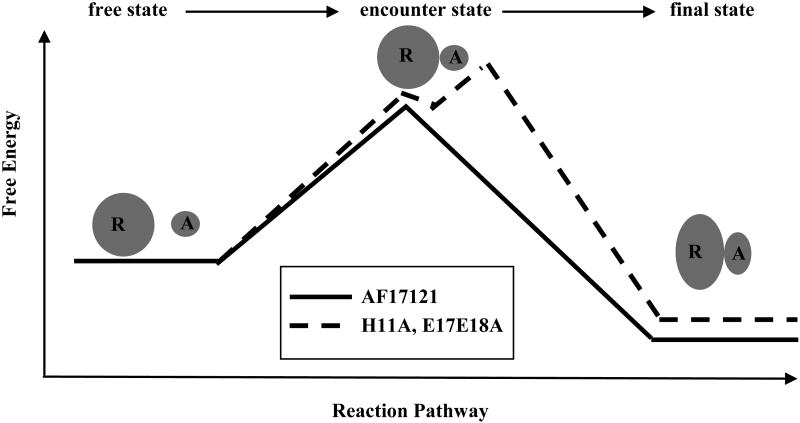
In this study, we performed the kinetic analysis of mutational variants of AF17121 using an SPR biosensor and a real-time binding assay. We found that the charged binding residues of AF17121 fall into three groups. The first is the critical epitope, Arg6, confirmed to be essential for the entire kinetic process, because its replacement completely abolished the interaction between AF17121 and IL5Rα. The second group, the ‘on-rate epitope’, consists of the N-terminal acidic cluster (Asp2 and Glu3) which preferentially modulates the association rate. The ‘off-rate epitope’, the third group of residues, consists of His11 and the C-terminal acidic cluster (Glu17 and Glu18) which preferentially modulates the dissociation rate. Interestingly, binding model analysis showed that the binding sensorgrams of third group mutants fit better to a two-step reaction model than a simple 1:1 binding model, whereas AF17121 and the D2A-E3A mutational variant showed a good fit to the 1:1 binding model (Fig. 2). This finding indicates that either an encounter state or an intermediate state could be observed with such mutations as H11A and E17A-E18A. Analysis of the H11A and E17A-E18A variants using the two-step reaction model showed that the k2 values were smaller than the k-1 values (Table 2). Figure 3 showed that the formation and breakdown of A:B complex are faster than those of AB complex, suggesting that the rate-limiting step of these mutational variants might be the transition to the final bound state (AB complex). Thus, according to the above Schreiber four-state kinetic model, the A:B complex would be an encounter state, which was observed upon such mutations as H11A and E17A-E18A, while an intermediate state could not be observed in this system. It seems that replacement of these residues would increase the transition state energy barrier between the encounter complex and final complex, in effect stabilizing the encounter complex as an observable intermediate state (Fig. 4). Hence, we assume that His11, Glu17 and Glu18 are involved in stabilizing the final complex and these residues are important to guide the encounter complex to the final one.
Figure 4.
Free energy profile describing a plausible kinetic pathway for the AF17121-IL5Rα interaction. The thick line shows the pathway of AF17121, while the dotted line shows the pathway of H11A and E17A-E18A variants. The encounter state of the peptide-receptor complex can be stabilized and observed upon such mutations as H11A and E17A-E18A.
We showed that Asp2 and Glu3 are the on-rate epitope, suggesting that these residues are more likely to be involved in stabilizing the encounter complex (Fig. 4). Although the nature of an encounter state is controversial, it seems that specific short-range interactions are not dominant in an encounter complex formation 31. The association rate constant can be enhanced by long-range attractive electrostatic forces. However, we could rule this out, because the kinetic parameters for the AF17121-IL5Rα interaction were not influenced by 1M NaCl (Table 1 and 3), which usually shields long-range electrostatic forces. Hence, these on-rate epitope residues might stabilize the encounter complex by relatively short range interactions, but in a non-specific fashion. Since Arg6 was found to be critical for receptor binding, we assume that Arg6 is involved in both the encounter and final complexes. In conclusion, we demonstrated an intriguing result that charged amino acids play different roles in the kinetics pathway of the interaction between AF17121 and IL5Rα. Based on the observations, we depict a kinetic model that Arg6 and the N-terminal acidic cluster are involved in formation of the encounter complex, and Arg6, His11 and the C-terminal acidic cluster are used for stabilization of the final complex. Although the negatively-charged clusters of AF17121 are probably distributed in the same structural regions relative to the disulfide bond bridge between Cys4 and Cys15, their impacts on the kinetics of interaction were asymmetric, that is to say, N- and C-terminal clusters are thought to be used asymmetrically in the kinetic pathway (Fig. 5). The finding that not all of the epitope residues of AF17121 are used for stabilization of the final complex leads to the possibility that a relatively small subset of structural elements ultimately could be utilized to design low molecular weight molecular antagonists.
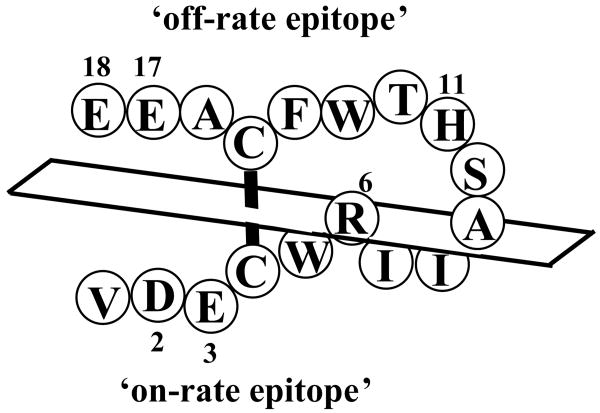
Figure 5.
Schematic diagram of the on-rate and off-rate epitope residues of AF17121. The positions of on-rate and off-rate epitope residues for IL5Rα are shown on the primary structure of AF17121. This diagram shows the association face comprised of Asp2 and Glu3, and the dissociation face comprised of His11, Glu17 and Glu18.
Mechanism of Receptor Deactivation by AF17121
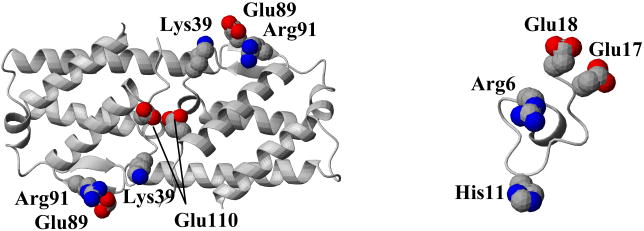
We have previously demonstrated that AF17121 can interact with Asp55, Arg188, and Arg297 of IL5Rα, which represent a significant subset of the key residues for IL5 binding 16. The fact that AF17121 mimics the receptor-binding capability of IL5 prompted us to examine whether the receptor epitope residues of AF17121 can be functionally correlated with those of IL5 in the three-dimensional structure. In order to interpret the mutational effect of AF17121 in terms of three dimensional structure, we performed MD simulation of the peptide. The receptor binding epitopes of AF17121 were mapped on a representative structure from the most populated cluster during the MD simulation. Figure 6 shows a spatial alignment of the receptor binding epitope of IL5 (Lys39, Glu89, Arg91 and Glu110) compared with the off-rate epitope of AF17121 (Arg6, His11, Glu17 and Glu18). Since IL5 is a homodimeric protein and the Glu110 is located close to the two-fold symmetric axis, the two Glu110 residues are in close proximity and constitute a cluster of negative charges, suggesting that the C-terminal negatively-charged cluster of AF17121 could correspond to the two Glu110 residues of IL5. We believe that Arg6 of AF17121 could be an equivalent residue of Arg91 in the IL5 molecule, because these arginine residues are the main receptor binding residues for AF17121 and IL5, respectively. By this view, His11 of AF17121 could correspond to Lys39 of IL5. Looking at the structures, however, the distances among these correlated residues are not consistent between IL5 and AF17121. One possible explanation for this inconsistency is that the structure of AF17121 is flexible enough to allow conformational adaptation upon binding to IL5Rα. Indeed, our previous circular dichroism data has indicated that cyclic AF17121 does not have significant α-helix or β-sheet structure 15. The activation of IL5 receptor is believed to depend on a sequence of interactions at the cell surface, starting with the recruitment of IL5 receptor α by IL5, then subsequent formation of oligomeric receptor complexes with receptor βc, disulfide bridge formation between receptor α and βc, and initiation of cytoplasmic phosphorylation events 4. The lack of agonist activity of AF17121 most likely relates to the inability of peptide complex with receptor α to form an activated complex with receptor βc. One possibility is that AF17121 lacks a critical binding epitope for βc, such as Glu13 in IL5, so as to be incapable of full receptor oligomerization of α and βc. Another possibility is that the conformational state of IL5Rα binding AF17121 may be different from that of IL5Rα binding IL5. This latter explanation is consistent with previous data from thermodynamic and kinetics analyses showing that considerable conformational rearrangement likely takes place in IL5Rα upon IL5 binding 6 13. By this view, AF17121 could be antagonizing IL5Rα by leading to an alternative conformational state of IL5Rα, perhaps due to a non-IL5-like spacing of charged residues, that stabilizes receptor α complexation (see above) and prevents effective βc recruitment and the resulting signal transduction. While the current study brings these possibilities into focus, deciding between them must await conformational and three dimensional structural elucidation of the IL5-IL5Rα and AF17121-IL5Rα complexes.
Figure 6. Comparison of receptor bindings epitope of IL5 and AF17121.
Crystal structure of homodimeric chains of human IL5 7 and a depiction of AF17121 (a MD structure simulated in this study) are shown. The receptor-binding residues of IL5 (Arg91, Lys39 and Glu110) and the off-rate epitope residues of AF17121 (Arg6, His11 Glu17 and Glu18) are labeled and expressed in CPK (Corey, Pauling and Kultun) models. Oxygen, nitrogen and sulfur atoms are colored as red, blue and yellow, respectively. Loop regions are represented as coils, α-strands as ribbons and β-strand as arrows. The molecular graphics figure was prepared with the program MOLMOL 32.
In summary, the present study reveals the essential role of charged residues in the kinetic pathway of the interaction of AF17121 with IL5Rα. We found that the N-terminal acidic residues compose an on-rate epitope, His11 and the C-terminal acidic residues compose an off-rate epitope and Arg6 is essential for both on- and off-rate process. Based on these findings, we propose a model in which Arg6 is essential for receptor binding, the N-terminal acidic residues modulate encounter complex formation, and His11 and the C-terminal acidic residues modulate stabilization of the final complex. These data argue that the non-Arg6 charged residues of AF17121 are utilized asymmetrically in the association and dissociation of the inhibitor-receptor complex. Revealing the key recognition elements of AF17121 and their kinetic roles can drive the future design of smaller compounds which can stabilize the functionally inactive IL5Rα state in a manner similar to AF17121 and therein would be viable antagonist leads for IL5-driven allergic inflammation.
Acknowledgments
We thank Dr. Takuya Yoshida (Osaka University, Osaka, Japan) for critically reading the manuscript.
Footnotes
This work was supported by National Institutes of Health Grants RO1-GM55648 and RO1-AI40462.
The abbreviations used are: IL, interleukin; IL5Rα, interleukin 5 receptor α chain; sIL5Rα, soluble form of interleukin 5 receptor α chain; trx, thioredoxin-fused; PBS, phosphate buffer saline; SPR, surface plasmon resonance; RU, resonance unit; kon, association rate constant; koff, dissociation rate constant; MD, molecular dynamics; RMSD, root mean square deviation; on-rate, association rate; off-rate, dissociation rate.
References
- 1.Karlen S, De Boer ML, Lipscombe RJ, Lutz W, Mordvinov VA, Sanderson CJ. Biological and molecular characteristics of interleukin-5 and its receptor. Int Rev Immunol. 1998;16:227–47. doi: 10.3109/08830189809042996. [DOI] [PubMed] [Google Scholar]
- 2.Sanderson CJ, Urwin D. Interleukin-5: a drug target for allergic diseases. Curr Opin Investig Drugs. 2000;1:435–41. [PubMed] [Google Scholar]
- 3.Foster PS, Hogan SP, Yang M, Mattes J, Young IG, Matthaei KI, Kumar RK, Mahalingam S, Webb DC. Interleukin-5 and eosinophils as therapeutic targets for asthma. Trends Mol Med. 2002;8:162–7. doi: 10.1016/s1471-4914(02)02302-x. [DOI] [PubMed] [Google Scholar]
- 4.Ishino T, Robertsonm N, Chaiken I. Cytokine Recognition by Human Interleukin-5 Receptor. In: Litwack G, editor. Vitamins and Hormones. Vol. 71. Academic Press; New York: 2005. pp. 321–344. [DOI] [PubMed] [Google Scholar]
- 5.McKinnon M, Banks M, Solari R, Robertson G. Interleukin-5 and the Interleukin-5 Receptor: Target for Drug Discovery in Asthma. In: Sanderson C, editor. Interleukin-5: From Molecule to Drug Target for Asthma. Marcel Dekker. Inc.; New York: 1999. [Google Scholar]
- 6.Johanson K, Appelbaum E, Doyle M, Hensley P, Zhao B, Abdel-Meguid SS, Young P, Cook R, Carr S, Matico R, et al. Binding interactions of human interleukin 5 with its receptor alpha subunit. Large scale production, structural, and functional studies of Drosophila-expressed recombinant proteins. J Biol Chem. 1995;270:9459–71. doi: 10.1074/jbc.270.16.9459. [DOI] [PubMed] [Google Scholar]
- 7.Milburn MV, Hassell AM, Lambert MH, Jordan SR, Proudfoot AE, Graber P, Wells TN. A novel dimer configuration revealed by the crystal structure at 2.4 A resolution of human interleukin-5. Nature. 1993;363:172–6. doi: 10.1038/363172a0. [DOI] [PubMed] [Google Scholar]
- 8.Dickason RR, Huston DP. Creation of a biologically active interleukin-5 monomer. Nature. 1996;379:652–5. doi: 10.1038/379652a0. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Cook R, Doyle ML, Hensley P, McNulty DE, Chaiken I. Monomeric isomers of human interleukin 5 show that 1:1 receptor recruitment is sufficient for function. Proc Natl Acad Sci U S A. 1997;94:6694–9. doi: 10.1073/pnas.94.13.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morton T, Li J, Cook R, Chaiken I. Mutagenesis in the C-terminal region of human interleukin 5 reveals a central patch for receptor alpha chain recognition. Proc Natl Acad Sci U S A. 1995;92:10879–83. doi: 10.1073/pnas.92.24.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graber P, Proudfoot AE, Talabot F, Bernard A, McKinnon M, Banks M, Fattah D, Solari R, Peitsch MC, Wells TN. Identification of key charged residues of human interleukin-5 in receptor binding and cellular activation. J Biol Chem. 1995;270:15762–9. doi: 10.1074/jbc.270.26.15762. [DOI] [PubMed] [Google Scholar]
- 12.Tavernier J, Tuypens T, Verhee A, Plaetinck G, Devos R, Van der Heyden J, Guisez Y, Oefner C. Identification of receptor-binding domains on human interleukin 5 and design of an interleukin 5-derived receptor antagonist. Proc Natl Acad Sci U S A. 1995;92:5194–8. doi: 10.1073/pnas.92.11.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishino T, Pasut G, Scibek J, Chaiken I. Kinetic interaction analysis of human interleukin 5 receptor alpha mutants reveals a unique binding topology and charge distribution for cytokine recognition. J Biol Chem. 2004;279:9547–56. doi: 10.1074/jbc.M309327200. [DOI] [PubMed] [Google Scholar]
- 14.England BP, Balasubramanian P, Uings I, Bethell S, Chen MJ, Schatz PJ, Yin Q, Chen YF, Whitehorn EA, Tsavaler A, Martens CL, Barrett RW, McKinnon M. A potent dimeric peptide antagonist of interleukin-5 that binds two interleukin-5 receptor alpha chains. Proc Natl Acad Sci U S A. 2000;97:6862–7. doi: 10.1073/pnas.110053997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruchala P, Varadi G, Ishino T, Scibek J, Bhattacharya M, Urbina C, Ryk DV, Uings I, Chaiken I. Cyclic peptide interleukin 5 antagonists mimic CD turn recognition epitope for receptor alpha. Biopolymers. 2004;73:556–68. doi: 10.1002/bip.20001. [DOI] [PubMed] [Google Scholar]
- 16.Ishino T, Urbina C, Bhattacharya M, Panarello D, Chaiken I. Receptor epitope usage by interleukin 5 mimetic peptide. J Biol Chem. 2005;280:22951–61. doi: 10.1074/jbc.M502341200. [DOI] [PubMed] [Google Scholar]
- 17.Dowd CS, Leavitt S, Babcock G, Godillot AP, Van Ryk D, Canziani GA, Sodroski J, Freire E, Chaiken IM. Beta-turn Phe in HIV-1 Env binding site of CD4 and CD4 mimetic miniprotein enhances Env binding affinity but is not required for activation of co-receptor/17b site. Biochemistry. 2002;41:7038–46. doi: 10.1021/bi012168i. [DOI] [PubMed] [Google Scholar]
- 18.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–23. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myszka DG, Morton TA. CLAMP: a biosensor kinetic data analysis program. Trends Biochem Sci. 1998;23:149–50. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 20.Morton TA, Myszka DG, Chaiken IM. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal Biochem. 1995;227:176–85. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 21.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 2000;323:325–40. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 22.van Gunsteren W, Billester S, Eising A, Hunenberger P, Kruger P, Mark A, Scott W, Tironi I. Biomolecular simulation: the GROMOS96 manual and user guide. Hochschulverlag AG an der ETH; Zurich, Switzerland: 1996. [Google Scholar]
- 23.Berendsen H, Postma J, van Gunsteren W, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular Forces. D. Reidel Publishing Co; Dordrecht, Netherlands: 1981. [Google Scholar]
- 24.Ryckaert J, Ciocitti G, Berendsen H. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 25.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 26.Joss L, Morton TA, Doyle ML, Myszka DG. Interpreting kinetic rate constants from optical biosensor data recorded on a decaying surface. Anal Biochem. 1998;261:203–10. doi: 10.1006/abio.1998.2744. [DOI] [PubMed] [Google Scholar]
- 27.Morton TA, Bennett DB, Appelbaum ER, Cusimano DM, Johanson KO, Matico RE, Young PR, Doyle M, Chaiken IM. Analysis of the interaction between human interleukin-5 and the soluble domain of its receptor using a surface plasmon resonance biosensor. J Mol Recognit. 1994;7:47–55. doi: 10.1002/jmr.300070107. [DOI] [PubMed] [Google Scholar]
- 28.Morton TA, Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Methods Enzymol. 1998;295:268–94. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- 29.McDonnell JM. Surface plasmon resonance: towards an understanding of the mechanisms of biological molecular recognition. Curr Opin Chem Biol. 2001;5:572–7. doi: 10.1016/s1367-5931(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 30.Schreiber G. Kinetic studies of protein-protein interactions. Curr Opin Struct Biol. 2002;12:41–7. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 31.Selzer T, Schreiber G. New insights into the mechanism of protein-protein association. Proteins. 2001;45:190–8. doi: 10.1002/prot.1139. [DOI] [PubMed] [Google Scholar]
- 32.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]