Summary
The Escherichia coli SOS response to DNA damage is modulated by the RecA protein, a recombinase that forms an extended filament on single-stranded DNA and hydrolyzes ATP. The RecA K72R (recA2201) mutation eliminates the ATPase activity of RecA protein. The mutation also limits the capacity of RecA to form long filaments in the presence of ATP. Strains with this mutation do not undergo SOS induction in vivo. We have combined the K72R variant of RecA with another mutation, RecA E38K (recA730). In vitro, the double mutant RecA E38K/K72R (recA730,2201) mimics the K72R mutant protein in that it has no ATPase activity. The double mutant protein will form long extended filaments on ssDNA and facilitate LexA cleavage almost as well as wild type, and do so in the presence of ATP. Unlike recA K72R, the recA E38K/K72R double mutant promotes SOS induction in vivo after UV treatment. Thus, SOS induction does not require ATP hydrolysis by the RecA protein, but does require formation of extended RecA filaments. The RecA E38K/K72R protein represents an improved reagent for studies of the function of ATP hydrolysis by RecA in vivo and in vitro.
Keywords: DNA repair, recombination, SOS Response, DNA replication, ATPase
Introduction
The RecA protein plays a central role in recombination, SOS induction and DNA repair in Escherichia coli and many other bacteria. Critical in these functions is the ability of RecA (bound to ATP) to polymerize onto single-stranded DNA (ssDNA) creating a helical nucleoprotein filament. This filament is a dynamic structure where RecA is added to the 3′-proximal end and dissociates through ATP hydrolysis from the 5′-proximal end. The ability to hydrolyze ATP in a DNA-dependent manner is crucial to RecA function in the cell. However, the way in which ATP hydrolysis contributes to RecA function(s) is not fully understood. This study focuses on the role of ATP hydrolysis in facilitating LexA auto-cleavage that is necessary for induction of the SOS response.
In vitro, RecA must first bind ATP or a non-hydrolyzable analog to adopt a high affinity conformation for ssDNA. This ATP-activated form of RecA constitutes an extended nucleoprotein filament documented by electron microscopic observation and analysis (Egelman and Stasiak, 1993; Yu and Egelman, 1992) and is almost certainly equivalent to the RecA* form mentioned in the earlier literature (Clark and Sandler, 1994; Roberts et al., 1978). Other nucleotides are available in vivo, and in vitro experiments show that RecA can bind several of them and retain at least some key functions (McEntee et al., 1981; Menge and Bryant, 1988, 1992a, b; Weinstock et al., 1981a, b, c). The nucleotide dATP substitutes best for ATP in vitro, and several RecA functions are enhanced when dATP replaces ATP in reaction mixtures (Ellouze et al., 1999; Menetski and Kowalczykowski, 1989; Robu et al., 2001; Shan et al., 1997).
Many recA mutants have advanced our understanding of the roles of RecA protein in the cell. A new double mutant combination, RecA E38K/K72R (recA730,2201), has been constructed and is the focus of the current report. The first mutation is RecA E38K (recA730). In vitro, a RecA protein with this single mutation is able to bind ssDNA coated with SSB better than is wild type RecA (Lavery and Kowalczykowski, 1992). In vivo, recA E38K mutant cells are Rec+, UVR and exhibit constitutive SOS (SOSC) expression (Cazaux et al., 1993; Ennis et al., 1995; Wang et al., 1993). This same mutation acts as a suppressor of recFOR mutations (Wang et al., 1993). The capacity of RecA E38K for unassisted displacement of SSB and binding to ssDNA may explain the constitutive SOS phenotype (Lavery and Kowalczykowski, 1992). The other mutant, RecA K72R (recA2201), has a change in a highly conserved lysine residue in the Walker A motif, and it binds but does not hydrolyze ATP. In vitro, this mutant protein functions poorly in the presence of ATP, but will form long filaments and promote DNA pairing in the presence of dATP (Rehrauer and Kowalczykowski, 1993; Shan et al., 1996). It is not clear if the enhancements to RecA function seen in the presence of dATP are physiologically relevant, since mixtures of dATP and ATP designed to approximate the in vivo ratios of these nucleotides (∼1/10) produce a RecA with the properties associated with ATP, not dATP binding (Shan et al., 1997). The LexA repressor of the SOS response undergoes autocatalytic cleavage in the presence of the RecA K72R protein, but at a much reduced rate (Shan et al., 1996). These observations lead to an expectation that RecA K72R should exhibit limited function and particularly should facilitate some SOS induction in vivo. However, in vivo the recA K72R (recA2201) mutant behaves like a null recA mutant with respect to sensitivity to DNA damaging agents and recombination (Konola et al., 1994). The recA K72R mutant does not promote SOS induction in vivo, suggesting that ATP hydrolysis is required for SOS induction (Renzette and Sandler, 2008). This conclusion is discrepant from the in vitro LexA cleavage properties of RecA K72R, and is complicated by the previously unaddressed observation that this mutant protein does not form extended filaments in the presence of ATP in vitro. Thus, in vivo results obtained with recA K72R, in particular its inability to induce SOS expression, could be due to its inability to hydrolyze ATP, its inability to adopt an extended conformation or due to other functional shortcomings when associated with ATP (Rehrauer and Kowalczykowski, 1993).
The RecA E38K/K72R was designed to improve the functional characteristics of the RecA K72R mutant protein, and to permit a more unambiguous determination of in vivo deficiencies that can be traced uniquely to a lack of ATP hydrolytic activity. This necessitates a thorough in vivo and in vitro characterization of this mutant protein.
Results
Experimental Rationale
In principle, the RecA E38K/K72R double mutant protein should retain an inability to hydrolyze ATP, while at the same time exhibiting improved DNA binding and filament formation activities in the presence of ATP. First, we thoroughly characterize the double mutant protein in vitro to document its properties relative to the wild type RecA protein and RecA K72R. We then explore the properties of the RecA E38K/K72R protein in vivo, along with the phenotypes of the cells that express it. Throughout, the major focus is the capacity of the mutant protein to hydrolyze ATP, form filaments on DNA in the extended conformation, facilitate LexA repressor cleavage and to induce the SOS response.
The RecA E38K/K72R Protein is Deficient in ATP and dATP Hydrolysis
First, it is critical to determine if the addition of the E38K mutation had any effect on the ATP hydrolytic function of the K72R mutant protein. The ATP or dATP hydrolytic activity of RecA E38K/K72R was examined and the results are depicted in Figure 1. The RecA E38K/K72R double mutant, like the K72R single mutant protein, exhibits a (d)ATPase activity best described as low to almost undetectable. Both the K72R single mutant and the E38K/K72R double mutant exhibited an apparent kcat between 0.15 min-1 and 0.30 min-1 in the presence of ATP or dATP. This finding is comparable to previous measurements of ATPase activity of the RecA K72R single mutant, with a reported kcat of 0.032 min-1 in the presence of ATP, and a kcat of 0.052 min-1 in the presence of dATP (Rehrauer and Kowalczykowski, 1993). Small differences may reflect the different reaction conditions employed. Wild type RecA, in the presence of either ATP or dATP, is included as a positive control and hydrolyzes ATP with a kcat of 32.10 (± 0.34) min-1 and dATP with a kcat of 42.24 (± 1.23) min-1, in good agreement with reported values (Brenner et al., 1987; Shan and Cox, 1997). The similarity of the ATPase properties of the two RecA mutants indicates that the E38K mutation does not improve the capacity of the RecA E38K/K72R double mutant to hydrolyze ATP or dATP when compared to the RecA K72R single mutant.
Figure 1. The ATPase activity of RecA E38K/K72R compared to the activity of RecA K72R and wild type RecA.
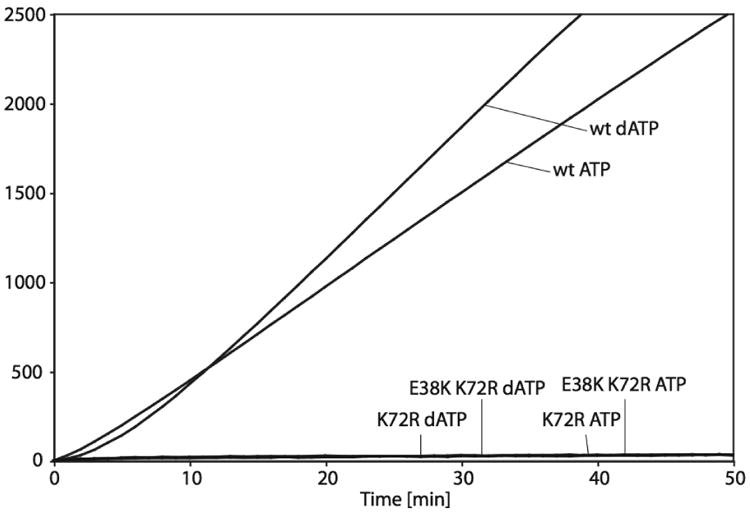
The RecA-catalyzed ATP and dATP hydrolysis in the presence of cssDNA was monitored over time. The reactions were performed as described in “Experimental Procedures.” The two top curves represent hydrolysis of ATP and dATP for wild type RecA protein, and the lower curves represent hydrolysis of ATP and dATP for RecA K72R and RecA E38K/K72R. Time zero corresponds to the time of ATP/SSB or dATP/SSB addition.
The RecA E38K/K72R Protein Forms Filaments Similar to Wild Type RecA on Circular Single Stranded DNA (cssDNA)
We next determined if the double mutant RecA protein can adopt the extended conformation in the presence of ATP. To test this, the capacity of RecA E38K/K72R and RecA K72R to form filaments on cssDNA in the presence of ATP and dATP was observed by electron microscopy (EM) and was compared to filaments of wild type RecA protein. In the first set of experiments, each sample was briefly incubated with ATPγS before spreading, a strategy that has been used previously to stabilize RecA filaments during the spreading process (Bork et al., 2001; Drees et al., 2004; Lusetti et al., 2003; Shan and Cox, 1997; Webb et al., 1997). Representative filaments of each protein are shown in Figure 2, and overall results are summarized in Table 1. The electron micrographs show that in the presence of ATP, the RecA E38K/K72R double mutant forms mostly fully extended filaments (Figure 2E-G), whereas the RecA K72R mutant protein mostly forms short filaments with large regions of cssDNA coated with SSB, referred to as gaps (Figure 2K-M). The RecA E38K/K72R protein formed about 61% full filaments without gaps and 39% filaments with 1 or more small gaps (total of 364 circular filaments counted). The average length (n = 57) of RecA E38K/K72R filaments was 2827 (± 564) nm, which is 78% of the average (n = 19) wild type RecA filament length of 3634 (± 62) nm. No full filaments were detected for RecA K72R and the average (n = 85) filament length was 780 ± 592 nm (Table 1). When dATP is present, the filament formation by both mutant proteins is enhanced. These results are consistent with the recA E38K mutation suppressing the inability of RecA to adopt an extended conformation and completely coat the cssDNA in the presence of ATP when the recA K72R mutation is also present.
Figure 2. RecA E38K K72R forms filaments similar to wild type RecA on cssDNA with ATP or dATP.
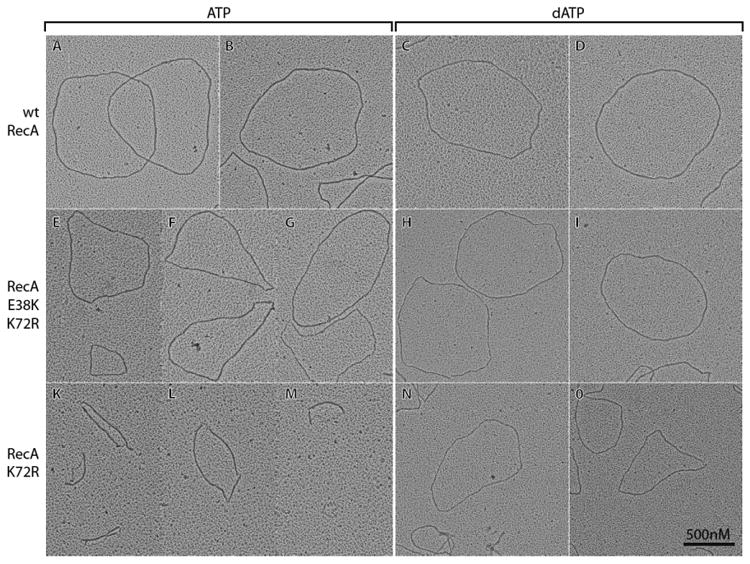
All samples were fixed by the addition of ATPγS prior to spreading. Due to the large variety of filaments observed, representative molecules are shown here. All pictures were taken at a magnification of 15000X and the scale bar shown in Panel O also applies to all other Panels. The contrast of individual images was adjusted uniformly using Adobe Photoshop.
Table 1. Summary of Filament Length and Filament Extension (Pitch) Measurements.
| Filament length | Pitch | Pitch | ||||
|---|---|---|---|---|---|---|
| 3 min ATPγS | 3 min ATPγS | no ATPγS | ||||
| ATP | dATP | ATP | dATP | ATP | dATP | |
| WT | 3634±62 | N.D. | 10.70±1.03 | 10.85±1.23 | 10.48±1.12 | 10.18±1.27 |
| E38K K72R | 2827±564 | N.D. | 10.52±1.03 | 10.41±0.96 | 10.79±1.57 | 10.36±1.27 |
| K72R | 780±592 | N.D. | 10.53±0.92 | 10.36±1.24 | N.A.* | 10.55±1.13 |
All measurements are on M13mp18 cssDNA and lengths are in nm. For filament length measurements n=19 for wt, n=57 for E38K K72R and n=85 for K72R. For pitch measurements 50 individual pitches from 6-10 different molecules were measured for all conditions.
N.A. is not available since RecA K72R did not form filaments under these conditions.
N.D. is not determined.
Further EM experiments were carried out without adding ATPγS prior to spreading, to determine if the ATPγS addition was affecting the results obtained with RecA K72R. The results are shown in Figure 3. The wild type, RecA K72R, and RecA E38K/K72R mutant proteins all exhibit good filament formation in the presence of dATP. In the presence of ATP, full filaments were observed for the wild type and RecA E38K/K72R mutant proteins, although the wild type RecA filaments showed some small discontinuities without the ATPγS addition. Filament extension measured by determining the pitch of the helical RecA filaments did not change significantly, for the wild type and double mutant proteins with ATP or dATP, or with RecA K72R in the presence of dATP (Table 1). Strikingly, filament formation was almost entirely absent when RecA K72R was incubated with ssDNA and ATP, with no ATPγS addition (Figure 3E). This suggests that the short RecA K72R filaments seen in Figure 2 were dependent on the added ATPγS, and that this mutant protein has limited or even no capacity to form filaments, extended or otherwise, in the presence of ATP.
Figure 3. Formation of extended filaments is abolished for RecA K72R in the presence of ATP when not fixed by ATPγS.
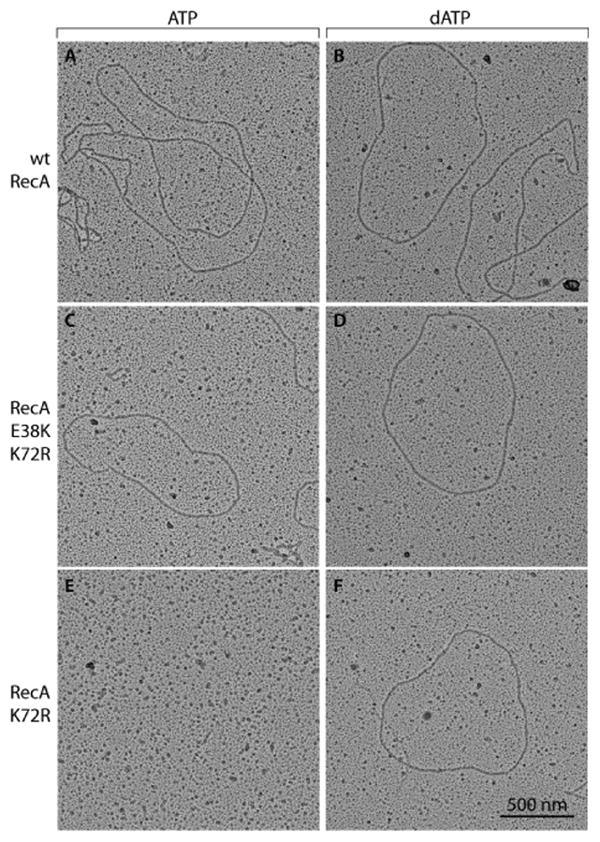
Filament fixation by ATPγS was omitted in all samples to prevent RecA binding to ATPγS from interfering with binding to ATP or dATP. Due to the large variety of filaments observed, representative molecules are shown here. All pictures were taken at a magnification of 15000X and the scale bar shown in Panel F also applies to all other Panels. The contrast of individual images was adjusted uniformly using Adobe Photoshop.
The RecA E38K/K72R Protein Facilitates Cleavage of the LexA Repressor Protein in the Presence of ATP or dATP
Since combining the recA E38K mutation with the recA K72R mutation did not increase the level of ATPase activity and allowed RecA E38K/K72R to adopt the extended conformation in the presence of ATP, we asked whether the E38K alteration would also increase the rate at which LexA could undergo auto-cleavage in the presence of ATP (vs. dATP). In the presence of ATP, RecA E38K/K72R mediates LexA cleavage almost to the extent of wild type RecA (Figures 4A and 4C). Proficient cleavage occurs 15 minutes after LexA addition, and complete cleavage is achieved at the 60-minute time point. In contrast, RecA K72R does not facilitate LexA cleavage when ATP is present under these conditions. In the presence of dATP both RecA mutants facilitate LexA cleavage to a similar level as the wild type RecA protein (Figures 4B and 4D). These results suggest that restoring the capacity to bind ATP and adopt an extended conformation, but not ATP hydrolysis, is central to the mechanism by which RecA E38K suppresses the inability of RecA K72R to induce the SOS response.
Figure 4. RecA E38K/K72R mediates LexA cleavage in the presence of ATP and dATP.
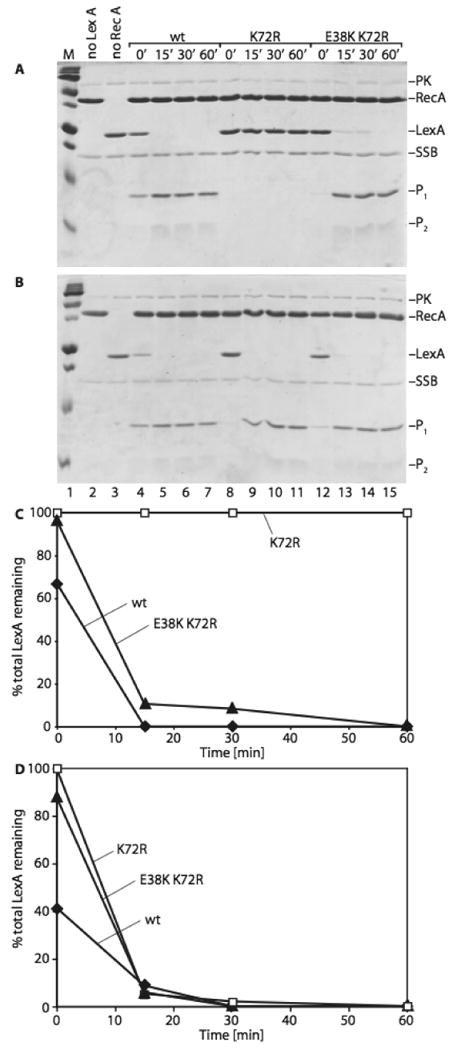
Reactions (50μl) were carried out as described under “Experimental Procedures.” Reactions for Figures 3A and 3B are set up the same way except ATP was used in the experiment depicted in 3A, and dATP was used in 3B. Lane 1 contains a Bio-Rad precision plus protein standard. Lane 2 shows a control reaction lacking LexA protein and lane 3 contains a control reaction without RecA protein. Figures 3C and 3D are quantitative analyses of the gels shown in 3A and 3B, respectively. The graphs show the percentage of uncleaved LexA protein remaining at the different time points. PK stands for pyruvate kinase, included in the reactions as part of the ATP regeneration system. P1 and P2 represent the cleaved LexA fragments.
The RecA E38K/K72R Protein Promotes Homologous Pairing and Formation of Joint Molecule Intermediates, but not Nicked Circular Product Formation in the Presence of ATP or dATP
Finally, to assess the in vitro recombination activity of the RecA E38K/K72R protein, its ability to carry out DNA 3-strand-exchange reactions was analyzed and compared to the strand-exchange activity of RecA K72R and wild type RecA (Figure 5). The RecA E38K/K72R double mutant converted some of the DNA substrates (S1 and S2) to joint molecule intermediates in the presence of either ATP or dATP. No nicked circular product (P) formation was observed in the reactions with RecA E38K/K72R under the conditions tested here. The RecA K72R mutant formed joint molecule intermediates only in the presence of dATP, not ATP. The results obtained for RecA K72R are consistent with previous findings (Rehrauer and Kowalczykowski, 1993; Shan et al., 1996). Included as a control, the nicked circular product of DNA strand-exchange was evident at 30 min for wild type RecA. The 30 minute point was the earliest time point taken in this experiment. The ability of RecA E38K/K72R to form joint molecules in the presence of ATP as well as dATP distinguishes this double mutant from RecA K72R. These results suggest that upon ATP binding, the RecA E38K mutation enables a conformational state that is not adopted in the RecA K72R single mutant in presence of ATP. This conformational state allows homologous pairing to take place.
Figure 5. The RecA E38K/K72R double mutant promotes homologous pairing in the presence of ATP and dATP.
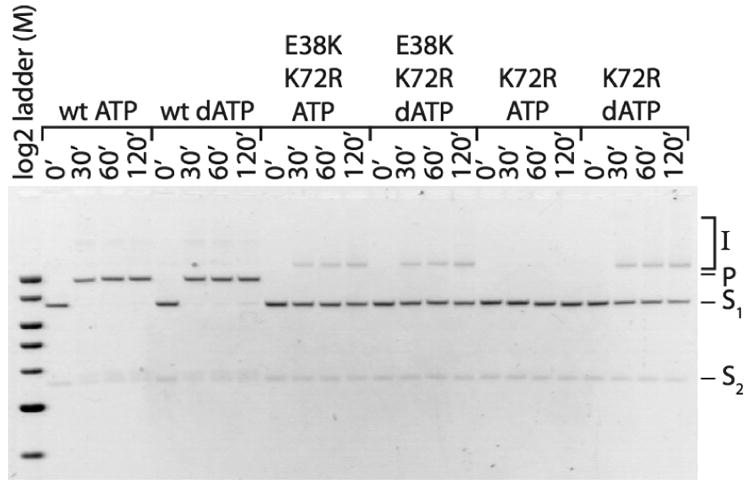
RecA E38K/K72R, RecA K72R and wild type RecA in vitro recombination activities were evaluated as follows. Reactions (50μl) were carried out as described under “Experimental Procedures.” Time points were taken at 0, 30, 60, and 120 minutes. The image was taken with a digital CCD camera utilizing GelExpert software (Nucleotech) and was inverted using Adobe Photoshop. The band labels mean the following: I: joint molecule intermediates; P: nicked circular product; S1: linear double-stranded substrate; S2: circular single-stranded substrate. The molecular weight marker used is a 2-Log DNA Ladder purchased from NEB.
recA E38K Suppresses the Defect in SOS Induction after UV Treatment of recA K72R
We also explored the phenotypes of the recA E38K/K72R double mutant in vivo. The recA E38K mutant exhibits an SOSC expression phenotype in un-irradiated cells and requires no other cofactors (Sommer et al., 1993; Witkin and Kogoma, 1984). To test if recA E38K can suppress recA K72R, the two mutations were combined on a plasmid and then transferred to the chromosome. The strain also contains a sulAp-gfp reporter system (McCool et al., 2004). Therefore, the Relative Fluorescence Intensity (RFI) is a measure of SOS expression. One can then measure the RFI in individual cells in a population and analyze the distribution of cells with a particular intensity in the population as well as the average RFI of the population (Figure 6).
Figure 6. SOS induction is expressed in recA K72R,E38K cells in a recF-dependent manner.
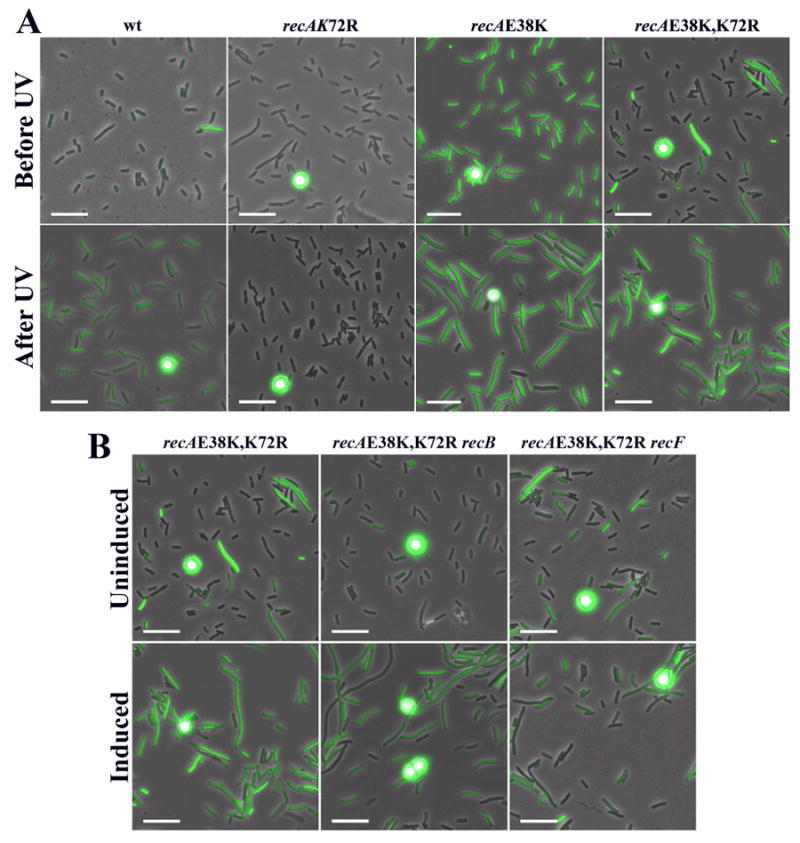
A sulA∷gfp reporter construct is used such that fluorescence intensity is correlated to SOS expression. All figures are merged images between phase contrast and fluorescence microscopy. A) SOS induction in cells expressing different RecA protein variants. Typical cell populations are shown before and after treatment with 5 J/m2 of UV light. B) SOS expression of a recAE38K,K72R strain with mutations in recB or recF before and after treatment with UV irradiation. The circular dots are calibration beads.
Table 2 shows that about 28% of the cells in the population expressing the recA double mutant have a significant level of SOSC expression (defined as six-fold above background) and the average across the population is about six-fold above the levels seen for wild type E. coli. This level of SOSC expression is decreased from recA E38K levels whereas no SOSC is seen for the recA K72R strain. The mechanism of how the double mutant attains SOSC will be tested below. Given this modest background of SOSC expression, it is possible to test if the SOS response is inducible in the remainder of the population. To do this, cells were grown in minimal media and treated with 5 J/m2 of UV light. Cells were then incubated for 90 minutes and the amount of fluorescence was measured. Table 2 shows that about 90% of the double mutant cells are now induced for SOS expression with an average RFI of 20. In comparison, about 70% of the wild type cells are induced to an average RFI of 8. Typical cell populations are shown in Figure 6A. This shows that SOS induction can occur in a timely manner in the recA E38K/K72R double mutant cells. Given the background levels, the final proportion of induced and uninduced cells in the population and the increase in average RFI is similar to wild type.
Table 2. The Level of SOS Expression before and after UV Treatment.
| SOS Expression | ||||||
|---|---|---|---|---|---|---|
| Genotype | Before UV Treatment | After UV Treatment | ||||
| Strain | recA | other | Relative Intensity | % Population High SOS | Relative Intensity | % Population High SOS |
| SS996 | + | 1.00 | 0.8 ± 0.4 | 8.1 ± 0.3 | 71.0 ± 4.0 | |
| SS1426 | del(recA) | 0.3 ± 0.4 | 0.0 ± 0.2 | 0.3 ± 0.2 | 0.00 ± 0.1 | |
| SS4629 | 730 (E38K) | 45.0 ± 1.3 | 99.0 ± 1.0 | N.D. | N.D. | |
| SS5602 | 2201(K72R) | 0.3 ± 0.2 | 0.0 ± 0.2 | 0.7 ± 0.2 | 0.00 ± 0.2 | |
| SS5690 | 730,2201 | 5.7 ± 0.9 | 28 ± 0.8 | 20 ± 1.9 | 91.0 ± 3.0 | |
| SS6418 | 803,2201 | 0.6 ± 0.8 | 0.0 ± 0.3 | 0.9 ± 0.7 | 0.1 ± 0.8 | |
| SS6441 | 730,2201 | recB268 | 1.3 ± 0.3 | 2.1 ± 0.3 | 9.6 ± 0.9 | 55.0 ± 2.0 |
| SS5691 | 730,2201 | recF4115 | 4.7 ± 0.4 | 25.0 ± 0.7 | 4.6 ± 0.9 | 24.0 ± 1.0 |
Shows the average of 1000-3000 total cells analyzed from three different experiments.
N.D. is not determined.
To further test if the mechanism of SOS induction of the recA E38K K72R double mutant is similar to that of wild type, the dependence of SOS induction on recF was tested. It is known that SOS induction after UV treatment is attenuated in a recF mutant (Hegde et al., 1995; McPartland et al., 1980; Thomas and Lloyd, 1983; Wackernagel, 1987). Therefore recA E38K/K72R was combined with recF4115. This recF4115 strain, in which the RecF protein is inactivated, showed approximately the same levels of SOSC expression as the recF+ strain (Table 2 and Fig. 6B). After UV treatment, no SOS induction was detected. This suggests that the recA E38K/K72R mutant is using a mechanism similar to that used by wild type recA to induce the SOS response after UV treatment.
Upon characterizing other phenotypes of the recA E38K/K72R mutant, Table 3 shows that conjugational recombination and resistance to UV irradiation are slightly enhanced relative to the null allele but are still well below the wild type levels (Table 3). This suggests that recA E38K is able to suppress the SOS deficiencies of the recA K72R mutant. However, it is limited in its abilities to suppress the recombinational and repair deficiencies of the strain, presumably because these functions require ATP hydrolysis.
Table 3. Summary of Sensitivity to UV light and Recombination Phenotypes of RecA Mutants.
| Strain | recA | Survival to UV a | Recombinationb |
|---|---|---|---|
| SS996 | + | 73 ± 2.33 | 8.44 ± .93 |
| SS1426 | del(recA) | < 10 -2 | < 10 -4 |
| SS4629 | 730 (E38K) | 75 ± 4.29 | 9.03 ± 1.02 |
| SS5602 | 2201 (K72R) | < 10 -2 | < 10 -4 |
| SS5690 | 730,2201 | 0.15 ± 0.18 | 0.012 ± 0.0073 |
| SS6418 | 803(V37M),2201 | < 10 -2 | < 10 -4 |
UV Resistance is measured by % survival of strains after 10J of UV irradiation
Conjugation frequency reported as % transconjugants/100 donors
The recA803 Mutation does not Suppress the recA K72R Mutant Phenotypes
As mentioned above, E. coli strains expressing RecA E38K are SOSC, and the RecA E38K protein binds ssDNA coated with SSB better than wild type RecA. To try to correlate which activity may be responsible for suppressing the inability of recA K72R to induce the SOS response, another mutation, recA803, was combined with recA K72R. RecA803 (V37M) is a mutation in the codon adjacent to the recA E38K mutation. Both RecA V37M and RecA E38K compete with SSB for binding to ssDNA (Lavery and Kowalczykowski, 1992; Madiraju et al., 1992). However, recA V37M does not display an SOSC phenotype like recA E38K (Madiraju et al., 1988; Madiraju et al., 1992). The effect of the RecA V37M mutation on extended filament formation is not known. It is presumed that the V37M and E38K mutations of RecA differ in their capacity to bind ssDNA and form extended filaments sufficient to generate constitutive SOS induction. It is thus interesting to determine if the V37M mutation can suppress recA K72R in the same manner as recA E38K does. However, the recA V37M/K72R double mutant has phenotypes essentially indistinguishable from recA K72R (Table 2 and 3). This experiment suggests that the E38K mutation confers a particularly robust capacity to bind ATP and/or to adopt an extended conformation sufficient to suppress the deficiencies in SOS induction of the recA K72R mutant.
The Mechanisms of SOSC Expression in recA E38K and recA E38K/K72R are Different
The decrease in SOSC expression in the recA E38K/K72R double mutant as compared to the recA E38K single mutant suggests that the ATPase activity is important for SOSC expression. In work to be published elsewhere, it is shown that recA E38K SOSC expression is recBCD-independent (Long et al., 2008). To further test if the mechanism of SOSC expression in recA E38K and a recA E38K/K72R strain is different, the recB-dependence was tested. Table 2 and Figure 6B show that a recA E38K/K72R recB268∷tet strain has four-fold less (essentially background levels) SOSC expression than recA E38K/K72R recB+. The recB mutation has little effect on the ability of the strain to induce SOS after UV treatment as expected (Keller et al., 2001). Thus, a recB mutation has a substantially greater effect on strains expressing the double mutant RecA E38K/K72R than it has on strains expressing the RecA E38K protein.
Discussion
There are four major conclusions arising from this work. First, ATP hydrolysis is not required for RecA to mediate SOS induction in E. coli. Second, efficient formation of RecA filaments in the extended conformation is necessary for SOS induction, even though ATP hydrolysis is not. Third, the filament formation deficiency of RecA K72R is suppressed by the E38K mutation. The suppression does not entail the uniformly constitutive SOS induction seen in cells expressing the E38K mutant protein. The fraction of cells expressing SOS increases substantially after UV irradiation in cells expressing the RecA E38K/K72R protein, and the expression depends on the function of RecF protein. The mechanism of SOS induction is thus largely preserved in this double mutant RecA protein. Even though SOS induction between the double mutant and wild type strains are very similar, it is not possible to rule out that SOS induction in the mutant may occur by a different mechanism. Finally, we present the RecA E38K/K72R double mutant protein as an improved reagent for the exploration of questions focused on the in vivo and in vitro role of RecA-mediated ATP hydrolysis. Unlike the K72R single mutant, the double mutant forms long extended filaments in the presence of ATP almost as well as does the wild type RecA protein. Given the preponderance of ATP over dATP in the intracellular environment, this allows an improvement of consistency between experiments carried out in vivo and in vitro.
ATP hydrolysis is used to enhance or promote a wide range of cellular processes. However, the role of an ATPase activity in the molecular function of a protein is not always obvious. Since the discovery of the Walker Motif (Walker et al., 1982) and the fact that a simple conservative change of a lysine to an arginine in a conserved GKT portion of the motif will usually eliminate the ATPase activity, hundreds of genes/proteins with putative ATPase activities have been altered in this way. For RecA protein, the K72R mutation has provided many insights (Rehrauer and Kowalczykowski, 1993; Shan et al., 1996), yet the precise role of the RecA ATPase activity in recombination and SOS induction has been elusive. This work shows the intragenic suppression of an ATPase mutant for a particular function of RecA, SOS induction, without restoration of the ATPase activity. It should be noted that while SOS induction is restored by the addition of E38K to the K72R mutation, the substantial deficiencies of recA K72R in the complicated recombination functions of RecA are altered very little by the same combination of mutations. The small amount of recombination ability and UV resistance seen the recA E38K/K72R double mutant, relative to the recA null mutant, may be due to the ability of the mutant protein to stabilize replication forks (Courcelle and Hanawalt, 2003), promote a limited strand-exchange in certain situations, or induce compensatory activities during SOS. In some ways, the recA E38K/K72R mutant can be considered a separation of function mutant, allowing investigators to more readily distinguish functions that do not need ATP hydrolysis from ones that do.
The mechanism of suppression almost certainly involves a structural contribution of the E38K mutation that provides the capacity to make long RecA filaments in the extended conformation in the presence of ATP. RecA K72R is not an inactive protein and can bind dATP and perform a variety of functions in vitro (Rehrauer and Kowalczykowski, 1993; Shan et al., 1996). But the capacity to form extended RecA filaments in the presence of ATP appears to be critical in vivo, where ATP is present at approximately 10-fold higher concentrations than dATP. The recA E38K/K72R double mutant demonstrates that the ATPase activity of RecA is not necessary for SOS induction as long as the active extended form of the RecA filament is accessible. An alternative possibility, that the ATPase activity is necessary for converting RecA+ to the extended conformation and that the recA E38K mutation allows the bypass of this requirement, is rendered unlikely by the capacity of ATPγS to induce extended filament formation in wild type RecA in vitro without significant hydrolysis (Weinstock et al., 1981d), a capacity seen in the current study.
RecA E38K/K72R is a hybrid that does not exhibit all of the properties of the RecA E38K single mutant protein. In a sense, the E38K mutation is enhancing the function of the K72R protein, and the K72R is weakening aspects of the function of E38K. The two mutations are not additive, but compensatory. The SOS response is not constitutively induced in the double mutant, as it is in strains expressing recA E38K. However, the ability of the E38K variant to suppress the inability of recA K72R to induce the SOS response is likely related to its capacity to form extended filaments, as exhibited by a constitutive SOS phenotype. Many other recA mutants have been shown to have an SOSC phenotype (Ennis et al., 1995; Lavery and Kowalczykowski, 1992; McCall et al., 1987; McGrew and Knight, 2003; Mirshad and Kowalczykowski, 2003; O'Reilly and Kreuzer, 2004; Tateishi et al., 1992). It is plausible that other recA variants that confer an SOSC phenotype may be able to suppress the SOS deficient phenotype of recA K72R, but it is also clear that not all of them do. In this study, a combination of recA803 (V37M) with the K72R mutation did not lead to substantial SOS induction. We hypothesize that there is some threshold of ssDNA binding and SSB displacement capacity in a RecA variant that is needed to observe constitutive SOS expression. There may be yet a higher threshold for ssDNA binding and SSB displacement that is required to suppress the considerable deficiency in those activities displayed by RecA K72R protein. The combination of recA803,4161 (V37M, ΔC17) exhibits SOSC expression in most cells whereas either single mutant showed very little if any SOSC expression (Long et al., 2008). This double mutant supplied some ability to suppress the SOS defect after UV exposure of recA K72R, but did not suppress as well as recA E38K (NR and SJS, unpublished result).
One of the roles of RecA ATPase activity is to allow RecA to dissociate from the DNA. If so, then how does the RecA E38K/K72R protein come off the DNA to allow replication and cell growth? One can imagine that a protein tightly bound to replication forks might be detrimental to replication restart, or could pose a block for a subsequent replication fork. This might explain why another recA ATPase-deficient mutant, recA E96D, is lethal (Campbell and Davis, 1999). Under certain circumstances, the UvrD helicase is critical for removing RecA from replication forks where RecA binding is detrimental (Flores et al., 2005; Veaute et al., 2005). It is possible that UvrD is essential for this purpose in the recA E38K/K72R mutant strain. Testing this has shown that uvrD and recA E38K/K72R are synthetically lethal (NR and SJS, unpublished results).
The recA E38K mutation acts as a suppressor of recF null mutations (Lavery and Kowalczykowski, 1992; Wackernagel, 1987; Wang et al., 1993). It was thus somewhat unexpected that recA E38K/K72R requires recF function for SOS induction after UV treatment. This again suggests that the double mutant does not retain all the DNA binding and filament formation capacity of the E38K mutant. A requirement for recF function is a hallmark of SOS induction in wild type cells. The similar requirement in strains expressing recA E38K/K72R provides another indication that this ATPase mutant is utilizing the normal SOS induction pathway.
Whereas this paper focuses mostly on the ability of RecA E38K to suppress aspects of the RecA K72R phenotypes, it also tests whether the RecA-ATPase function is required for any of the RecA E38K phenotypes. We found that ATP hydrolysis is very important for RecA to carry out recombination after conjugation and to repair DNA after UV treatment. Both of these functions are reduced in RecA E38K K72R by almost 500-fold compared to wild type, but are not quite reduced to the level of a null mutant (Table 3).
Is there any role for the RecA ATPase activity in SOS induction in a wild type cell? It is clear that RecA E38K K72R protein can respond to UV treatment in a manner very similar to the wild type protein and induce the SOS response without ATPase activity. Thus, while it is not required for SOS induction, it is may be possible that ATP hydrolysis has an indirect role in negatively regulating the SOS Responseresponse. This idea stems from the observations that in a population of wild type cells under normal growth conditions, approximately 15% of the cells have a RecA-DNA filament (Renzette et al., 2005), yet less than 1% of the cells are induced for SOS (McCool et al., 2004; Pennington and Rosenberg, 2007). Thus, not all RecA-DNA filaments are inducing the SOS Response. It is possible that if the default function of RecA bound to DNA in cells is recombinational repair, then active engagement in recombinational repair may inhibit SOS induction. To induce SOS, one might then require the temporary suspension of functions requiring the ATPase so that LexA cleavage could occur. This idea is supported by experiments that show a competition between LexA and a homologous duplex to interact with a RecA-DNA filament (Rehrauer et al., 1996). Some of the many proteins now being shown to regulate RecA function (Cox, 2007) may play a role in determining which pathway is undertaken.
The enhancement of RecA function in vitro with dATP might suggest that dATP plays some role in RecA function in vivo. This would require a locally high concentration of dATP near the sites of recombinational DNA repair. The capacity of RecA K72R/E38K (which can utilize ATP) to function in SOS induction in vivo, coupled with the lack of in vivo function of RecA K72R (which requires dATP), argues that no such local concentration of dATP exists in the cell. The role of dATP in RecA function, if any, remains a mystery.
In conclusion, this work shows that RecA E38K/K72R can serve as useful reagent to study RecA filament dynamics since the protein has many properties similar to wild type RecA. For example, RecA E38K/K72R could be used in studies of RecA filaments bound to ATP, as was previously performed using RecA E96D (VanLoock et al., 2003). However, RecA E96D retains significant residual ATPase activity (VanLoock et al., 2003). The near complete inactivation of ATPase activity in the RecA E38K/K72R protein offers a new tool to study this important process in RecA biology. Furthermore, recA E96D is lethal to host cells (Campbell and Davis, 1999) while recA E38K/K72R is not (when coupled with a sulB103 mutation).
Experimental Procedures
Bacterial strains
All bacterial strains used in this work are derivatives of E. coli K-12 and are described in Table 4. The protocol for P1 transduction has been described elsewhere (Willetts and Mount, 1969). Protocols for assaying UV sensitivity and conjugal recombination have been described elsewhere (Clark and Margulies, 1965; Sandler et al., 1996).
Table 4. Strains Used in this Work.
| Strain | recA | Other relevant genotype | Origin or Reference |
|---|---|---|---|
| GY8382 | 938∷cat | (Marsh and Walker, 1987) | |
| JC13509 a | + | Lab Stock | |
| JC18825 | + | recF4115 tnaA300∷tn10 | (Sandler, 1994) |
| JJC315 | + | recB268∷tn10 | (Michel et al., 1997) |
| SS996 | + | Ωgfp b | (McCool et al., 2004) |
| SS1426 | + | recF4115 Ωgfp | JC18825 → SS996 |
| SS1433 | + | recB268∷tn10 Ωgfp | JJC315 → SS996 |
| SS1436 | 938∷cat | Ωgfp | GY8383 → SS996 |
| SS4629 | 730 | Ωgfp | (Long et al., 2008) |
| SS4660 | + | recB268∷tn10 Ωgfp | pKD46 → SS1433 c |
| SS5602 | 2201 | ygaD1∷kan Ωgfp | (Renzette and Sandler, 2008) |
| SS5687 | 730,2201 | ygaD1∷kan | This Work |
| SS5690 | 730,2201 | ygaD1∷kan Ωgfp | SS5687 → SS996 |
| SS5691 | 730,2201 | ygaD1∷kan recF4115 tnaA300∷tn10 Ωgfp | SS5687 → SS1426 |
| SS6408 | 803,2201 | ygaD1∷kan | This Work |
| SS6418 | 803,2201 | ygaD1∷kan Ωgfp | SS6408 → SS996 |
| SS6441 | 730,2201 | ygaD1∷kan recB268∷tn10 Ωgfp | SS5687 → SS4660 d |
JC13509 has the following partial genotype: sulB103- lacMS286 Φ80dIIlacBK1 argE3 his-4 thi-1 xyl-5 mtl-1 rplS31 tsx. The lacMS286 Φ80dIIlacBK1 codes for two partial non-overlapping deletions of the lac operon. See (Konrad, 1977; Zieg and Kushner, 1977).
attλ∷sulApΩgfp-mut2 is abbreviated Ωgfp.
pKD46 expresses the Lambda Red proteins (Datsenko and Wanner, 2000).
P1 transduction with SS4660 as a recipient was carried out as normal with the exception that the strain was grown in Luria broth with ampicillin (50 μg/mL) and induced with 0.2% arabinose 1 hour before harvesting the culture.
Plasmid Construction
recA E38K/K72R was cloned into the NdeI and HindIII sites of pET21a (Novagen), creating pEAW483. pNR114 was created by replacing the NcoI and BlpI fragment of pSJS1373 with the homologous fragment of pEAW483.
Strain Construction
The recA E38K/K72R mutation was placed on the chromosome similarly to other recA mutations (Renzette and Sandler, 2008). Briefly, pNR114 was digested with BamHI and the 4.5kb linear fragment was purified. This fragment encodes ygaD1∷kan recA E38K/K72R recX and was used to transform cells expressing the Red recombinase (SS1153) (Datsenko and Wanner, 2000). Transformants were selected on Luria broth and kanamycin and screened for UV sensitivity. All strains in this study were derived from JC13509, which contains a sulB103 mutation. SulB103 is an allele of ftsZ that causes FtsZ to be insensitive to the action of the SulA cell division inhibitor (Bi and Lutkenhaus, 1990).
Buffers
P buffer contained 20mM potassium phosphate (pH 6.8), 1 mM DTT, 0.1 mM EDTA, 10% (w/v) glycerol. R buffer contained 20 mM Tris-HCl, 80% cation (pH 7.5), 1 mM DTT, 0.1 mM EDTA, 10% (w/v) glycerol.
Proteins and Biochemicals
The E. coli wild type RecA and single-strand DNA binding (SSB) proteins were purified as described (Lohman et al., 1986; Shan et al., 1996). The concentration of the purified SSB protein was determined from the absorbance at 280 nm using the extinction coefficient of 2.83 × 104 M-1 cm-1 (Lohman and Overman, 1985).
E. coli LexA repressor protein was purified using a combination of published protocols (Little et al., 1994; Schnarr et al., 1985) from an E. coli strain STL327/pT7pol26 transformed with the pEAW329 plasmid that has the lexA gene under the control of an isopropyl-1-thio-β-D-galactopyranoside-inducible promoter. Cell growth and harvesting was performed as described previously (Lusetti et al., 2003). About 22.9 g of cells were thawed in 100 ml Buffer A (50 mM Tris-HCl pH 8.0, 0.5 mM EDTA, 10% (w/v) sucrose, 1 mM DTT, 200 mM NaCl). Cell lysis by addition of 5 mg/ml lysozyme in Buffer A, and subsequent sonication were performed. The cell debris was pelleted by centrifugation at 16,000 rpm for 60 minutes. Contaminants were precipitated by adding 0.07 ml of 5% polyethyleneimine per ml of solution and pelleted by centrifugation at 12,000 rpm for 10 minutes. Solid ammonium sulfate at a final concentration of 0.4 g/ml was added to the supernatant and the precipitated LexA protein was washed twice with Buffer A plus 0.49 g/ml ammonium sulfate. The final pellet was resuspended in 10 ml of Buffer B (20 mM potassium phosphate pH 7.2, 0.1 mM EDTA, 10% (w/v) glycerol, 80 mM NaCl) plus 500 mM NaCl. Column chromatography was carried out as described in (Little et al., 1994) up through the hydroxyapatite purification step. The concentration of LexA protein was determined using an extinction coefficient of 7300 M-1 cm-1.
The RecA K72R protein was purified as described (Shan et al., 1996) with the following changes. Briefly, 16 g of cell paste were thawed on ice overnight. Cell lysis, polyethyleneimine precipitation, and ammonium sulfate extraction and precipitation were performed as described (Shan et al., 1996). The resulting pellet was resuspended in 70 ml of R buffer plus 1M ammonium sulfate and loaded onto a Phenyl-Sepharose low sub 6 FF column. The column was washed with three column volumes of the same buffer and a 10-column volume gradient was run in R buffer, with a gradient of ammonium sulfate from 1.0 M to 0 M. SDS-PAGE analysis revealed that the RecA mutant protein was in the flow through peak fractions, which were dialyzed versus R buffer. The RecA K72R protein then was loaded onto a Source 15 Q column, washed with three column volumes of R buffer and eluted by a 10-column volume gradient from R buffer to R buffer plus 1 M KCl. The peak fractions were analyzed by SDS-PAGE, revealing that the RecA K72R protein eluted at 500 mM KCl. The RecA mutant containing fractions were pooled, dialyzed against P buffer and loaded onto a ceramic hydroxyapatite column, washed with three column volumes of P buffer and eluted by a 10 column volume gradient from 20 mM phosphate to 500 mM phosphate. The peak fractions were analyzed by SDS-PAGE, pooled and concentrated by ammonium sulfate precipitation (0.35 mg/ml). The pellet was resuspended in R buffer, dialyzed against R buffer, frozen in liquid N2 and stored at -80°C. The RecA K72R protein was free of detectable nuclease activity. The concentration of the RecA K72R protein was determined using the wild type RecA protein extinction coefficient (2.23×104 M-1 cm-1).
RecA E38K/K72R protein was purified using modifications to previously described protocols (Cox et al., 1981; Lusetti et al., 2003). All the steps were carried out at 4 °C. Thirteen and a half g of cell paste were thawed on ice overnight in 45 ml of 25% (w/v) sucrose and 250 mM Tris-HCl (80% cation, pH 7.5). Cells were lysed by incubation with a solution containing 250 mM Tris-HCl (80% cation, pH 7.5) and 2.5 mg/ml lysozyme (final concentration) followed by the addition of 0.4 ml per ml solution of 25 mM EDTA, six cycles of sonication for 1 minute (0.5 s on 0.5 s off) at output 60% (Fischer Scientific Sonic Dismembrator Model 500), and 60 minutes of centrifugation at 16,000 rpm in a Beckman JLA-16 rotor. The RecA was precipitated by drop wise addition of 0.111 ml of 5% (w/v) polyethyleneimine (pH 7.5) per ml of solution. The pellet was resuspended in 130 ml of R buffer and the RecA protein was extracted two times with 110 ml of R buffer plus 300 mM ammonium sulfate. The RecA was precipitated by slow addition of ammonium sulfate to a final concentration of 0.28 g/ml. The resulting pellet was washed three times with 100 ml of R buffer plus 0.28 g/ml ammonium sulfate. The pellet was then resuspended in 45 ml R buffer plus 1M ammonium sulfate and loaded onto a Phenyl-Sepharose column, washed with two column volumes of R buffer plus 1M ammonium sulfate and then eluted with three column volumes of R buffer. The flow-through peak fractions contained the RecA as determined by SDS-PAGE analysis. These fractions were pooled and dialyzed versus P buffer. The protein was then loaded onto a ceramic hydoxyapatite column, washed with two column volumes of P buffer and eluted with a linear gradient from 20 mM to 1M phosphate buffer over 10 column volumes. RecA eluted at 120 mM phosphate, as determined by SDS-PAGE, and was pooled and precipitated by adding 0.35 g/ml of solid ammonium sulfate. The resulting pellet was resuspended in 20 ml of R buffer plus 1M arginine hydrochloride, loaded onto a Sephacryl S-100 Hi Prep 10/60 column (purchased from GE Healthcare) and washed with two column volumes of R buffer plus 1M arginine hydrochloride. The peak fractions were analyzed by SDS-PAGE. Those fractions containing RecA were pooled, dialyzed into R buffer, flash-frozen in liquid N2 and stored at -80°C. The concentration of the RecA E38K/K72R protein (37,896 Da) was determined from the absorbance at 280 nm using the extinction coefficient for wild type RecA (2.23 × 104 M-1 cm-1). The protein was free of detectable nuclease activity.
DNA Substrates
Circular single-stranded DNA (cssDNA) from bacteriophage M13mp18 (7229 nucleotides) was prepared as described (Neuendorf and Cox, 1986). The concentration of cssDNA was determined by absorbance at 260 nm, using 36 μg ml-1 A260-1 as the conversion factor. All DNA concentrations are given in μM nucleotides. Linear double stranded DNA (lds DNA) used in the DNA strand-exchange experiments was generated by complete digestion of M13mp18 circular double-stranded DNA (cds DNA) using PstI according to the manufacturer's instructions.
DNA Three-strand-exchange Reactions
Three-strand-exchange reactions were carried out in standard strand-exchange buffer containing 25mM Tris-OAc (80% cation, pH 7.4), 1mM DTT, 5% (w/v) glycerol, 3mM potassium glutamate, and 10 mM Mg(OAc)2. An ATP regeneration system consisting of 10 units/ml pyruvate kinase and 2 mM phosphoenolpyruvate was included. All incubations were carried out at 37 °C, and all listed reagent concentrations are final. The wild type RecA or RecA mutant protein (4 μM) were preincubated with 10 μM M13mp18 circular single stranded DNA for 10 min. SSB protein (1 μM) and ATP (3 mM) were then added, followed by another 10-min of incubation. The reactions were initiated by the addition of 10 μM M13mp18 linear double stranded DNA (ldsDNA). The reactions were incubated for 120 min and time points were taken as indicated in the figure legends. To stop the reaction, a 10-μl aliquot was removed and added to 5 μl of a solution containing 15% Ficoll, 0.25% xylene cyanol, 72 mM EDTA, and 4% SDS. Samples were subjected to electrophoresis in 0.8% agarose gels with TAE buffer, stained with ethidium bromide, and exposed to ultraviolet light. Gel images were captured with a digital CCD camera utilizing GelExpert software (Nucleotech) and inverted using Adobe Photoshop.
ATPase Assay
A coupled spectrophotometric enzyme assay (Lindsley and Cox, 1990; Morrical et al., 1986) was used to measure the DNA-dependent ATPase activities of the wild type RecA, the RecA E38K/K72R and the RecA K72R proteins. The regeneration of ATP from PEP and ADP was coupled to the oxidation of NADH and was observed as a decrease in absorbance at 380 nm (the maximal absorbance for NADH is at 340 nm, but 380 nm wavelength was used so that the signal remained within the linear range of the spectrophotometer for the duration of the experiment). The assays were carried out in a Varian Cary 300 dual beam spectrophotometer equipped with a temperature controller and a 12-position cell changer. The cell path length and band pass were 1 cm and 2 nm, respectively. The NADH extinction coefficient at 380 nm of 1.21 mM-1 cm-1 was used to convert the amount of NADH oxidized to the amount of ATP hydrolyzed. Reaction mixtures (400 μl) contained 5 μM circular M13mp18 ssDNA, 3 μM of wild type or mutant RecA protein, and a coupling system of 1.5 mM NADH and 10 units/ml lactate dehydrogenase in standard strand-exchange reaction buffer. The reactions were started after a 10-minute incubation of the reaction mixtures by the addition of a mixture of ATP and SSB protein at 3 mM and 0.4 μM final concentrations, respectively.
LexA Repressor Cleavage Assay
Reactions were carried out at 37°C and contained 25 mM Tris(OAc) (80% cation, pH 7.5), 3mM Mg(OAc)2, 5% (w/v) glycerol, 3 mM potassium glutamate, 1mM DTT, an ATP regeneration system (2 mM phosphoenolpyruvate, 10 units/ml pyruvate kinase), 9 μM circular M13mp18 ssDNA, and 3 μM wild type or mutant RecA protein. After 6 min incubation, 3 mM ATP or dATP and 1.5 μM SSB protein were added, followed by another 6 min of incubation. To initiate the reactions, 3 μM LexA repressor protein was added. Time points were taken as indicated in the figure legends by adding 10 μl of the reaction solutions to 5 μl Laemmli buffer. The samples were analyzed by SDS-PAGE on 17% polyacrylamide gels. Totallab TL100 v2006c software was used to quantify the bands on the gels.
Electron Microscopy
A modified Alcian method was used to visualize RecA filaments. Activated grids were prepared as previously described (Lusetti et al., 2003).
Samples for electron microscopy analysis were prepared as follows. All incubations were carried out at 37°C. Wild type RecA protein, RecA E38K/K72R, or RecA K72R protein (3 μM) was preincubated with 6 μM M13mp18 circular ssDNA, 25 mM Tris-OAc (80% cation; pH 7.5) buffer, 5% (w/v) glycerol, 3 mM potassium glutamate, and 10 mM Mg(OAc)2 for 10 min. An ATP regeneration system of 10 units/ml creatine phosphokinase and 12 mM phosphocreatine was also included in the incubation. ATP and SSB protein were added to 3 mM and 0.6 μM, respectively, and the reaction was incubated for another 10 min. Where indicated, ATPγS was then added to 3 mM to stabilize the filaments, followed by another 3 min incubation.
A 3 μl sample of the reaction mixture described above was diluted 8-fold with 200 mM ammonium acetate, 10 mM Hepes (pH 7.5), and 10% glycerol and adsorbed to an activated carbon grid (Alcian grid) for 3 min. The grid was then touched to a drop of the same buffer, followed by floating on a second drop of the buffer for 1 min. The sample was then stained by touching to a drop of 5% uranyl acetate followed by floating on a fresh drop of 5% uranyl acetate solution for 30 s. Finally, the grid was washed by touching to a drop of double distilled water followed by successive immersion in two 10-ml beakers of double distilled water and one beaker of 100% ethyl alcohol. After the sample was dried, it was rotary-shadowed with platinum. This protocol is designed for visualization of complete reaction mixtures, and no attempt was made to remove unreacted material. Although this approach should yield results that provide insight into reaction components, it does lead to samples with a high background of unreacted proteins.
To determine the proportion of the molecules that were either fully or partially coated by RecA E38K K72R protein, or bound only by the SSB protein, in the presence of ATP, 423 molecules from three separate regions of the grid were counted at an identical magnification. A RecA filament was considered gapped if the ssDNA molecule it was bound to had a detectable region of SSB-coated DNA of any size.
Imaging and photography were carried out with a TECNAI G2 12 Twin Electron Microscope (FEI Co.) equipped with a GATAN 890 CCD camera. Digital images of the nucleoprotein filaments were taken at 15,000 X magnification. Filament lengths were measured using MetaMorph analysis software. Nineteen filaments from wild type RecA protein, 51 filaments from RecA K72R E38K, and 85 filaments from RecA K72R were measured. Each filament was measured 3 times, and the average length was calculated. To measure the helical pitch of RecA filaments MetaMorph analysis software was used. The distance between 50 individual striations for each condition was measured and averaged. A Cytochrome C method was used to visualize M13mp18 circular ssDNA to check the purity of the DNA. Samples were prepared as described previously (Schnos and Inman, 2000) except that the spreading solutions were made by combining 32 μl of 1 M Na2CO3, 40 μl of 0.126 M Na2EDTA, 400 μl of 37% HCHO, 292 μl of 100% formamide, and 10 μl of 5 M KOH. The final DNA concentration was 0.004 μg/μl.
Preparation of Cells for Microscopy
To measure SOS expression in either log phase or UV treated cells using quantitative fluorescence microscopy, the protocol is as described previously (McCool et al., 2004). For all UV treatment experiments, cultures were irradiated with 5 J/m2 of UV light at 254 nm using a germicidal lamp. Processing of images and quantitation of relative fluorescence intensity was as described previously (McCool et al., 2004).
Acknowledgments
NR, EL, and SJS were supported by AI059027 from the National Institutes of Health. MCG, SC-P, and MMC were supported by GM32335 from the National Institutes of General Medical Sciences of the National Institutes of Health. We would like to thank the Martin lab and the Weibel lab at UW Madison for the use of their MetaMorph software. We also thank Elizabeth Wood for constructing the strains overexpressing the RecA E38K K72R, RecA K72R and LexA proteins, and Shelley Lusetti and Dennis Harris for purifying LexA and RecA K72R.
References
- Bi E, Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA) J Bacteriol. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork JM, Cox MM, Inman RB. RecA protein filaments disassemble in the 5′ to 3′ direction on single- stranded DNA. J Biol Chem. 2001;276:45740–45743. doi: 10.1074/jbc.M109247200. [DOI] [PubMed] [Google Scholar]
- Brenner SL, Mitchell RS, Morrical SW, Neuendorf SK, Schutte BC, Cox MM. RecA protein-promoted ATP hydrolysis occurs throughout RecA nucleoprotein filaments. J Biol Chem. 1987;262:4011–4016. [PubMed] [Google Scholar]
- Campbell MJ, Davis RW. Toxic mutations in the recA gene of E. coli prevent proper chromosome segregation. J Mol Biol. 1999;286:417–435. doi: 10.1006/jmbi.1998.2456. [DOI] [PubMed] [Google Scholar]
- Cazaux C, Mazard AM, Defais M. Inducibility of the SOS response in a recA730 or recA441 strain is restored by transformation with a new recA allele. Mol Gen Genet. 1993;240:296–301. doi: 10.1007/BF00277070. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Margulies AD. Isolation and characterization of recombination deficient mutants of Escherichia coli K-12. Proc Natl Acad Sci, USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Sandler SJ. Homologous genetic recombination: the pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Ann Rev Genetics. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- Cox MM, McEntee K, Lehman IR. A simple and rapid procedure for the large scale purification of the RecA protein of Escherichia coli. J Biol Chem. 1981;256:4676–4678. [PubMed] [Google Scholar]
- Cox MM. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci, USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. A RecA filament capping mechanism for RecX protein. Mol Cell. 2004;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Egelman EH, Stasiak A. Electron microscopy of RecA-DNA complexes: two different states, their functional significance and relation to the solved crystal structure. Micron. 1993;24:309–324. [Google Scholar]
- Ellouze C, Selmane T, Kim HK, Tuite E, Norden B, Mortensen K, Takahashi M. Difference between active and inactive nucleotide cofactors in the effect on the DNA binding and the helical structure of RecA filament - Dissociation of RecA-DNA complex by inactive nucleotides. Eur J Biochem. 1999;262:88–94. doi: 10.1046/j.1432-1327.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- Ennis DG, Levine AS, Koch WH, Woodgate R. Analysis of recA mutants with altered SOS functions. Mutation Res. 1995;336:39–48. doi: 10.1016/0921-8777(94)00045-8. [DOI] [PubMed] [Google Scholar]
- Flores MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol Micro. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- Hegde S, Sandler SJ, Clark AJ, Madiraju MV. recO and recR mutations delay induction of the SOS response in Escherichia coli. Mol Gen Genetics. 1995;246:254–258. doi: 10.1007/BF00294689. [DOI] [PubMed] [Google Scholar]
- Keller KL, Overbeck-Carrick TL, Beck DJ. Survival and induction of SOS in Escherichia coli treated with cisplatin, UV-irradiation, or mitomycin C are dependent on the function of the RecBC and RecFOR pathways of homologous recombination. Mutation Res. 2001;486:21–29. doi: 10.1016/s0921-8777(01)00077-5. [DOI] [PubMed] [Google Scholar]
- Konola JT, Logan KM, Knight KL. Functional characterization of residues in the P-loop motif of the RecA protein ATP binding site. J Mol Biol. 1994;237:20–34. doi: 10.1006/jmbi.1994.1206. [DOI] [PubMed] [Google Scholar]
- Konrad EB. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977;130:167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery PE, Kowalczykowski SC. Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J Biol Chem. 1992;267:20648–20658. [PubMed] [Google Scholar]
- Lindsley JE, Cox MM. Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J Biol Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- Little JW, Kim B, Roland KL, Smith MH, Lin LL, Slilaty SN. Cleavage of LexA Repressor. Proteolytic Enzymes: Serine and Cysteine Peptidases. 1994;244:266–284. doi: 10.1016/0076-6879(94)44022-0. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- Long E, Renzette N, Richard CC, Sandler SJ. Determining mechanisms of constitutive SOS expression in Escherichia coli K-12 using fluorescence microscopy and single cell analysis. PLoS Genetics. 2008 submitted. [Google Scholar]
- Lusetti SL, Wood EA, Fleming CD, Modica MJ, Korth J, Abbott L, Dwyer DW, Roca AI, Inman RB, Cox MM. C-terminal deletions of the Escherichia coli RecA protein - Characterization of in vivo and in vitro effects. J Biol Chem. 2003;278:16372–16380. doi: 10.1074/jbc.M212917200. [DOI] [PubMed] [Google Scholar]
- Madiraju MV, Templin A, Clark AJ. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc Natl Acad Sci USA. 1988;85:6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju MV, Lavery PE, Kowalczykowski SC, Clark AJ. Enzymatic properties of the RecA803 protein, a partial suppressor of recF mutations. Biochemistry. 1992;31:10529–10535. doi: 10.1021/bi00158a016. [DOI] [PubMed] [Google Scholar]
- Marsh L, Walker GC. New phenotypes associated with mucAB: alteration of a MucA sequence homologous to the LexA cleavage site. J Bacteriol. 1987;169:1818–1823. doi: 10.1128/jb.169.5.1818-1823.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall JO, Witkin EM, Kogoma T, Roegner MV. Constitutive expression of the SOS response in recA718 mutants of Escherichia coli requires amplification of RecA718 protein. J Bacteriol. 1987;169:728–734. doi: 10.1128/jb.169.2.728-734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Micro. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- McEntee K, Weinstock GM, Lehman IR. DNA and nucleoside triphosphate binding properties of recA protein from Escherichia coli. Prog Nucleic Acid Res and Mol Biol. 1981;26:265–279. doi: 10.1016/s0079-6603(08)60411-0. [DOI] [PubMed] [Google Scholar]
- McGrew DA, Knight KL. Molecular design and functional organization of the RecA protein. Crit Rev Biochem and Mol Biol. 2003;38:385–432. doi: 10.1080/10409230390242489. [DOI] [PubMed] [Google Scholar]
- McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- Menetski JP, Kowalczykowski SC. Enhancement of Escherichia coli RecA protein enzymatic function by dATP. Biochemistry. 1989;28:5871–5881. doi: 10.1021/bi00440a025. [DOI] [PubMed] [Google Scholar]
- Menge KL, Bryant FR. ATP-stimulated hydrolysis of GTP by RecA protein: kinetic consequences of cooperative RecA protein-ATP interactions. Biochemistry. 1988;27:2635–2640. doi: 10.1021/bi00407a055. [DOI] [PubMed] [Google Scholar]
- Menge KL, Bryant FR. Effect of nucleotide cofactor structure on recA protein-promoted DNA pairing. 1. Three-strand exchange reaction. Biochemistry. 1992a;31:5151–5157. doi: 10.1021/bi00137a009. [DOI] [PubMed] [Google Scholar]
- Menge KL, Bryant FR. Effect of nucleotide cofactor structure on recA protein-promoted DNA pairing. 2. DNA renaturation reaction. Biochemistry. 1992b;31:5158–5165. doi: 10.1021/bi00137a010. [DOI] [PubMed] [Google Scholar]
- Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirshad JK, Kowalczykowski SC. Biochemical basis of the constitutive coprotease activity of RecA P67W protein. Biochemistry. 2003;42:5937–5944. doi: 10.1021/bi027232q. [DOI] [PubMed] [Google Scholar]
- Morrical SW, Lee J, Cox MM. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry. 1986;25:1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- Neuendorf SK, Cox MM. Exchange of RecA protein between adjacent RecA protein-single-stranded DNA complexes. J Biol Chem. 1986;261:8276–8282. [PubMed] [Google Scholar]
- O'Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186:7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer WM, Kowalczykowski SC. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J Biol Chem. 1993;268:1292–1297. [PubMed] [Google Scholar]
- Rehrauer WM, Lavery PE, Palmer EL, Singh RN, Kowalczykowski SC. Interaction of Escherichia coli RecA protein with LexA repressor. I. LexA repressor cleavage is competitive with binding of a secondary DNA molecule. J Biol Chem. 1996;271:23865–23873. [PubMed] [Google Scholar]
- Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, Centore R, Boonsombat R, Sandler SJ. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol Microbiol. 2005;57:1074–1085. doi: 10.1111/j.1365-2958.2005.04755.x. [DOI] [PubMed] [Google Scholar]
- Renzette N, Sandler SJ. Requirements for ATP binding and hydrolysis in RecA function in Escherichia coli. Mol Micro. 2008;67:1347–1359. doi: 10.1111/j.1365-2958.2008.06130.x. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Roberts CW, Craig NL. Escherichia coli recA gene product inactivates phage lambda repressor. Proc Natl Acad Sci U S A. 1978;75:4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Inman RB, Cox MM. RecA protein promotes the regression of stalled replication forks in vitro. Proc Natl Acad Sci USA. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler SJ. Studies on the mechanism of reduction of UV-inducible sulAp expression by recF overexpression in Escherichia coli K-12. Mol Gen Genet. 1994;245:741–749. doi: 10.1007/BF00297281. [DOI] [PubMed] [Google Scholar]
- Sandler SJ, Samra HS, Clark AJ. Differential suppression of priA2∷kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnarr M, Pouyet J, Grangerschnarr M, Daune M. Large-Scale Purification, Oligomerization Equilibria, and Specific Interaction of the LexA Repressor of Escherichia coli. Biochemistry. 1985;24:2812–2818. doi: 10.1021/bi00332a032. [DOI] [PubMed] [Google Scholar]
- Schnos M, Inman RB. New insights into protein-DNA interactions obtained by electron microscopy. Mol Biotech Appl Biochem Biotech. 2000;16:77–86. doi: 10.1385/MB:16:1:77. [DOI] [PubMed] [Google Scholar]
- Shan Q, Cox MM, Inman RB. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- Shan Q, Cox MM. RecA filament dynamics during DNA strand exchange reactions. J Biol Chem. 1997;272:11063–11073. doi: 10.1074/jbc.272.17.11063. [DOI] [PubMed] [Google Scholar]
- Sommer S, Knezevic J, Bailone A, Devoret R. Induction of only one SOS operon, umuDC, is required for SOS mutagenesis in Escherichia coli. Mol Gen Genet. 1993;239:137–144. doi: 10.1007/BF00281612. [DOI] [PubMed] [Google Scholar]
- Tateishi S, Horii T, Ogawa T, Ogawa H. C-terminal truncated Escherichia coli RecA protein RecA5327 has enhanced binding affinities to single- and double-stranded DNAs. J Mol Biol. 1992;223:115–129. doi: 10.1016/0022-2836(92)90720-5. [DOI] [PubMed] [Google Scholar]
- Thomas A, Lloyd RG. Control of recA dependent activities in Escherichia coli: a possible role for the recF product. J Gen Microbiol. 1983;129:681–686. doi: 10.1099/00221287-129-3-681. [DOI] [PubMed] [Google Scholar]
- VanLoock MS, Yu X, Yang S, Lai AL, Low C, Campbell MJ, Egelman EH. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:1–20. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmas P, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackernagel WTB. Regulatory role of recF in the SOS response of Escherichia coli: impaired induction of SOS genes by UV irradiation and nalidixic acid in a recF mutant. J Bacteriol. 1987;169:1731. doi: 10.1128/jb.169.4.1731-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α and β subunits of the ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TC, Chang HY, Hung JL. Cosuppression of recF, recR and recO mutations by mutant recA alleles in Escherichia coli cells. Mutat Res. 1993;294:157–166. doi: 10.1016/0921-8777(93)90024-b. [DOI] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Characterization of ATP hydrolysis. J Biol Chem. 1981a;256:8829–8834. [PubMed] [Google Scholar]
- Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Hydrolysis of UTP. J Biol Chem. 1981b;256:8856–8858. [PubMed] [Google Scholar]
- Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli. Steady state kinetic analysis of ATP hydrolysis. J Biol Chem. 1981c;256:8845–8849. [PubMed] [Google Scholar]
- Weinstock GM, McEntee K, Lehman IR. Interaction of the recA protein of Escherichia coli with adenosine 5′-O-(3-thiotriphosphate) J Biol Chem. 1981d;256:8850–8855. [PubMed] [Google Scholar]
- Willetts NS, Mount DW. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J Bacteriol. 1969;100:923–934. doi: 10.1128/jb.100.2.923-934.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM, Kogoma T. Involvement of the activated form of RecA protein in SOS mutagenesis and stable DNA replication in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7539–7543. doi: 10.1073/pnas.81.23.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- Zieg J, Kushner SR. Analysis of genetic recombination between two partially deleted lactose operons of Escherichia coli K-12. J Bacteriol. 1977;131:123–132. doi: 10.1128/jb.131.1.123-132.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


