Abstract
The 70 kDa heat shock proteins (Hsp70) are molecular chaperones that assist in folding of newly synthesized polypeptides, refolding or denaturation of misfolded proteins, and translocation of proteins across biological membranes. In addition, Hsp70 play regulatory roles in signal transduction, cell cycle, and apoptosis. Here we present a novel assay platform based on fluorescence polarization that is suitable for investigating the yet elusive molecular mechanics of human Hsp70 allosteric regulation.
Heat shock protein 70 (Hsp70) is a ubiquitous family of protein chaperones that assists protein folding and is involved in protein biogenesis, degradation and transport across membranes, and in the dynamics of macromolecular protein assembly.1 To perform these diverse functions, Hsp70s interact with a wide repertoire of protein co-chaperones. These regulate the activity of the Hsp70 chaperone or aid in the folding of specific substrate proteins. Co-chaperones can harness the ATP-dependent mechanisms of Hsp70 to do conformational work in diverse functional contexts, including vesicle secretion and recycling, protein transport and the regulated assembly and/or disassembly of protein complexes. The profound utilization of co-chaperones and resulting flexibility of functional specification are unique to the cytosolic Hsp70 and heat shock protein 90 (Hsp90) chaperone system. 1
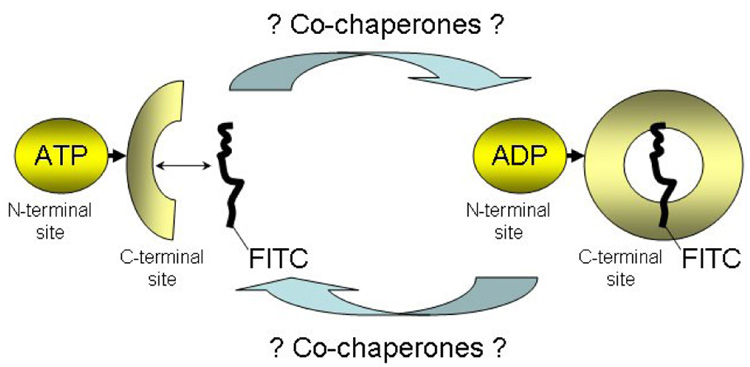
Hsp70 is composed of two functional domains: the N-terminal ATPase domain and the C-terminal peptide-binding domain (Figure 1).1,2 The interplay between these domains creates a ligand-activated, bidirectional molecular switch. ATP and inhibitors of the chaperone’s ATPase activity drive Hsp70 into the “open” conformation, which results in high rates of peptide binding and release. However, peptide binding induces a conformational change that is propagated back to the ATPase domain, and that stimulates the rate of ATP hydrolysis. This, in turn, leads to the “closed” conformation and facilitates peptide-capture.
Figure 1.
The Hsp70 cycle and design of the Hsp70 assay. In the ADP-conformation, Hsp70 binds to an unfolded peptide, here labeled with FITC, leading to an increase in the fluorescence polarization signal. This state interconverts with an ATP-bound state which leads to peptide release and a decrease of the FP signal. The full spectrum of co-chaperones and other proteins that regulate the Hsp70 cycle in mammalian cells is still being characterized.
The molecular mechanisms underlying of the ATPase and substrate binding/release cycles have been analyzed in detail only for only a few Hsp70 homologs, including E. coli DnaK and HscA (Hsc66), Thermus thermophilus DnaK, S. cerevisiae Ssa1, and bovine and hamster Hsc70.3 Even though these proteins are highly conserved, distinct attributes have been noted in their mechanism of action. In particular, differences in their cycles, with implications for their chaperone activities have been noted, suggesting that the analysis of other members of this family will prove essential. Surprisingly, the human Hsp70 (hHsp70) cycle has not been carefully dissected. The molecular mechanics and kinetics of the hHsp70 allosteric regulation, as well as the spectrum of interacting co-chaperones, are largely unknown, and represent an open area of research in the field. To this end, we have developed an assay platform that is based on fluorescence polarization, and that allows for kinetic measurement of the human Hsp70 cycle.
Fluorescence polarization (FP) is an assay with wide applicability in the discovery of novel protein modulators and in measuring real-time interactions between proteins or proteins and their ligands.4 The principle of FP is based on the observation that when a relatively small, fast-tumbling fluorescent-labeled compound is excited with plane-polarized light, the emitted light is random with respect to the plane of polarization, resulting in a lower mP value. When the compound is bound to a molecule with greater mass (in this case Hsp70 or Hsp70 in complex with its co-chaperones), the complex tumbles much slower and the emitted light is polarized, resulting in a higher mP value. Thus, the change of mP reflects the interaction between the fluorescent-labeled compound and the protein. Moreover, the mP value is proportional to the fractions of bound ligand, and thus FP assays can measure protein–inhibitor interactions in solution and in real-time.4
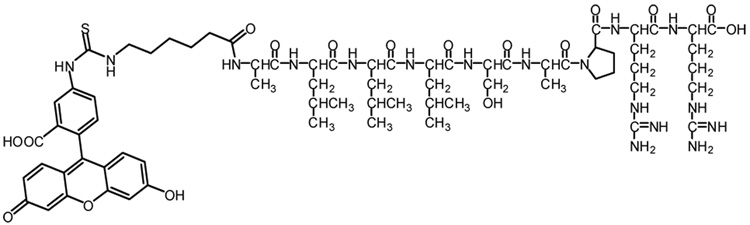
To establish an Hsp70 FP assay, we probed whether the peptide alap5 (ALLLSAPRR) binds to the C-terminal domain of hHsp70 and induces the characteristic conformational switch. This peptide was reported to bind with high affinity in the absence of ATP to DnaK, the E. coli Hsp70 (Kd=400 nM).5 We therefore synthesized a fluorescein isothiocyanate (FITC)-labeled alap5 (FITC-alap5) (Figure 2) for use in the FP assay, and the chemical integrity and purity of FITC-alap5 were confirmed by mass spectrum and high-pressure liquid chromatography (HPLC).6 The peptide was >90% pure as determined by HPLC and demonstrated the correct molecular mass of 1,500 Da (M+H).
Figure 2.
Chemical structure of FITC-alap5
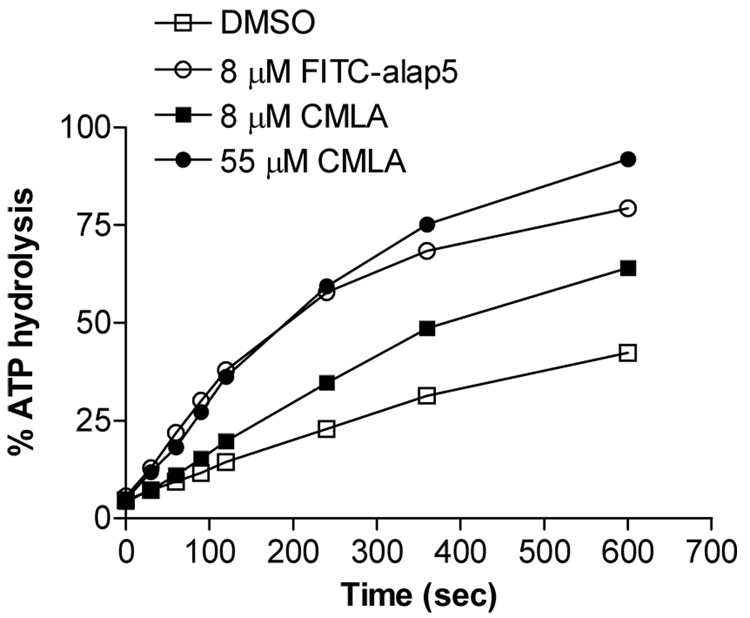
Next, the binding of this peptide to hHsp70 was confirmed by an independent, established method. As shown in Figure 3, the peptide enhanced the single turnover ATPase activity of the chaperone (KCAT),7 and the magnitude of stimulation was equivalent to that when a ~7-fold higher concentration of a known Hsp70-interacting unfolded polypeptide, carboxymethyl lactalbumin (CMLA),8 was used. These data indicate that FITC-alap5 binds productively to Hsp70, and should therefore serve as a valid reporter in the FP assay.
Figure 3.
The peptide FITC-alap5 interacts functionally with hHsp70. At a final concentration of 8 µM, the peptide activates the single-turnover ATPase activity of hHsp70 as well as a permanently unfolded polypeptide, CMLA, at a final concentration of 55 µM. The data are from a single-turnover ATPase assay such that the peptide or DMSO are present at t = 0. Data were plotted in Prism 4.0.
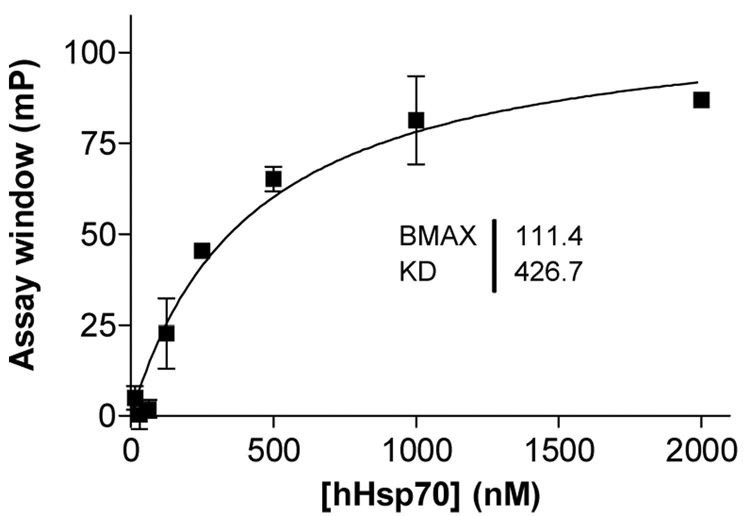
Next, using the FP assay, we determined that FITC-alap5 (at a final concentration of 5 nM) binds recombinant hHsp70 in a dose-dependent manner (Figure 4).9 To provide a valuable biological read-out, the FP ligand has to bind specifically and with good affinity to the protein of interest. To determine the affinity of FITC-alap5 for hHsp70, we titrated hHsp70 in reactions containing fluorescent alap5.
Figure 4.
hHsp70 at the indicated concentrations was incubated with FITC-alap5 (5 nM) at 40°C and the response was measured at 24 h. Fluorescence polarization was read with an Analyst GT instrument. The assay window data were obtained by subtracting free ligand values from values recorded in the presence of specified protein concentrations. Each data point is the average of two experiments. Data were plotted and analyzed in Prism 4.0. Error bars, s.d.
As the amount of hHsp70 increased, a greater fraction of fluorescent alap5 was protein-bound and polarization progressively increased to reach saturation (Figure 4). The signal remained stable for over 24 h. FITC-alap5 bound well to hHsp70 with an affinity resembling its binding to DnaK (Kd=426 nM). The assay window reached approximately 110 mP.
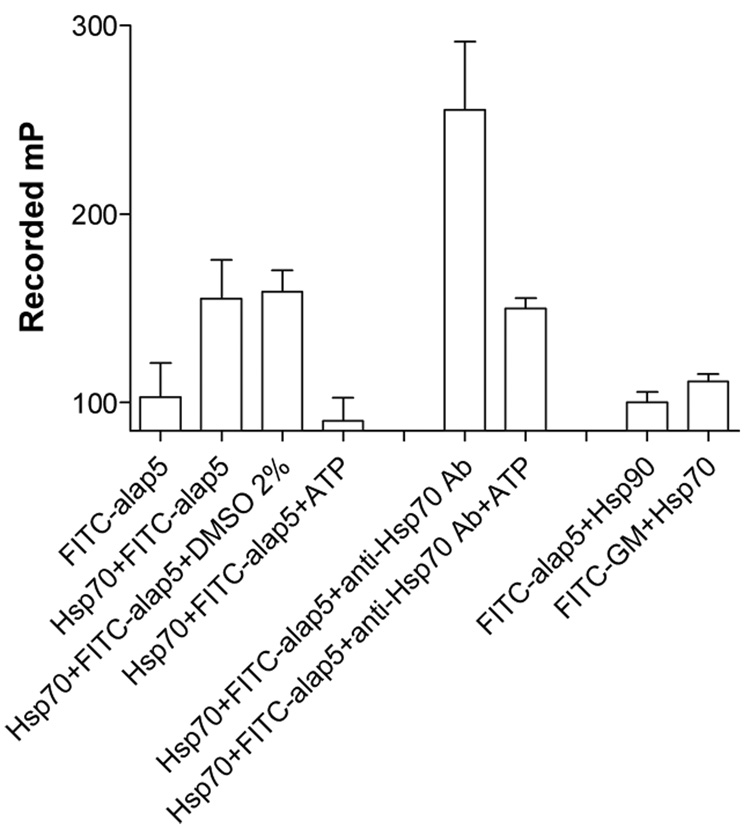
To investigate whether the assay can be employed to detect conformational changes in hHsp70, we included ATP in the reaction and showed that FITC-alap5 binding was significantly reduced (Figure 5, column 4). To probe whether the assay can also be used to identify interactors that might augment the affinity of hHsp70 for peptide substrates, we screened several antibodies raised specifically against Hsp70. We found that alap5 binding to hHsp70 was augmented by an anti-Hsp70 monoclonal antibody (clone C92F3A-5),10 prepared against human Hsp70 protein, suggesting that antibody binding may stabilize the closed, high-affinity peptide-binding conformation (Figure 5, column 5). Importantly, the effect was reversible because ATP reverted the antibody-induced conformational change to the low-affinity state (Figure 5, column 6).
Figure 5.
Binding of hHsp70 (500 nM) to FITC-alap5 (5 nM) was analyzed by FP. DMSO (2% v/v) had no effect on peptide-binding to hHsp70 (Hsp70+FITC-alap5+DMSO 2%). Binding of peptide to Hsp70 was competed by 1 mM ATP (Hsp70+FITC-alap5+ATP). An anti-Hsp70 antibody (Stressgen#SPA-810) increases the binding of Hsp70 to alap5 by 150mP (Hsp70+FITC-alap5+anti-Hsp70 Ab), an effect that. is reverted by 1 mM ATP (Hsp70+FITC-alap5+anti-Hsp70 Ab+ATP). The antibody was tested at a 1:100 dilution. The peptide FITC-alap5 (5 nM) does not interact with Hsp90 (200 nM) (FITC-alap5+Hsp90), and conversely, Hsp70 (500 nM) fails to interact with a FITC-labeled ligand that is specific for Hsp90 (FITC-GM, 5nM).
The assay was insensitive to low concentrations of DMSO, an organic solvent often used to dissolve small molecule modulators that could be screened to identify effectors of Hsp70 cycling (Figure 5, column 3). Furthermore, binding of FITC-alap5 to Hsp70 was specific, because the ligand failed to bind another molecular chaperone, Hsp9011 (Figure 5, column 7). Conversely, FITC-GM could not bind Hsp70 (Figure 5, column 8). FITC-GM12 is a FITC labeled geldanamycin (GM), an inhibitor of Hsp90 that interacts with the chaperone through direct interaction with its ATP-binding regulatory pocket.13
Collectively, these data indicate that FITC-alap5 is a suitable reporter for an hHsp70-specific FP assay. Moreover, the assay can be used to detect ATP-dependent conformational changes in the chaperone. Thus, we propose that FITC-alap5 can be co-opted to report on co-chaperone and protein-mediated effects on hHsp70 peptide binding and release. Because the protein and ligand are added simultaneously in solution, and a response is read directly, the assay can also provide important information on the kinetics of Hsp70 cycling and the effect of co-chaperones or other protein modulators on the mechanism of cycling. This is a great advantage over other methods that require immobilization of reagents or radioactive ligands. Our FP assay also utilizes a low amount of protein and ligand in comparison to other biochemical methods, and is thus, more cost effective. In consequence, we have designed a flexible assay platform that can be used to conduct real-time biochemical studies on hHsp70 cycling, and moreover, can be implemented towards the discovery of small molecule hHsp70 modulators.
Acknowledgements
The work was funded in part by grants from the National Institutes of Health (R01 CA119001) and National Institute of Aging (1R21AG028811), and a generous donation from the Byrne Fund of Memorial Sloan-Kettering Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.(a) Hartl FU, Hayer-Hartl M. Science. 2002;295:1852. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]; (b) Young JC, Agashe VR, Siegers K, Hartl FU. Nat. Rev. Mol. Cell. Biol. 2004;5:781. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]; (c) Mayer MP, Bukau B. Cell Mol. Life Sci. 2005;62:670. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) James P, Pfund C, Craig EA. Science. 1997;275:387. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Prasad K, Lafer EM, Sousa R. Mol. Cell. 2005;50:513. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Karzai AW, McMacken R. J. Biol. Chem. 1996;271:11236. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]; (b) Ha J-H, Johnson ER, McKay DB, Sousa MC, Takeda S, Wilbanks SM. Structure and mechanism of Hsp70 proteins. In: Bukau B, editor. Molecular Chaperones and Folding Catalysts Regulation, Cellular Function and Mechanism. Amsterdam: Harwood Academic Publishers; 1999. pp. 573–607. [Google Scholar]
- 4.Lakowitz JR. Fluorescence anisotropy. In: Lakowitz JR, editor. Principles of Fluorescence Spectroscopy. 2nd ed. New York: Plenum; 1999. pp. 291–319. [Google Scholar]
- 5.Han W, Christen P. FEBS Lett. 2004;563:146. doi: 10.1016/S0014-5793(04)00290-X. [DOI] [PubMed] [Google Scholar]
- 6.The FITC-alap5 peptide was synthesized at the Rockefeller University Proteomics Resource Center on a Symphony Peptide Synthesizer (Protein Technologies Inc.) using standard solid phase synthesis cycles with Fmoc-L-amino acids. Aminohexanoyl linker (Ahx) was added on to the N-terminus by standard coupling method, followed by fluoresceination with FITC (1.1 eq, 16h) in a mixture of pyridine, dimethyl formamide and dichloromethane (12:7:5). The tagged peptide was then cleaved from the solid support with a cocktail of TFA/water/triisopropyl silane (95:2.5:2.5), and purified on a Biocad preparative HPLC system using reverse phase C18 column (Vydac/WRGrace) (0.1% aqueous TFA/Acetonitrile: linear gradient of 0–60% in 40 min). The respective fractions were analyzed for final purity on Waters Millennium analytical HPLC and their molecular weight determined on an Applied Biosystems Voyager MALDI mass spectrometer. Pure fractions were lyophilized to yield the desired fluoresceinated peptide.
- 7.Single turnover ATPase assays with hHsp70 were performed as described in Srinivasan A, McClellan AJ, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky JL, Pipas JM. Mol. Cell. Biol. 1997;17:4761. doi: 10.1128/mcb.17.8.4761.
- 8.Cyr DM, Lu X, Douglas MG. J. Biol. Chem. 1992;267:20927. [PubMed] [Google Scholar]
- 9.FP assays were conducted in 25 mM HEPES-K pH 7.4, 20 mM NaCl, 2 mM Mg(OAc)2, 110 mM KOAc, 100 µM CaCl2, 4 mM DTT. Recombinant human Hsp70 and Hsp90 were purchased from Stressgen (NSP-555 and SPP-776, www.assaydesigns.com). FP measurements were performed on an Analyst GT instrument (Molecular Devices, Sunnyvale, CA). Measurements were taken in black 96-well microtiter plates (Corning # 3650) where both the excitation and the emission occur from the top of the wells. In the Analyst GT, a xenon arc lamp provides excitation light that passes through an excitation filter and then a polarizer filter. A beam-splitter filter directs the polarized excitation light into the well and emitted fluorescence transmits back through the same beam-splitter filter, through a polarizer filter, then through the emission filter for detection. In this study, read time was 0.1 s per well. The excitation polarizer filter is fixed in the parallel position, while the emission polarizer filter is changed for measuring parallel and perpendicular emission fluorescence intensity. All polarization values are expressed in millipolarization (mP) units (mP). The mP values were calculated using the equation mP=1000 * [(IS-ISB)-(IP-IPB)]/[(IS-ISB)+(IP-IPB)], where IS is the parallel emission intensity measurement and IP is the perpendicular emission intensity sample measurement, while ISB and ISP are the corresponding measurements for background (buffer). Total fluorescence was be determined as 2*IP+IS. For FITC labeled alap5, measurements were made with excitation at 485 nm and emission at 530 nm using a 505 nm dichroic filter. Data were imported into SoftMaxPro4 and analyzed in GraphPad Prism4.
- 10.The anti-Hsp70 antibody was purchased from Stressgen (catalog number SPA-810).
- 11.(a) Neckers L, Neckers K. Expert Opin. Emerg. Drugs. 2005;1:137. doi: 10.1517/14728214.10.1.137. [DOI] [PubMed] [Google Scholar]; (b) Whitesell L, Lindquist SL. Nat. Rev. Cancer. 2005;10:761. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]; (c) Chiosis G. Expert Opin. Ther. Targets. 2006;10:37. doi: 10.1517/14728222.10.1.37. [DOI] [PubMed] [Google Scholar]
- 12.Llauger-Bufi L, Felts SJ, Huezo H, Rosen N, Chiosis G. Bioorg. Med. Chem. Lett. 2003;13:3975. doi: 10.1016/j.bmcl.2003.08.065. [DOI] [PubMed] [Google Scholar]
- 13.Neckers L, Schulte TW, Mimnaugh E. Invest. New Drugs. 1999;17:361. doi: 10.1023/a:1006382320697. [DOI] [PubMed] [Google Scholar]