Abstract
The design, synthesis and biochemical characterization of a mechanism-based aryl carrier protein (ArCP) affinity probe that selectively modifies the terminal thiol of the aryl carrier protein phosphopantethein (Ppant) prosthetic group is described. Labeling of the aryl carrier protein was shown to require the cognate adenylating enzyme to channel the affinity probe onto the Ppant cofactor. The selective labeling was established by observation of the phosphopantetheinyl ejection ion via MS/MS and the probe was also found to stabilize an interaction between an aryl carrier protein and adenylating enzyme by an electrophoretic mobility shift assay.
Carrier proteins (CPs) play a central role in the biosynthesis of polyketide synthase (PKS), nonribosomal peptide synthetase (NRPS), and fatty acid synthase (FAS) derived natural products. CPs, also referred to as thiolation domains, are responsible for transporting the substrate and chain intermediates to the catalytic centers of the PKS, NRPS, and FAS assembly lines. The biosynthetic chain intermediates are tethered as thioesters on the terminal thiol of a phosphopantetheine (Ppant) prosthetic group that is covalentiy attached to an invariant serine residue of the CP.2 CPs are either freestanding or embedded in these multifunctional proteins and exist as three variants: an acyl carrier protein (ACP) found in PKSs and FASs, a peptidyl carrier protein (PCP) found in NRPS systems, and an aryl carrier protein (ArCP) commonly found in siderophore NRPS synthetases.2
An understanding of how CPs recognize their upstream and downstream partner proteins is essential to “deciphering the logic” for assembly of these natural products.1 CPs also constitute a potential target for the development of a new class of antibiotics since they are involved in the synthesis of several bacterial virulence factors and essential primary metabolites. For example, the natural antibiotic platensimycin acts by disrupting interactions between a CP and a ketosynthase (KS) domain in the bacterial type n FAS.2 The groups of Burkart, Walsh and Johnsson demonstrated the utility of CPs as low molecular weight protein fusion tags that can be easily modified by exploiting the promiscuity of phosphopantetheinyl transferases (PPTase) to incorporate fluorescent and affinity tags onto the conserved serine residue of the carrier domain.3 Mechanism-based affinity probes that specifically modify the terminal thiol of the Ppant prosthetic group of CPs could serve as new reagents for the site specific labeling of proteins, lead to the development of novel antibacterial agents, and provide powerful chemical probes to study the interactions of CPs with their partner enzyme domains. In this latter regard, Burkart and co-workers recently detailed a pantetheine analogue, which cross-links KS and ACP domains.4 We report herein our complementary efforts at the design and biochemical characterization of a mechanism-based CP affinity probe that selectively modifies ArCPs.
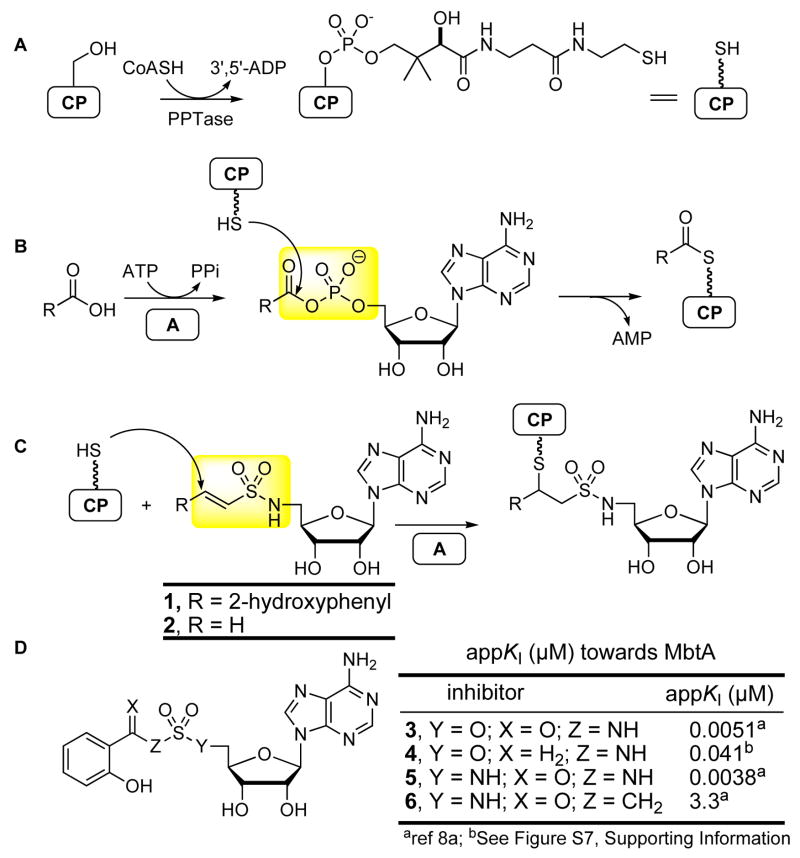
The ArCP affinity probe design is based on the reaction mechanism for ArCP loading, catalyzed by an adenylation enzyme illustrated in Figure 1B. In the first half-reaction, the substrate aryl acid and ATP are condensed to form a tightly bound acyladenylate with the release of pyrophosphate (see Figure 1B). In the second half-reaction, the adenylation enzyme binds ArCP and transfers the acyl group onto this protein. Consequently, analogues that mimic the essential features of the acyladenylate intermediate to confer sufficient binding to the adenylating enzyme, but incorporate a reactive functional group that can either reversibly or irreversibly bind the terminal thiol of the Ppant prosthetic group of the CP, are expected to provide selective mechanism-based inactivators. We conceived of a vinylsulfonamide as an acyladenylate surrogate that contains a Michael acceptor at the precise position of the incoming nucleophile (Figure 1C). Roush and co-workers recently ranked the relative activities of Michael acceptors: enone > vinylsulfone > vinylsulfonate > enoate > vinylsulfonamide.5 The vinylsulfonamide was chosen initially since this is the least reactive member in the series to minimize nonspecific thiol addition. Additionally, molecular modeling showed that the vinylsulfonamide adopted the required folded conformation of the bound acyladenylate (Figure S11, Supporting Information).
Figure 1.
Carrier Protein (CP) Modifications. (A) The Ppant prosthetic group on an invariant serine residue is installed post-translationally by a PPTase. (B) Loading of CPs mediated by adenylation domains. (C) A mechanism-based CP affinity probe for CP domains incorporates a Michael acceptor. (D) Reversible bisubstrate inhibitors of MbtA.8
As a model system we selected the ArCP domain in MbtB and its cognate adenylating enzyme MbtA that are responsible for incorporating salicylic acid into mycobactin, a siderophore produced by Mycobacterium tuberculosis.6 MbtA and MbtB are independent proteins enabling us to dissect the specificity and mechanism of the affinity probe. In order to facilitate subsequent MS-analysis, the N-terminal ArCP domain (~10 kDa) of mbtB was subcloned into pET28b and co-expressed in E. coli BL21 (DE3) with sfp7 to afford holo MbtB-ArCP as a C-terminal His6-tagged fusion protein. Synthesis of affinity probe 1 and 2 (Figure 1C) are described in the Supporting Information. Since the mechanism-based inactivator 1 requires binding to MbtA, we first demonstrated that 1 is able to interact with this protein using an ATP-[32P]PPi exchange assay.8 Affinity probe 1 displayed competitive inhibition with respect to both ATP (appKi(ATP) = 288 ± 56μM) and salicylic acid (apptKi(Sal) = 143 ± 9μM) (Figures Sl–S2, Supporting Information). Previously, we designed and synthesized analogues 3–6 as potent reversible inhibitors of MbtA which provided important comparative SAR data (Figure 1D).9 The approximately five orders of magnitude difference in activity between 1 and 5 can be attributed to the removal of the carbonyl group which interacts with Lys519 of MbtA (~100-fold loss, compare 3 vs. 4 in Figure 1D) and the central nitrogen atom (~1000-fold loss, compare 5 vs. 6 in Figure 1D).9a However, the modest micromolar activity of 1 towards MbtA was deemed adequate to ensure binding to MbtA before channeling onto MbtB-ArCP.
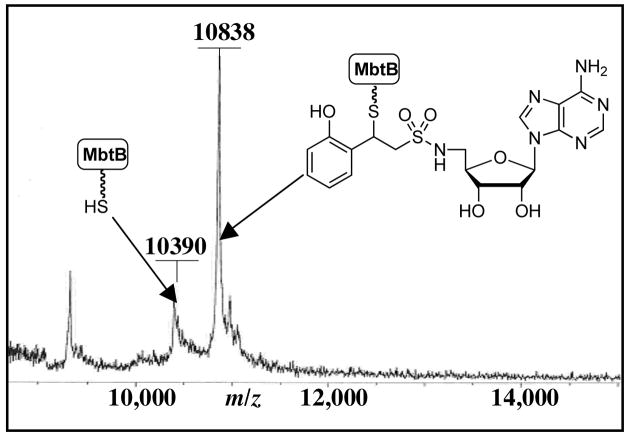
Incubation of 1 with MbtB-ArCP (1.0 mM 1, 10 μM MbtB-ArCP, 50 mM Tris, pH 8.5, 1.0 mM TCEP, 10 mM MgCl2, 37 °C, 24 h) did not lead to any covalent modification demonstrating that the affinity probe does not react nonspecifically. However, addition of catalytic MbtA (2 μM MbtA) afforded a molecular ion peak corresponding to covalent modification at m/z 10838 [MbtB-ArCP + 1]+ as determined by MALDI-TOF (Figure 2). Labeling of MbtB-ArCP by 1 could be completely suppressed by addition of 3, which is a reversible nanomolar inhibitor of MbtA. By contrast, unsubstituted vinylsulfonamide 2 reacted nonspecifically with MbtB-ArCP consistent with the greater reactivity of this unconjugated Michael acceptor. These results validate the design strategy and serve to highlight the requirement for the adenylating enzyme that must first bind 1 then channel this inactivator onto the ArCP. The second order rate constant for reaction of 1 with N-acetylcysteamine was found to be (1.85 ± 0.8)× 10−3 min−1 at 25 °C and pH 8.0 illustrating the low intrinsic reactivity of 1 towards thiols.
Figure 2.
MALDI-TOF of the ArCP domain of MbtB modified with affinity probe 1. MS: calcd for [C7S holo-ArCP] 10376; calcd for [C7S holo-ArCP+1] 10843.
In order to confirm that modification of MbtB-ArCP occurred on the Ppant prosthetic group of S39 the protein was subjected to trypsin digestion. After proteolysis, the modified peptide fragment ADALHPGANLVGQGLDS*IR (A23–R41) with m/z 2692.0 [A23–R41 + Ppant + 1]* was observed (Figure S9, Supporting Information). Tandem mass sequencing (ESI+) of the 4+ charge state of this modified peptide yielded two major ions due to Ppant elimination at m/z 992.4788 [A23–R41 + PO3]2+ and 355.1137 [Ppant + 1 − PO3]2+, which unequivocally proved the affinity probe 1 was linked to Ppant (Figure S10 and Schemes S1–S2, Supporting Information).10
The ability of other non-cognate adenylation domains to transfer affinity probe 1 to MbtB-ArCP was assessed to determine the specificity of this process. The adenylating enzymes YbtE and EntE were evaluated since these are responsible for transferring salicylic acid and 2,3-dihydroxybenzoic acid (2,3-DHBA) to their cognate aryl carrier domains YbtB and EntB involved in biosynthesis of the siderophores yersinabactin and enterobactin respectively.11 Incubation of either YbtE or EntE with MbtB-ArCP and 1 did not result in any detectable modification of MbtB-ArCP. We confirmed that vinylsulfonamide 1 is a competitive inhibitor with respect to salicylic acid (YbtE: appKi(Sal) = 133 ± 18) and 2,3-DHBA (EntE: appKi(2,3-DHBA) = 639 ± 114), but a noncompetitive inhibitor with respect to ATP of both YbtE (appKi(ATP) = 91 ± 43, α = 7.9) and EntE (appKi(ATP) = 535 ± 236 μM, α = 1.7) (Figures S3–S6, Supporting Information). The attenuated activity of 1 towards EntE is consistent with the preference of EntE for a 3-hydroxy group on the aryl ring. Thus, the lack of in trans modification of MbtB-ArCP by either YbtE or EntE is likely a result of improper protein-protein interactions between these heterologous protein pairs. This establishes another level of selectivity of affinity probe 1, which requires not only binding to the adenylating enzyme, but also proper interdomain recognition between the ArCP and cognate adenylation enzyme.12
Finally, we investigated the ability of affinity probe 1 to promote an interaction between the adenylating enzyme MbtA and MbtB-ArCP using an electrophoretic mobility shift assay with a nondenaturing polyacrylamide gel. Western blot analysis of MbtB revealed that 1 indeed did stabilize the protein-protein interaction between MbtA and MbtB-ArCP as the His6-tagged MbtB-ArCP coalesced to a sharp band only in the presence of MbtA and affinity probe 1 (Figure S8, Supporting Information).
In summary, we have designed, synthesized, and biochemically characterized an exquisitely selective aryl carrier protein affinity probe. The ability to cross-link CPs with adenylation domains located in cis may provide a means to crystallize such didomain pairs and provide insight into the elusive second half-reaction catalyzed by adenylation domains. Finally, this affinity probe of MbtB-ArCP could serve as a prototype for a new class of antitubercular agents since a mbtB disruption mutant of M. tuberculosis exhibited attenuated virulence in macrophages, the primary site of infection in pulmonary tuberculosis.13
Supplementary Material
Experimental procedures, supplementary data and the complete reference 2. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This research was supported by a grant from the NIH (R01AI070219) and funding from the Center for Drug Design, University of Minnesota to C.C.A. We thank the Minnesota Supercomputing Institute VWL lab for computer time.
References
- 1.Lai JR, Koglin A, Walsh CT. Biochemistry. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, et al. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 3.(a) La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Chem Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]; (b) Yin J, Liu F, Li X, Walsh CT. J Am Chem Soc. 2004;126:7754–7755. doi: 10.1021/ja047749k. [DOI] [PubMed] [Google Scholar]; (c) George N, Pick H, Vogel H, Johnsson N, Johnsson K. J Am Chem Soc. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 4.Worthington AS, Rivera H, Jr, Torpey JW, Alexander MD, Burkart MD. ACS Chem Biol. 2006;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- 5.Reddick JJ, Cheng J, Roush WR. Org Lett. 2003;5:1967–1970. doi: 10.1021/ol034555l. [DOI] [PubMed] [Google Scholar]
- 6.Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 7.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 8.Linne U, Marahiel MA. Methods Enzymol. 2004;388:293–315. doi: 10.1016/S0076-6879(04)88024-8. [DOI] [PubMed] [Google Scholar]
- 9.(a) Vannada J, Bennett EM, Wilson DJ, Boshoff HI, Barry CE, III, Aldrich CC. Org Lett. 2006;8:4707–4710. doi: 10.1021/ol0617289. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Somu RV, Boshoff HI, Qiao C, Bennett EM, Barry CE, III, Aldrich CC. J Med Chem. 2006;49:31–34. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 10.Dorrestein PC, Bumpus SB, Calderone CT, Garneau-Tsodikova S, Aron ZD, Straight PD, Kolter R, Walsh CT, Kelleher NL. Biochemistry. 2006;45:12756–12766. doi: 10.1021/bi061169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosa JH, Walsh CT. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehmann DE, Shaw-Reid CA, Losey HC, Walsh CT. Proc Natl Acad Sci USA. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE., 3rd Proc Natl Acad Sci USA. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures, supplementary data and the complete reference 2. This material is available free of charge via the Internet at http://pubs.acs.org.