Abstract
Peroxisome proliferator activated receptor-γ (PPARγ) regulates metabolic homeostasis and adipocyte differentiation, and it is activated by oxidized and nitrated fatty acids. Here we report the crystal structure of the PPARγ ligand binding domain bound to nitrated linoleic acid, a potent endogenous ligand of PPARγ. Structural and functional studies of receptor-ligand interactions reveal the molecular basis of PPARγ discrimination of various naturally occurring fatty acid derivatives.
PPARγ is a nuclear receptor that regulates adipocyte differentiation and glucose homeostasis1. The synthetic PPARγ ligands rosiglitazone (Avandia) and pioglitazone (Actos) are thiazolidinedione derivatives (TZDs) used for the treatment of type 2 diabetes. Despite the importance of PPARγ in human physiology and drug discovery, the identity of physiological ligands remains elusive. The first implicated natural ligand of PPARγ was 15-deoxy-Δ12,14-prostaglandin J2 (PGJ2), an oxidized fatty acid2,3. Later, 9-hydroxyoctadecadienoic acid (9-HODE) and 13-hydroxyoctadecadienoic acid (13-HODE), two oxidized fatty acids present in oxidized low-density lipoprotein, were shown to activate PPARγ in macrophages4. Despite the low receptor affinity of these oxidized fatty acids, they are more potent than native fatty acid counterparts. Recent studies provide functional evidence that nitrated linoleic acid (LNO2) and nitrated oleic acid (OA-NO2) potently activate PPARγ at nanomolar concentrations5–7, with net concentrations of nitrated fatty acid species exceeding 1 μM in human plasma lipids, supporting the idea that nitrated fatty acids are high-affinity, endogenous PPARγ ligands. This is also relevant to clinical therapeutics, as PPARγ, lipid and nitric oxide signaling pathways have been implicated in diabetes, obesity and cardiovascular diseases8,9. Moreover, nitric oxide–mediated oxidative and nitrative reactions induce fatty acid nitration, thus representing a convergence of lipid and nitric oxide–mediated signaling. The discovery of nitrated fatty acids as potent PPARγ ligands suggests further functional linkages between these seemingly disparate signaling pathways. The inactivity of linoleic acid as a PPARγ ligand4 raises questions as to why the oxidation and nitration of linoleic acid can yield much more potent PPAR activators.
Although TZDs improve insulin sensitivity and lower plasma glucose levels, they have adverse side effects, including weight gain, fluid retention and hepatotoxicity10. In addition, the recently identified cardiovascular risks of rosiglitazone11 may jeopardize further development and clinical application of TZD-based PPARγ ligands. A drug-design strategy based on endogenous PPARγ ligands may yield more efficacious PPARγ-targeted drugs, but little is known about how PPARγ interacts with natural ligands such as LNO2.
To unravel the biochemical mechanism of human PPARγ activation by nitrated fatty acids, we determined the ability of LNO2 to promote recruitment of coactivator LXXLL motifs by PPARγ using the AlphaScreen biochemical assay12 (Supplementary Methods online). Both LNO2 and rosiglitazone strongly enhanced the interaction of PPARγ with various coactivator LXXLL motifs from the family of steroid receptor coactivators (SRC2−3 and SRC1−2), CREB binding protein (CBP), TRAP220 and PGC-1α, indicating that LNO2 functions as a PPARγ agonist (Supplementary Fig. 1a online). Similarly to rosiglitazone, LNO2 also potently promoted the interaction of PPARγ with coactivator motifs in a concentration-dependent manner (Supplementary Fig. 1b,c). PPARγ showed only weak interaction with coactivators upon addition of 20 μM of either 9-HODE or 13-HODE (Supplementary Fig. 1a), in agreement with their in vivo effects4. These results reaffirm that nitrated fatty acids are potent PPARγ ligands.
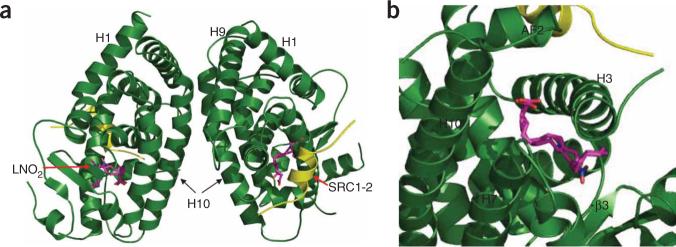
To determine the molecular basis of the high-affinity binding of LNO2 by PPARγ, we solved the crystal structure of PPARγ complexed with LNO2 and the SRC1−2 LXXLL motif at 2.4 Å-resolution (Supplementary Table 1 online). The PPARγ ligand binding domain (LBD) forms a dimer, with helix 10 from each monomer forming a dimer interface that resembles that of the PPARγ–RXRα heterodimer complex13 (Fig. 1a). The binding mode of LNO2 is similar to that of rosiglitazone, with one LNO2 molecule occupying roughly 40% of the ligand binding pocket (Fig. 1b). The existence of two distinct C10 and C12 LNO2 regioisomers in the PPARγ pocket was apparent from the highly revealing electron density map (Fig. 2). The acidic head group of both LNO2 isomers forms several hydrogen bonds with the surrounding PPARγ residues, including Gln286 from helix 3 (Fig. 2a). These interactions are observed for both LNO2 and rosiglitazone, and they support a crucial conserved mechanism for ligand-mediated activation of PPARγ. Indeed, the Q286A mutation that disrupts these interactions decreased the activation of PPARγ by both LNO2 and rosiglitazone in cell-based assays using a PPARγ-response reporter (Fig. 3).
Figure 1.
The structure of the PPARγ LBD and LNO2 complex. (a) Overall structure of the PPARγ–LNO2–SRC1−2 complex in ribbon representation. (b) A closer view of the binding of LNO2 to PPARγ. PPARγ is in green, the SRC1−2 peptide is in yellow and the bound LNO2 is shown in stick representation with carbon, nitrogen and oxygen atoms depicted in pink, blue and red, respectively.
Figure 2.
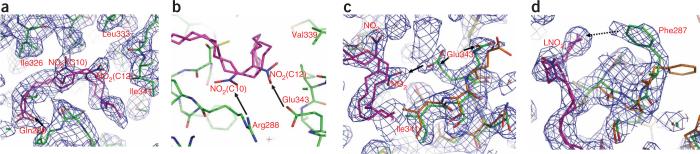
The structural determinants of the PPARγ LBD and LNO2 complex. (a) 2Fo – Fc electron density map (1.0σ) showing two bound LNO2 isomers and the surrounding PPARγ residues. LNO2 is shown in stick representation with carbon, nitrogen and oxygen atoms depicted in pink, blue and red, respectively. The key residues that determine PPARγ selectivity are noted, and hydrogen bonds are indicated by arrows. (b) Charged interactions of PPARγ residues with specific nitro groups in the LNO2 ligand as determinants of LNO2 selectivity. Hydrogen bonds are indicated by arrows. (c,d) The conformational changes of PPARγ induced by LNO2. Overlays of the PPARγ–LNO2 structure with the PPARγ–rosiglitazone structure, where LNO2-bound PPARγ is in green and rosiglitazone-bound PPARγ is in gold. The conformational shifts of Glu343 toward the nitro group and the shift of Phe287 toward the LNO2 backbone are indicated. The hydrophobic interaction between Phe287 and the LNO2 backbone is shown with a dashed line.
Figure 3.
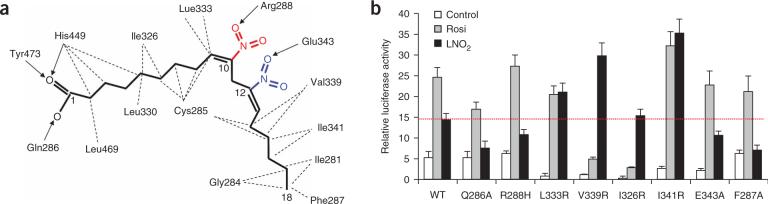
Functional correlation of the LNO2–PPARγ interactions. (a) Schematic representation of PPARγ–LNO2 interactions. The two isomers of LNO2 found in crystals with the PPARγ LBD are shown in red and blue, respectively, whereas the black indicates the identical conformation and structure shared by those LNO2 isomers. Hydrophobic interactions are indicated by dashed lines, and hydrogen bonds are indicated by arrows from proton donors to acceptors. Key hydrocarbon positions are indicated. (b) Effects of mutations of key PPARγ residues on LNO2-mediated PPARγ transcriptional activity in cell-based assays. The cells were co-transfected with a 3×-PPRE luciferase reporter together with plasmids encoding either full-length wild-type PPARγ or the mutants, as indicated in the figure. The cells were treated with 1 μM rosiglitazone or LNO2, respectively. The red dashed line indicates the activation level of wild-type PPARγ by LNO2. Error bars indicate s.d.
The selectivity of PPARγ for binding LNO2 is explained by the specific interactions of the two nitro groups with the PPARγ pocket residues (Fig. 3a). The C10 nitro group forms a hydrogen bond with Arg288, and the C12 nitro group interacts with Glu343 (Fig. 2b). Mutation of either residue decreased PPARγ activation by LNO2 but had no effect on PPARγ activation by rosiglitazone (Fig. 3b), suggesting that nitro group interactions with Arg288 and Glu343 are required for stable binding of LNO2. Notably, modeling studies indicate that the hydroxyl group in 9-HODE and 13-HODE occupies the same position as the nitro groups of the two LNO2 isomers (Supplementary Fig. 2a,b online), suggesting that these hydrogen-bond interactions should be preserved in the complex between PPARγ and these oxidized fatty acid regioisomers. Notably, the R288H mutation detected in colon cancers induced decreased binding of PGJ2 (ref. 14). Furthermore, the Arg288 residue has been implicated in the binding of the endogenous PPARγ ligand alkylglycerophosphate (AGP)15. Together, these data help to explain why PPARγ binds nitrated and oxidized linoleic acid but not unmodified linoleic acid4,5.
To further validate the specific impact of charged residues on PPARγ–LNO2 binding, we mutated several hydrophobic residues to arginine in the ligand binding pocket surrounding the nitro groups. These mutations should favor hydrogen bonding between the PPARγ pocket residues and the bound LNO2. Accordingly, all these mutations substantially increased PPARγ activation by LNO2 but not by rosiglitazone (Fig. 3b).
The binding of LNO2 is further facilitated by the two cis double bonds of LNO2, which introduce a 90° bend around the C9 to C13 position that allows the rest of the hydrocarbon chain to fit into a hydrophobic pocket of PPARγ (Fig. 2a). Conformational changes in two pocket residues (Glu343 and Phe287) are evidenced when the LNO2–PPARγ complex is overlaid on the rosiglitazone–PPARγ structure (Fig. 2c,d). In response to LNO2 binding, the charged side chain of Glu343 adopts a second conformation, allowing the receptor to form a hydrogen bond with the nitro group. The hydrophobic side chain of Phe287 also shifts from its rosiglitazone-bound conformation toward the hydrophobic tail (C18) of LNO2, thus stabilizing LNO2 binding by making additional hydrophobic interactions with the LNO2 backbone. To validate the differential roles of Phe287 and Glu343 in binding to rosiglitazone versus LNO2, we determined the in vitro ligand binding properties of PPARγ mutants using Alpha-Screen assays (Supplementary Fig. 3 online). The binding potency of LNO2 was decreased by both mutations, whereas the binding potency of rosiglitazone was not affected. Mutations in Phe287 or Glu343 also reduced the activation of PPARγ by LNO2 but not by rosiglitazone in cell-based assays, further affirming that these interactions are specific for LNO2 (Fig. 3b).
The three PPAR subtypes (α, β/δ and γ) are characterized as fatty acid receptors with distinct selectivity for ligands2,3,16. PPARγ has a preference for unsaturated or hydrophilic fatty acids, whereas PPARα can also bind saturated fatty acids16. Together with the structure of PPARβ/δ-bound eicosapentaenoic acid, the structure of LNO2 bound to PPARγ presented herein reveals a molecular basis for PPAR selectivity toward various fatty acids. Despite the high degree of homology among the three PPARs, the charged residues Arg288 and Glu343 of PPARγ, which make specific contacts with the nitro groups, are not conserved (Supplementary Fig. 2c), indicating that the PPARγ ligand binding pocket has unique hydrophilic properties that dictate the discrimination of native fatty acids and those modified by oxidation and nitration reactions.
The specific interactions between crucial LBD residues of PPARγ and LNO2 thus provide an important perspective regarding the recognition of nitrated and oxidized fatty acids by PPARγ. On the basis of the above structural observations, PPARγ contains two electrostatic binding epitopes: one epitope, in part comprising the C-terminal activation helix (AF2, Fig. 1b), is common for binding of the acidic group of LNO2 and TZDs; the other epitope, comprising Arg288 and Glu343, is specific for the nitrated or oxidized groups of modified fatty acids. The differential binding modes of endogenous ligand versus the therapeutic drug rosiglitazone may allow the differential modulation of PPARγ ligand binding selectivity and affinity, thereby affecting the physiological outcome of different PPARγ ligands. As such, the structural mechanism may provide a more rational template for designing PPARγ ligands that would better mimic nitro group–receptor interactions by the endogenous PPARγ ligand LNO2. On the basis of the structural insight into LNO2–PPARγ interactions gained herein, we expect that new compounds can be developed that have a more favorable pharmacological impact than current TZD-based synthetic PPARγ ligands.
Supplementary Material
ACKNOWLEDGMENTS
We thank W.D. Tolbert and Z. Wawrzak for assistance in data collection at the DND-CAT of the Advanced Photon Source. Use of the Advanced Photon Source was supported by the US Office of Science of the US Department of Energy. This work was supported in part by the Jay and Betty Van Andel Foundation (H.E.X.), US National Institutes of Health Grants DK071662 and DK066202 (H.E.X.), HL089301 (H.E.X. and Y.L.), HL68878, HL089544 and HL75397 (Y.E.C.), HL58115 and HL64937 (B.A.F.), American Diabetes Association (P.R.S.B.) and awards from the American Heart Association (Y.L., F.J.S.).
Footnotes
Accession code. Protein Data Bank: Coordinates for PPARγ–LNO2 have been deposited with accession code 3CWD.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Lehrke M, Lazar MA. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Forman BM, et al. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 3.Kliewer SA, et al. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 4.Nagy L, et al. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 5.Schopfer FJ, et al. Proc. Natl. Acad. Sci. USA. 2005;102:2340–2345. doi: 10.1073/pnas.0408384102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker PR, et al. J. Biol. Chem. 2005;280:42464–42475. doi: 10.1074/jbc.M504212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman BA, et al. J. Biol. Chem. 2008;283:15515–15519. doi: 10.1074/jbc.R800004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans RM, Barish GD, Wang YX. Nat. Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 9.Straus DS, Glass CK. Trends Immunol. 2007;28:551–558. doi: 10.1016/j.it.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Yki-Jarvinen H. N. Engl. J. Med. 2004;351:1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 11.Nissen SE, Wolski K. N. Engl. J. Med. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, et al. Mol. Cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Gampe RT, et al. Mol. Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 14.Sarraf P, et al. Mol. Cell. 1999;3:799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- 15.Tsukahara T, et al. J. Biol. Chem. 2006;281:3398–3407. doi: 10.1074/jbc.M510843200. [DOI] [PubMed] [Google Scholar]
- 16.Xu HE, et al. Mol. Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.