Abstract
Background and purpose:
Most antiviral therapies directed against herpes simplex virus (HSV) infections are limited to a small group of nucleoside analogues that target the viral polymerase. Extensive clinical use of these drugs has led to the emergence of resistant viral strains, mainly in immunocompromised patients. This highlights the need for the development of new anti-herpesviral drugs with novel targets. Herein the effects of a plant anthraquinone, emodin, on the HSV-1 alkaline nuclease activity and virus yields were investigated.
Experimental approach:
HSV-1 alkaline nuclease activity was examined by nuclease activity assay. Inhibition of virus yields was measured by plaque reduction assay and immunohistochemical staining. Interaction between emodin and alkaline nuclease was analysed by docking technology.
Key results:
Emodin specifically inhibited the nuclease activity of HSV-1 UL12 alkaline nuclease in a biochemical assay. Plaque reduction assay revealed that emodin reduced the plaque formation with an EC50 of 21.5±4.4 μM. Immunohistochemical staining using the anti-nucleocapsid protein antibody demonstrated that emodin induced the accumulation of viral nucleocapsids in the nucleus in a dose-dependent manner. Docking analysis further suggested that the inhibitory effect of emodin on the UL12 activity may result from the interaction between emodin and critical catalytic amino acid residues of UL12.
Conclusions and implications:
Our findings suggest that emodin is a potent anti-HSV agent that inhibits the yields of HSV-1 via the suppression of a novel target, UL12.
Keywords: herpes simplex virus type 1, emodin, alkaline nuclease
Introduction
Herpes simplex virus (HSV) causes herpes labialis, herpes keratitis, genetic herpes and life-threatening herpes encephalitis. HSV infections are more severe in immunocompromised patients, which are characterized by chronic and extensive lesions of the mucous membranes (Whitley, 2001). Most therapies directed against HSV infections are nucleotides, nucleosides or pyrophosphate analogues, such as acyclovir, valacyclovir, penciclovir and famciclovir. After uptake by virus-infected cells, these drugs are phosphorylated by virus-encoded thymidine kinase, compete with the nucleotides to inhibit the viral DNA polymerase and subsequently cause the termination of growing viral DNA chains (Field, 2001). Although these drugs are effective in the treatment of many acute infections, the intensive use of these drugs has led to the emergence of resistant viral strains, mainly in immunocompromised patients (Field, 2001). Therefore, there is a need to provide other drugs with distinct mechanisms as alternatives to existing therapies.
Alkaline nuclease, which is encoded by the UL12 gene of HSV-1, possesses both endonuclease and exonuclease activities under alkaline pH conditions (Hoffmann and Cheng, 1978; McGeoch et al., 1986; Knopf and Weisshart, 1990). Null mutants incapable of expressing UL12 are able to synthesize near wild-type levels of viral DNA, suggesting that UL12 is not essential for viral DNA replication in culture (Weller et al., 1991). Although UL12 is not essential for viral DNA synthesis, UL12 mutant viral yields are 0.1–1% of wild-type yields (Shao et al., 1993; Martinez et al., 1996a). The analysis of UL12 null mutants has shown that the decrease in virus yield results from the reduction of capsids exiting from the nucleus (Martinez et al., 1996b; Goldstein and Weller, 1998). Analysis of replicating DNA from UL12 mutant-infected cells has shown that UL12 is implicated in resolving branched structures of HSV-1 replicative intermediates prior to encapsidation (Martinez et al., 1996a; Porter and Stow, 2004a, 2004b). Therefore, these results indicate that, even though UL12 is not essential for either viral DNA synthesis or packaging, UL12 is required for full efficiency of these processes. Additionally, these findings suggest that HSV-1 UL12 can be a novel target for anti-herpes viral drugs.
Increasing the emergence of resistant viral strains has highlighted the crucial need for the development of new anti-herpes virus drugs with different mechanisms. Several potential viral targets, such as helicase–primase complex and DNA polymerase, have been known to be involved in HSV-1 infection and for which specific inhibitors with anti-HSV activity, at least in cell cultures, have been identified (Crute et al., 2002; Thomsen et al., 2003; Greco et al., 2007). In the present study, we analysed the potent inhibitor of HSV-1 that targeted viral UL12. Our findings indicated that emodin (1,3,8-trihydroxy-6-methylanthraquinone), the naturally occurring anthraquinone present in the root and bark of numerous plants of the genera Rheum and Polygonum, inhibited HSV-1 UL12 activity, leading to the accumulation of nucleocapsids in the nucleus and the subsequent reduction of HSV-1 yields in Vero cells.
Methods
Materials
All chemicals, except where indicated, were purchased from Sigma (St Louis, MO, USA). Plant materials were purchased from Sun Ten Pharmaceutical Corporation (Taipei, Taiwan). Plant samples were ground to fine powders with homogenizers and extracted with methanol, as described previously (Chen et al., 2007a). Emodin and its analogues were dissolved in dimethyl sulphoxide (DMSO). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was dissolved in phosphate-buffered saline (PBS) (137 mM NaCl, 1.4 mM KH2PO4, 4.3 mM Na2HPO4, 2.7 mM KCl, pH 7.2). Bovine pancreatic DNase I was purchased from New England BioLabs (Beverly, MA, USA). Mouse anti-HSV-1 nucleocapsid protein monoclonal antibody and fluorescein (FITC)-conjugated goat anti-mouse antibody were purchased from USBiological (Swampscott, MA, USA) and Jackson ImmunoResearch Laboratories (West Grove, PA, USA), respectively.
Cells and viruses
African green monkey kidney cells (Vero cells), which were purchased from Bioresource Collection and Research Center (Hsinchu, Taiwan), were cultured in Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (HyClone, Logan, UT, USA) and grown at 37 °C in a humidified CO2 atmosphere. Laboratory strain (KOS) of HSV-1 was used, and the viral stock was prepared and titrated in Vero cells.
Cloning, expression and purification of recombinant HSV-1 UL12
To clone the HSV-1 UL12 gene, viral genomic DNA was extracted from HSV-1-infected Vero cells as described previously and amplified for 35 cycles with UL12-P (5′-TCGGATCCATGGAGTCCACGGTAGGCCCAGC-3′) and UL12-M (5′-CGAATTCGGTCAGCGAGACGACCTCCCCG-3′) primers (Hsiang et al., 1996; Wu et al., 1998). The 1897-bp UL12 gene fragment was inserted into EcoR I and BamH I sites of histidine-tagged expression vector pET-28a(+) (Novagen, Madison, WI, USA) to create the pET-UL12. Recombinant UL12 protein was expressed in Escherichia coli BL21 (DE3) pLysS strain by transforming the pET-UL12 to produce an N-terminal fusion with six histidine residues. The protein was purified by affinity chromatography as described previously (Hsiang et al., 1998; Ho et al., 2000). Purified protein was analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis, quantified with a Bradford assay (Bio-Rad, Hercules, CA, USA), and stored at −70 °C until further assays.
Nuclease activity assay
Plasmid pUC18 dsDNA, prepared by Qiagen Plasmid Midi Kit (Qiagen, Valencia, CA, USA), was mixed with purified UL12 in DNase buffer (50 mM Tris-HCl, pH 9.0, 2 mM MgCl2, 10 mM 2-mercaptoethanol) and incubated at 37 °C. The reaction was then stopped by the addition of stop solution (25% glycerol, 0.5% sodium dodecyl sulphate, 0.05% bromophenol blue, 50 mM EDTA), and the resulting products were analysed by electrophoresis on 1.2% agarose gels. The intensities of substrates on the gel were measured by Gel-Pro Analyzer (Media Cybernetics, Silver Spring, MD, USA). Nuclease activity (%) was calculated by ((intensity of untreated substrate−intensity of nuclease-treated substrate)/intensity of untreated substrate) × 100%.
Plaque reduction assay
Plaque reduction assay was performed as described previously with a slight modification (Hodinka et al., 2000). Cell monolayers, cultured in 24-well culture plates, were infected with 30 plaque-forming units (PFU) of HSV-1 for 1 h at room temperature and subsequently for 30 min at 37 °C. The viruses were then discarded, and the cells were overlaid with 1 mL of 1% methylcellulose medium containing emodin and incubated at 37 °C in a humidified CO2 atmosphere. Three days later, cells were fixed and stained by 0.5% crystal violet in 50% methanol, and the number of plaques was counted (Hsiang et al., 2001). EC50 value was determined as the quantity of emodin required to reduce the plaque number by 50%.
MTT assay
Cell viability was monitored by MTT colorimetric assay as described previously (Lee et al., 2007). Briefly, cells were treated with emodin for 16 h. One-tenth volume of 5 mg mL−1 MTT was then added to the culture medium. After a 4-h incubation at 37 °C, equal cell culture volume of 0.04 N HCl in isopropanol was added to dissolve the MTT formazan, and the absorbance value was measured at 570 nm using an ELISA plate reader. Cell viability (%) was calculated by (OD of emodin-treated cells/OD of solvent-treated cells) × 100.
Immunohistochemical staining
Vero cells (104 cells) were seeded in 24-well plates containing glass coverslips and incubated at 37 °C. One day later, cells were infected with 30 PFU of HSV-1 for 1 h at room temperature and subsequently for 30 min at 37 °C. The viruses were then discarded and the cells were overlaid with medium containing various amounts of emodin at 37 °C for indicated time. The coverslips were then rinsed with PBS, fixed with 3.7% PBS-buffered formaldehyde at room temperature for 30 min and blocked with 1% BSA at 37 °C for 1 h. After four washes with PBS, diluted mouse anti-HSV-1 nucleocapsid monoclonal antibody was added to each coverslip and incubated at 4 °C overnight. After four washes with PBS, diluted FITC-conjugated secondary antibody was added and incubated at 37 °C for 90 min in the dark. The coverslips were then washed four times with PBS, placed onto glass slides, mounted with fluoromount G (Electron Microscopy Sciences, Hatfield, PA, USA), and observed under a confocal microscope (Leica, Wetzlar, Germany).
Protein structure prediction and docking technology
UL12 protein structure was generated via the Meta Server (http://bioinfo.pl/). (Ginalski et al., 2003). The MEDock (Maximum Entropy based Docking) web server (http://bioinfo.mc.ntu.edu.tw/medock/) was used for the prediction of ligand-binding sites (Chang et al., 2005; Chen et al., 2007b). The input file was in the PDBQ format, which is an extension of the PDB format. The PDBQ format for emodin has been generated by Dundee's PRODRG server (http://davapc1.bioch.dundee.ac.uk/programs/prodrg/) (Schuttelkopf and van Aalten, 2004).
Statistical analysis
Data are presented as mean±s.e.mean. Student's t-test was used for comparisons between two experiments. A value of P<0.05 was considered statistically significant.
Results
Nuclease activity of recombinant HSV-1 UL12
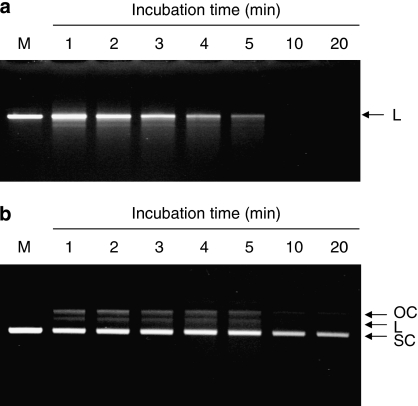
The nuclease activity of HSV-1 UL12 was analysed on different forms of pUC18 dsDNA and observed by agarose electrophoresis. When linear pUC18 dsDNA was treated with UL12, a smear was visible after 2 min of digestion and pUC18 dsDNA was totally degraded after 10 min (Figure 1a). When supercoiled pUC18 dsDNA was treated with UL12, it was firstly converted into an open circular form and then converted into full-length linear dsDNA (Figure 1b). With increasing incubation time, the supercoiled form of pUC18 dsDNA was gradually degraded, and the open circular and linear forms of pUC18 dsDNA were completely degraded. These results indicated that recombinant HSV-1 UL12 exhibited both exonuclease and endonuclease activities, which are consistent with previous studies (Bronstein and Weber, 1996).
Figure 1.
Nuclease activity of recombinant HSV-1 UL12. UL12 (1 pmol) was mixed with 0.1 μg of EcoR I-linearized (a) or supercoiled pUC18 dsDNA (b) in the reaction buffer. The reaction mixtures were incubated at 37 °C for the indicated periods, and the resulting products were analysed by electrophoresis on 1.2% agarose gels. Lane M represents the reaction performed in the absence of UL12. Arrowheads denote the different topological forms of pUC18 plasmids. OC, open circular; L, linear; SC, supercoiled; HSV-1, herpes simplex virus type 1.
Rheum officinale inhibits the nuclease activity of HSV-1 UL12
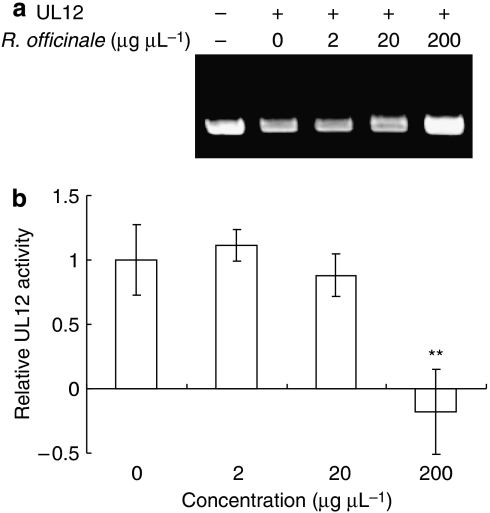
In a previous study, we found that Rheum officinale, Paeonia suffruticosa, Melia toosendan, and Sophora flavescens are able to inhibit HSV-1 productions in Vero cells through prevention of viral attachment or penetration (Hsiang et al., 2001). We are interested to know whether these herbs also inhibit the UL12 activity. Therefore, the methanolic extracts of these herbs were mixed with HSV-1 UL12 and the nuclease activity was analysed. As shown in Figure 2, the methanolic extract of R. officinale inhibited the UL12 activity in a dose-dependent manner. Three other herbs did not show the inhibitions on UL12 activity (data not shown). Methanol alone did not affect the UL12 activity (data not shown). Therefore, these results indicated that, in addition to virus attachment, R. officinale exhibited an anti-UL12 activity.
Figure 2.
Effect of Rheum officinale on the nuclease activity of HSV-1 UL12. (a) Agarose gel electrophoresis. The reaction mixtures, containing 10 pmol of UL12, 0.5 μg of supercoiled pUC18 dsDNA and various amounts of R. officinale, were incubated at 37 °C for 5 min. The resulting products were then analysed by 1.2% agarose gels and visualized with ethidium bromide. (b) Quantitative analysis. Results are expressed as relative UL12 activity, which is presented as a comparison, with the nuclease activity of R. officinale-treated UL12 relative to solvent-treated UL12. Values are mean±s.e.mean of three independent experiments. **P<0.01, compared with solvent-treated UL12 activity. HSV-1, herpes simplex virus type 1.
Emodin inhibits the nuclease activity of HSV-1 UL12 with specificity
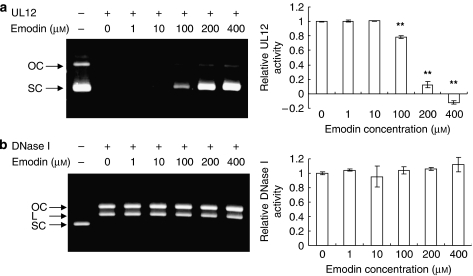
Emodin is the naturally occurring anthraquinone present in R. officinale (Koyama et al., 2003). Therefore, we are interested to know whether emodin inhibits the nuclease activity of HSV-1 UL12. As shown in Figure 3a, the input DNA was totally degraded in the absence of emodin. However, with increasing concentrations, the nuclease activity of UL12 was gradually inhibited by emodin. DMSO alone did not affect the UL12 activity (data not shown). To further analyse the specificity of emodin, pUC18 dsDNA was mixed with emodin-treated bovine pancreatic DNase I. As shown in Figure 3b, the input DNA (supercoiled pUC18 dsDNA) was converted into open circular and linear forms in the presence of DNase I. With increasing concentrations, the endonuclease activity of DNase I was consistent. Therefore, these findings indicated that emodin is likely to be the active compound of R. officinale, which inhibited the UL12 activity with specificity.
Figure 3.
Effects of emodin on the nuclease activities of HSV-1 UL12 and bovine pancreatic DNase I. Various amounts of emodin were mixed with 0.5 μg of supercoiled pUC18 dsDNA and 20 pmol of UL12 (a) or 0.01 pmol of DNase I (b). The reaction mixtures were incubated at 37 °C for 10 or 5 min. The resulting products were then analysed by 1.2% agarose gels and visualized with ethidium bromide. Arrowheads denote the different topological forms of pUC18 plasmids. OC, open circular; L, linear; SC, supercoiled. Relative UL12 or DNase I activity, which is presented as a comparison with the nuclease activity of emodin-treated nuclease relative to solvent-treated nuclease, is shown on the right panel. Values are mean±s.e.mean of three independent experiments. **P<0.01, compared with solvent-treated UL12 activity. HSV-1, herpes simplex virus type 1.
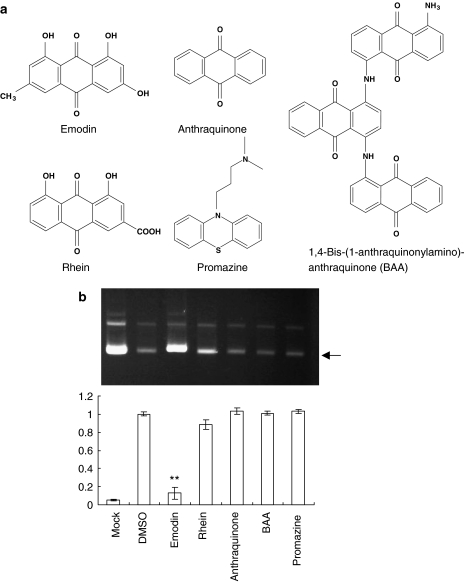
Emodin is an anthraquinone compound consisting of three cyclic rings. We wonder whether the other emodin analogues exhibit better anti-UL12 abilities than emodin. Similar to emodin, rhein (1,8-dihydroxy-3-carboxyl-9-10-anthraquinone) and anthraquinone consist of three cyclic rings (Figure 4a). In contrast to emodin, they consist of different functional groups. 1,4-Bis-(1-anthraquinonylamino)-anthraquinone consists of nine cyclic rings. The anti-psychotic drug promazine shares a similar structure with emodin. Although the structural similarity is observed among these emodin analogues, emodin was the only compound that significantly inhibited the nuclease activity of HSV-1 UL12 (Figure 4b).
Figure 4.
Effects of emodin analogues on the nuclease activities of HSV-1 UL12. (a) Chemical structures of emodin and its analogues. (b) Nuclease activity assay. The reaction mixtures, containing 20 pmol of UL12, 0.5 μg of supercoiled pUC18 dsDNA and 200 μM of compounds, were incubated at 37 °C for 10 min. The resulting products were then analysed by 1.2% agarose gels and visualized with ethidium bromide. Lane Mock represents the reaction performed in the absence of UL12. Arrowhead denotes the supercoiled form of pUC18 plasmid. Relative UL12 activity, which is presented as the comparison with the nuclease activity of compound-treated UL12 relative to DMSO-treated UL12, is shown on the bottom. Values are mean±s.e.mean of four independent experiments. **P<0.01, compared with solvent-treated UL12 activity. HSV-1, herpes simplex virus type 1.
Emodin reduces the plaque formation by the accumulation of nucleocapsids in the nucleus
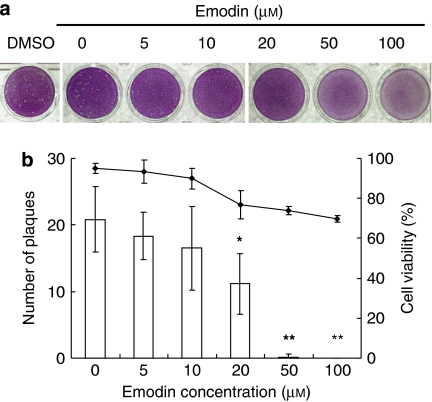
To test whether emodin inhibited HSV-1 yields, Vero cells were infected with HSV-1 and then overlaid with methylcellulose medium containing various amounts of emodin. As shown in Figure 5, DMSO alone did not affect the number of plaques. Emodin decreased the number and the size of plaques in a dose-dependent manner. The EC50 of emodin was 21.5±4.4 μM. Moreover, no significant loss of mitochondrial function was detected by MTT assay. Therefore, these findings indicated that emodin reduced the plaque formation by the inhibition of UL12 activity.
Figure 5.
Effect of emodin on the HSV-1 yields in Vero cells. (a) Plaque reduction assay. Vero cells were infected with HSV-1 for 2 h. Methylcellulose media, containing various amounts of emodin, were then added to wells and incubated at 37 °C for 72 h. After the removal of methylcellulose medium, cells were stained with crystal violet and the number of plaques was counted. Solvent control (DMSO) is shown at the left. (b) Quantitative analysis. Cell viability was determined by MTT and is represented by the symbols. The columns represent the number of plaques. Values are mean±s.e.mean of three independent experiments. *P<0.05 and **P<0.01, compared with Mock. HSV-1, herpes simplex virus type 1.
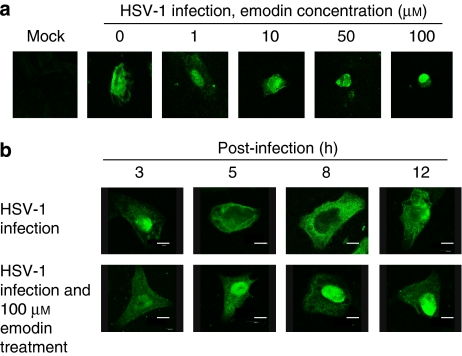
Previous studies indicated that HSV-1 UL12 is involved in viral DNA processing and capsid egression (Martinez et al., 1996b; Goldstein and Weller, 1998). We wondered whether emodin induces the accumulation of nucleocapsids in the nucleus by the inhibition of UL12 activity. Immunohistochemical staining, using anti-HSV-1 nucleocapsid protein antibody, was therefore performed to analyse the localization of viral nucleocapsids during emodin treatment. No fluorescent signal was observed in mock cells (Figure 6a). As expected, the nucleocapsids were localized diffusely in both the nucleus and the cytoplasm at 16 h post-infection because the HSV-1 progenies are assembled and released from cells at 16 h post-infection (Roizman and Knipe, 2001). In contrast, emodin induced the accumulation of nucleocapsid protein in the nucleus in a dose-dependent manner at 16 h post-infection. Time course assay showed that, in the absence of emodin, nucleocapsids mainly remained in the nucleus at 3 h post-infection, diffused to cytoplasm at 5 h post-infection, and mainly localized in cytoplasm at 8 h post-infection. In contrast, the fluorescent signal mainly remained in the nucleus during emodin treatment. These findings suggest that emodin inhibited HSV-1 UL12 activity, leading to the accumulation of nucleocapsids in the nucleus and the subsequent reduction of HSV-1 yields. Our findings are also consistent with previous studies showing that UL12 is involved in the egression of capsid from the nucleus (Martinez et al., 1996b; Goldstein and Weller, 1998).
Figure 6.
Inhibitory effect of emodin on the yield of HSV-1 progenies by IHC. (a) Dose response. Vero cells were cultured on glass coverslips, infected with 30 PFU of HSV-1 for 2 h, and treated without or with various amounts of emodin. After a 16-h infection period, cells were fixed and stained with anti-HSV-1 nucleocapsid protein antibody and FITC-conjugated secondary antibody. Results were evaluated under a confocal microscope. (b) Time course of the response. Vero cells were cultured on glass coverslips, infected with 30 PFU of HSV-1 for 2 h, and treated without (top panel) or with 100 μM of emodin (bottom panel). Cells were fixed at indicated periods and then stained with anti-HSV-1 nucleocapsid protein antibody and FITC-conjugated secondary antibody. Scale bar=8 μm. FITC, fluorescein; HSV-1, herpes simplex virus type 1; IHC, immunohistochemical staining.
Emodin docks into HSV-1 UL12 with complementarity
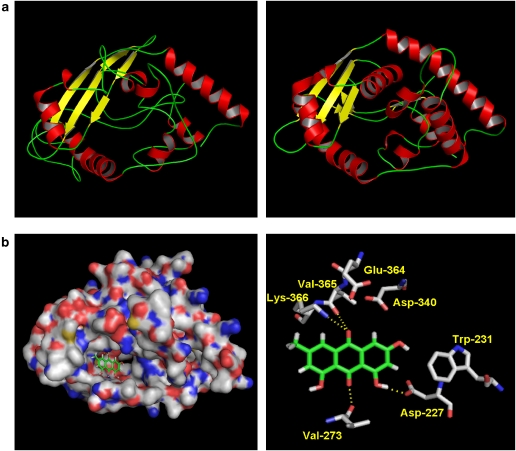
We further investigated the binding site of emodin in UL12 by docking technology. To achieve this, we modelled the three-dimensional structure of HSV-1 UL12. The modelling of HSV-1 UL12 was performed using the FFAS03 and SWISS-MODEL Workspace (http://swissmodel.expasy.org/workspace/index.php) (Jaroszewski et al., 2005; Arnold et al., 2006). A significant similarity, with the FFAS03 score of −19.2, was found between UL12 and phage λ exonuclease. A full atom three-dimensional structure of HSV-1 UL12 was, therefore, modelled using the phage λ exonuclease (PDB code 1avq) as the reference protein (Figure 7a). Emodin wholly docked into the pocket of UL12, with the predicted binding energy score of −76.67 kcal mol−1. Emodin exhibited critical hydrogen bonds with Asp-227, Val-273, Val-365, and Lys-366 residues of UL12 (Figure 7b). Hydrophobic interactions with Trp-231, Asp-340, and Glu-364 residues of UL12 were also found.
Figure 7.
Docking structure between HSV-1 UL12 and emodin. (a) Homology modelling. The three-dimensional structure of UL12 (left panel) was modelled using phage λ exonuclease (PDB code 1avq) (right panel) as the reference protein. (b) Docking analysis. Surface representation of UL12 complexed with emodin (left panel). Close-up view of UL12 complexed with emodin (right panel). Emodin and the side chains of selected residues are represented by sticks. Hydrogen bonds between UL12 and emodin are represented by yellow dash lines. HSV-1, herpes simplex virus type 1.
Discussion and conclusions
Antiviral drugs have been used for the treatment of HSV infections for over 45 years (Field, 2001). Acyclovir is of significant therapeutic value and is considered as the ‘gold standard' in HSV therapy. However, approximately 5% of the isolates from immunocompromised patients, which receive a long-term prophylactic treatment with acyclovir, have experienced the emergence of resistant strains (Danve-Szatanek et al., 2004). Even in immunocompetent populations, the prevalence of resistance ranges from 0.32 to 3.5% by large-scale studies (Fife et al., 1994; Danve-Szatanek et al., 2004). Therefore, the development of antiviral drugs with different mechanisms is an alternative approach to the control of HSV infections.
Viral proteins, that are known to be involved in HSV infection, have been used as the targets for chemotherapy. For examples, viral glycoproteins together with the cell membrane receptors are involved in viral attachment and penetration (Rajcáni and Vojvodová, 1998). Sulphated polymer-based inhibitors, which interact directly with viral envelope glycoproteins and prevent viral attachment, are now being tested in Phase II or III clinical trials (Keller et al., 2005). Helicase–primase complex is essential for the unwinding of dsDNA and the generation of primers for DNA synthesis (Boehmer and Lehman, 1997). Aminothiazolyl-phenyl compounds (BILS 179 BS and BILS 45 BS) and thiazolyl sulphonamide compound (BAY 57-1293), that prevent the propagation of helicase–primase catalytic cycle and inhibit its ATPase activity, respectively, display potent anti-HSV effects in mice (Crute et al., 2002; Duan et al., 2003; Biswas et al., 2007). Viral DNA polymerase is essential for DNA replication (Boehmer and Lehman, 1997). 4-Hydroxyquinoline-3-carboxamides (PNU-182171 and PNU-183792), that compete with incoming nucleotides and dislodge the template from the active site, display anti-herpes virus activities in preclinical animal studies (Thomsen et al., 2003; Liu et al., 2006). In principle, all the replication-essential viral proteins can be considered as potential targets for chemotherapy. This has raised the question. Is UL12 a possible candidate for anti-herpes virus therapy? Although UL12 mutants are able to synthesize near wild-type levels of viral DNA, the yields of mutant virus are reduced by 100- to 1000-fold (Shao et al., 1993; Martinez et al., 1996a). UL12 mutants display the failure of DNA-containing capsids to migrate into the cytoplasm and the more complex structure of replicative intermediates with an increased frequency of branches (Martinez et al., 1996b; Goldstein and Weller, 1998; Porter and Stow, 2004a, 2004b). Additionally, antisense phosphorothioate oligonucleotides, targeting an internal start codon of HSV-1 UL12 mRNA, inhibit HSV-1 replication in Vero cells (Chiba et al., 2000). Furthermore, emodin, that inhibited UL12 activity in vitro, displayed the reduction of HSV-1 yields in Vero cells in this study. These findings indicated that UL12, which is conserved in all species of Herpesviridae, can be considered as the target for the anti-herpes virus therapy.
Emodin, the active principle of herbal medicine derived from genera Rheum and Polygonum, has demonstrated antiviral effects to some enveloped viruses, such as hepatitis B virus, HSV, human cytomegalovirus and severe acute respiratory syndrome-coronavirus, and non-enveloped viruses, such as poliovirus (Barnard et al., 1992; Cohen et al., 1996; Semple et al., 2001; Dang et al., 2006; Ho et al., 2007). Several studies have revealed that the antiviral activity of emodin is through casein kinase 2 (CK2) inhibition, which is exploited by viruses for the phosphorylation of proteins that are essential for viral life cycle (Yim et al., 1999; Battistutta et al., 2000). Moreover, emodin has affinity for phospholipid membrane and is effective in weakening hydrophobic interactions between hydrocarbon chains in phospholipid bilayers, contributing to the antiviral capacity of emodin against enveloped viruses (Sydiskis et al., 1991; Alves et al., 2004). In this study, we demonstrated that emodin can exert its antiviral activity by the third mechanism, the inhibition of HSV-1 UL12 alkaline nuclease activity. These findings suggest that emodin may be a potential anti-HSV-1 candidate with a broad spectrum of antiviral activities.
Our results indicate that emodin inhibits HSV-1 UL12 activity, leading to the reduction of HSV-1 yields in Vero cells. How did emodin inhibit nuclease activity of HSV-1 UL12? To answer this question, we modelled the three-dimensional structure of UL12 using phage λ exonuclease as the template protein. Although HSV-1 UL12 exhibits a low amino acid sequence similarity (13.6%) with λ exonuclease, HSV-1 UL12 shares similar enzyme activities and biological functions with λ exonuclease. For example, both proteins preferentially degrade DNA from double-stranded end in the 5′–3′ direction (Cassuto and Radding, 1971; Knopf and Weisshart, 1990). Moreover, they mediate DNA strand exchange by interacting with ssDNA-binding protein and participate in initiating viral recombination events (Muniyappa and Radding, 1986). The recognizable homology suggests that using λ exonuclease as the template for the modelling of UL12 is reasonable. The interaction of emodin with UL12 was predicted by docking analysis. Results showed that emodin docked into UL12 but not bovine pancreatic DNase I (data not shown). Emodin interacted with Asp-227, Trp-231, Val-273, Asp-340, Glu-364, Val-365 and Lys-366 of UL12 via hydrogen bonds or hydrophobic interactions. Interestingly, some of these amino acid residues may be critical for the nuclease activity. Site-directed mutagenesis on the HSV-1 UL12 homologue, Epstein–Barr virus DNase, has revealed that Asp-203, Glu-225 and Lys-227 of Epstein–Barr virus DNase, corresponding to Asp-340, Glu-364 and Lys-366 of UL12, respectively, play important roles in catalysis (Liu et al., 2003). Glu-225 of Epstein–Barr virus DNase, corresponding to Glu-364 of UL12, is involved in metal binding. The docking of emodin into UL12 may affect or occupy the catalytic site of UL12, leading to the inhibition of nuclease activity. Therefore, the interaction between emodin and critical amino acid residues of UL12 may explain why emodin inhibited the nuclease activity of HSV-1 UL12.
In conclusion, emodin significantly reduced the plaque formation in Vero cells. Serum profiles after oral administration of emodin at a dosage of 2 g kg−1 in mice showed that the peak serum concentration of emodin is 700 μM (Mengs et al., 1997). We revealed that emodin at a concentration of 21.5 μM was sufficient to reduce 50% virus yields without cytotoxic effect. Moreover, there is no evidence or equivocal evidence of carcinogenic activity of emodin in rats or mice (National Toxicology Program, 2001). Therefore, we speculate that the antiviral effect of emodin measured in vitro may occur in vivo. Furthermore, in addition to the inhibition of UL12, emodin possesses antiviral activities via the disruption of phospholipid bilayer and the inhibition of CK2. Therefore, these results suggest that emodin may be a potent herpes viral inhibitor with a broad spectrum of antiviral activities.
Acknowledgments
This work was supported by grants from the National Research Program for Genomic Medicine, National Science and Technology Program for Agricultural Biotechnology, National Science Council, Committee on Chinese Medicine and Pharmacy (CCMP 96-RD-201 and CCMP 97-RD-201) and China Medical University (CMU95-053, CMU95-055, and CMU95-067), Taiwan.
Abbreviations
- CK2
casein kinase 2
- DMSO
dimethyl sulphoxide
- FITC
fluorescein
- HSV
herpes simplex virus
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS
phosphate-buffered saline
- PFU
plaque-forming unit
Conflict of interest
The authors state no conflict of interest.
References
- Alves DS, Pérez-Fons L, Estepa A, Micol V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem Pharmacol. 2004;68:549–561. doi: 10.1016/j.bcp.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Barnard DL, Huffman JH, Morris JL, Wood SG, Hughes BG, Sidwell RW. Evaluation of the antiviral activity of anthraquinones, anthrones and anthraquinone derivatives against human cytomegalovirus. Antiviral Res. 1992;17:63–77. doi: 10.1016/0166-3542(92)90091-i. [DOI] [PubMed] [Google Scholar]
- Battistutta R, Sarno S, De Moliner E, Papinutto G, Zanotti G, Pinna LA. The replacement of ATP by the competitive inhibitor emodin induces conformational modifications in the catalytic site of protein kinase CK2. J Biol Chem. 2000;275:29618–29622. doi: 10.1074/jbc.M004257200. [DOI] [PubMed] [Google Scholar]
- Biswas S, Jennens L, Field HJ. The helicase primase inhibitor, BAY 57-1293 shows potent therapeutic antiviral activity superior to famciclovir in BALB/c mice infected with herpes simplex virus type 1. Antiviral Res. 2007;75:30–35. doi: 10.1016/j.antiviral.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- Bronstein JC, Weber PC. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J Virol. 1996;70:2008–2013. doi: 10.1128/jvi.70.3.2008-2013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E, Radding CM. Mechanism for the action of lambda exonuclease in genetic recombination. Nat New Biol. 1971;229:13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- Chang DT, Oyang YJ, Lin JH. MEDock: a web server for efficient prediction of ligand binding sites based on a novel optimization algorithm. Nucleic Acids Res. 2005;33:W233–W238. doi: 10.1093/nar/gki586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Chang YS, Wu SL, Chao DC, Chang CS, Li CC, et al. Inhibition of Escherichia coli heat-labile enterotoxin-induced diarrhea by Chaenomeles speciosa. J Ethanopharmacol. 2007a;113:233–239. doi: 10.1016/j.jep.2007.05.031. [DOI] [PubMed] [Google Scholar]
- Chen JC, Huang LJ, Wu SL, Kuo SC, Ho TY, Hsiang CY. Ginger and its bioactive component inhibit enterotoxigenic Escherichia coli heat-labile enterotoxin-induced diarrhea in mice. J Agric Food Chem. 2007b;55:8390–8397. doi: 10.1021/jf071460f. [DOI] [PubMed] [Google Scholar]
- Chiba A, Ogasawara M, Yoshida I, Knox YM, Suzutani T. Herpesvirus alkaline deoxyribonuclease; a possible candidate as a novel target for anti-herpesvirus therapy. Tohoku J Exp Med. 2000;192:141–149. doi: 10.1620/tjem.192.141. [DOI] [PubMed] [Google Scholar]
- Cohen PA, Hudson JB, Towers GH. Antiviral activities of anthraquinones, bianthrones and hypericin derivatives from lichens. Experientia. 1996;52:180–183. doi: 10.1007/BF01923366. [DOI] [PubMed] [Google Scholar]
- Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, et al. Herpes simplex virus helicase–primase inhibitors are active in animal models of human disease. Nat Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- Dang S, Zhang Z, Chen Y, Zhang X, Wang B, Yuan L, et al. Inhibition of the replication of hepatitis B virus in vitro by emodin. Med Sci Monit. 2006;12:BR302–BR306. [PubMed] [Google Scholar]
- Danve-Szatanek C, Aymard M, Thouvenot D, Morfin F, Agius G, Bertin I, et al. Surveillance network for herpes simplex virus resistance to antiviral drugs: 3-year follow-up. J Clin Microbiol. 2004;42:242–249. doi: 10.1128/JCM.42.1.242-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Liuzzi M, Paris W, Liard F, Browne A, Dansereau N, et al. Oral bioavailability and in vivo efficacy of the helicase–primase inhibitor BILS 45 BS against acyclovir-resistant herpes simplex virus type 1. Antimicrob Agents Chemother. 2003;47:1798–1804. doi: 10.1128/AAC.47.6.1798-1804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HJ. Herpes simplex virus antiviral drug resistance—current trends and future prospects. J Clin Virol. 2001;21:261–269. doi: 10.1016/s1386-6532(00)00169-4. [DOI] [PubMed] [Google Scholar]
- Fife KH, Crumpacker CS, Mertz GJ, Hill EL, Boone GS. Recurrence and resistance patterns of herpes simplex virus following cessation of > or =6 years of chronic suppression with acyclovir. Acyclovir Study Group. J Infect Dis. 1994;169:1338–1341. doi: 10.1093/infdis/169.6.1338. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Goldstein JN, Weller SK. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology. 1998;244:442–457. doi: 10.1006/viro.1998.9129. [DOI] [PubMed] [Google Scholar]
- Greco A, Diaz JJ, Thouvenot D, Morfin F. Novel targets for the development of anti-herpes compounds. Infect Disord Drug Targets. 2007;7:11–18. doi: 10.2174/187152607780090766. [DOI] [PubMed] [Google Scholar]
- Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TY, Wu SL, Hsiang CH, Chang TJ, Hsiang CY. Identification of a DNA-binding domain and an active-site residue of pseudorabies virus DNase. Biochem J. 2000;346:441–445. [PMC free article] [PubMed] [Google Scholar]
- Hodinka R, Swierkosz E, Lancaster D, Moore BM, Sacks S, Scholl D, Wright DK. Antiviral Susceptibility Testing. Proposed Standard M33-P. National Committee for Clinical Laboratory Standards: Pennsylvania; 2000. [Google Scholar]
- Hoffmann PJ, Cheng YC. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J Biol Chem. 1978;253:3557–3562. [PubMed] [Google Scholar]
- Hsiang CY, Ho TY, Chang TJ. Identification of a pseudorabies virus UL12 (deoxyribonuclease) gene. Gene. 1996;177:109–113. doi: 10.1016/0378-1119(96)00285-5. [DOI] [PubMed] [Google Scholar]
- Hsiang CY, Ho TY, Hsiang CH, Chang TJ. Recombinant pseudorabies virus DNase exhibits a RecBCD-like catalytic function. Biochem J. 1998;330:55–59. doi: 10.1042/bj3300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang CY, Hsieh CL, Wu SL, Lai II, Ho TY. Inhibitory effect of anti-pyretic and anti-inflammatory herbs on herpes simplex virus replication. Am J Chin Med. 2001;29:459–467. doi: 10.1142/S0192415X01000472. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MJ, Tuyama A, Carlucci MJ, Herold BC. Topical microbicides for the prevention of genital herpes infection. J Antimicrob Chemother. 2005;55:420–423. doi: 10.1093/jac/dki056. [DOI] [PubMed] [Google Scholar]
- Knopf CW, Weisshart K. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur J Biochem. 1990;191:263–273. doi: 10.1111/j.1432-1033.1990.tb19119.x. [DOI] [PubMed] [Google Scholar]
- Koyama J, Morita I, Kawanishi K, Tagahara K, Kobayashi N. Capillary elelctrophoresis for simultaneous determination of emodin, chrysophanol, and their 8-beta-D-glucosides. Chem Pharm Bull (Tokyo) 2003;51:418–420. doi: 10.1248/cpb.51.418. [DOI] [PubMed] [Google Scholar]
- Lee CH, Chen JC, Hsiang CY, Wu SL, Wu HC, Ho TY. Berberine suppresses inflammatory agents-induced interleukin-1β and tumor necrosis factor-α productions via the inhibition of IκB degradation in human lung cells. Pharmacol Res. 2007;56:193–201. doi: 10.1016/j.phrs.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Liu MT, Hu HP, Hsu TY, Chen JY. Site-directed mutagenesis in a conserved motif of Epstein–Barr virus DNase that is homologous to the catalytic centre of type II restriction endonucleases. J Gen Virol. 2003;84:677–686. doi: 10.1099/vir.0.18739-0. [DOI] [PubMed] [Google Scholar]
- Liu S, Knafels JD, Chang JS, Waszak GA, Baldwin ET, Deibel MR, Jr, et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J Biol Chem. 2006;281:18193–18200. doi: 10.1074/jbc.M602414200. [DOI] [PubMed] [Google Scholar]
- Martinez R, Shao L, Bronstein JC, Weber PC, Weller SK. The product of 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology. 1996a;215:152–164. doi: 10.1006/viro.1996.0018. [DOI] [PubMed] [Google Scholar]
- Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996b;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Dolan A, Frame MC. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the exonuclease gene and neighbouring genes. Nucleic Acids Res. 1986;14:3435–3448. doi: 10.1093/nar/14.8.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengs U, Krumbiegel G, Völkner W. Lack of emodin genotoxicity in the mouse micronucleus assay. Mutat Res. 1997;393:289–293. doi: 10.1016/s1383-5718(97)00113-7. [DOI] [PubMed] [Google Scholar]
- Muniyappa K, Radding CM. The homologous recombination system of phage lambda. Pairing activities of beta protein. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- National Toxicology Program NTP toxicology and carcinogenesis studies of emodin (CAS NO. 518-82-1) feed studies in F344/N rats and B6C3F1 mice. Natl Toxicol Program Tech Rep Ser. 2001;493:1–278. [PubMed] [Google Scholar]
- Porter IM, Stow MD. Replication, recombination and packaging of amplicon DNA in cells infected with the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12. J Gen Virol. 2004b;85:3501–3510. doi: 10.1099/vir.0.80403-0. [DOI] [PubMed] [Google Scholar]
- Porter IM, Stow ND. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J Gen Virol. 2004a;85:583–591. doi: 10.1099/vir.0.19657-0. [DOI] [PubMed] [Google Scholar]
- Rajcáni J, Vojvodová A. The role of herpes simplex virus glycoproteins in the virus replication cycle. Acta Virol. 1998;42:103–118. [PubMed] [Google Scholar]
- Roizman B, Knipe DM.Herpes simplex viruses and their replication Field's Virology 2001Lippincott Williams and Wilkins: Philadelphia; 2399–2459.In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B (eds).4th edn. [Google Scholar]
- Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Semple SJ, Pyke SM, Reynolds GD, Flower RL. In vitro antiviral activity of the anthraquinone chrysophanic acid against poliovirus. Antiviral Res. 2001;49:169–178. doi: 10.1016/s0166-3542(01)00125-5. [DOI] [PubMed] [Google Scholar]
- Shao L, Rapp LM, Weller SK. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–162. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- Sydiskis RJ, Owen DG, Lohr JL, Rosler KH, Blomster RN. Inactivation of enveloped viruses by anthraquinones extracted from plants. Antimicrob Agents Chemother. 1991;35:2463–2466. doi: 10.1128/aac.35.12.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen DR, Oien NL, Hopkins TA, Knechtel ML, Brideau RJ, Wathen MW, et al. Amino acid changes within conserved region III of the herpes simplex virus and human cytomegalovirus DNA polymerases confer resistance to 4-oxo-dihydroquinolines, a novel class of herpesvirus antiviral agents. J Virol. 2003;77:1868–1876. doi: 10.1128/JVI.77.3.1868-1876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller SK, Seghatoleslami MR, Shao L, Rowse D, Carmichael EP. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J Gen Virol. 1991;71:2941–2952. doi: 10.1099/0022-1317-71-12-2941. [DOI] [PubMed] [Google Scholar]
- Whitley RJ.Herpes simplex viruses Field's Virology 2001Lippincott Williams and Wilkins: Philadelphia; 2461–2509.In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B (eds).4th edn. [Google Scholar]
- Wu SL, Hsiang CY, Ho TY, Chang TJ. Identification, expression, and characterization of the pseudorabies virus DNA-binding protein gene and gene product. Virus Res. 1998;56:1–9. doi: 10.1016/s0168-1702(98)00051-3. [DOI] [PubMed] [Google Scholar]
- Yim H, Lee YH, Lee CH, Lee SK. Emodin, an anthraquinone derivative isolated from the rhizomes of Rheum palmatum, selectively inhibits the activity of casein kinase II as a competitive inhibitor. Planta Med. 1999;65:9–13. doi: 10.1055/s-1999-13953. [DOI] [PubMed] [Google Scholar]