Abstract
In the fission yeast Schizosaccharomyces pombe, passage from G1 to S-phase requires the execution of the transcriptional factor complex that consists of the Cdc10 and Res1/2 molecules. This complex activates the MluI cell cycle box cis-element contained in genes essential for S-phase onset and progression. The rep2+ gene, isolated as a multicopy suppressor of a temperature-sensitive cdc10 mutant, has been postulated to encode a putative transcriptional activator subunit for the Res2–Cdc10 complex. To identify the rep2+ function and molecularly define its domain organization, we reconstituted the Res2–Cdc10 complex-dependent transcriptional activation in Saccharomyces cerevisiae. Reconstitution experiments, deletion analyses using one and two hybrid systems, and in vivo Res2 coimmunoprecipitation assays show that the Res2–Cdc10 complex itself can recognize but cannot activate MluI cell cycle box without Rep2, and that consistent with its postulated function, Rep2 contains 45-amino acid Res2 binding and 22-amino acid transcriptional activation domains in the middle and C terminus of the molecule, respectively. The functional essentiality of these domains is also demonstrated by their requirement for rescue of the cold-sensitive rep2 deletion mutant of fission yeast.
INTRODUCTION
The periodic expression of genes required for cell cycle progression is a common feature of cell cycle regulation in eukaryotes. A number of genes required for the onset of DNA synthesis are expressed during the G1–S transition. In yeast a few such genes contain a cis-regulatory element called MluI cell cycle box (MCB) (ACGCGTNA) in their promoter (Lowndes et al., 1991; McIntosh et al., 1991; for review, see Johnston et al., 1991; Andrews, 1992). A factor complex that specifically binds to the MCB cis-element was initially identified in budding yeast by gel shift assay and called DSC (Lowndes et al., 1991). Subsequently, a DSC-like factor was also detected from the fission yeast Schizosaccharomyces pombe (Lowndes et al., 1992b). DSC consists of at least two distinct molecules, which in S. pombe are Cdc10, a structural homologue of Saccharomyces cerevisiae Swi6, and Res2 or Res1, homologues of budding yeast Swi4 or Mbp1. Cdc10 forms a complex with Res2 or Res1. The Res and Mbp1 subunits possess an MCB binding domain in their N-terminal region (Lowndes et al., 1992b; Tanaka et al., 1992; Caligiuri and Beach, 1993; Koch et al., 1993; Reymond et al., 1993; Miyamoto et al., 1994; Zhu et al., 1994; Ayte et al., 1995; Zhu et al., 1997; for review, see Moll et al., 1993). Although Cdc10 and Swi6 are essential for the DSC activity (Dirick et al., 1992; Lowndes et al., 1992a,b; Verma et al., 1992; Reymond et al., 1993), their role remains unknown. DSC was initially thought to be a transcriptional factor complex that activates MCB, but recent analysis indicates that it is rather a transcriptionally inactive complex responsible for transcriptional repression in late S–G2 for fission yeast (McInerny et al., 1995; Baum et al., 1997). Although the active transcriptional complex requires the same components (Tanaka et al., 1992; Caligiuri et al., 1993; Reymond et al., 1993; Miyamoto et al., 1994; Zhu et al., 1994, 1997), its biochemical nature is little understood.
Recently we identified a new component for the active Res2–Cdc10 complex that functions to start the mitotic cell cycle. It is a zinc-finger protein encoded by rep2+, which was isolated as a multicopy suppressor of a cdc10 mutant (Nakashima et al., 1995). rep2+ suppresses growth defects of cells lacking res1+. The Rep2 molecule binds Res2 in vitro and forms a complex with Res2–Cdc10 in vivo (Nakashima et al., 1995). In the cells deleted for rep2+, MCB is only partially activated, as evident from reduced induction of cdc18+, a key target gene for Res–Cdc10, yet additional deletion of res2+ restores the activation of MCB (Baum et al., 1997). Consequently, these genetic and biochemical data strongly suggested that Rep2 is a transcriptional activator subunit for the Res2–Cdc10 complex that functions as an MCB binding complex.
To obtain definitive evidence and identify the functional domains of Rep2, we reconstituted Res2–Cdc10-dependent transcriptional regulation in the budding yeast S. cerevisiae and carried out deletion analysis of the Rep2 molecule. In this article we provide solid evidence that Rep2 is a transcriptional activator subunit for Res2–Cdc10 and show that the Rep2 molecule contains a Res2 binding and a transcriptional activation domain in the C-terminal half, both of which are essential for Rep2 function.
MATERIALS AND METHODS
Strain and Media
The strains of S. cerevisiae and S. pombe used in this study are listed in Table 1. Media were prepared as described (Guthrie and Fink, 1991; Sturm and Okayama, 1996).
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| YPH499 | MATa ura3-52 ade2-101 leu2-1 trp1-Δ63 his3-Δ200 lys2-Δ1 |
| YPH-ls | MATa ura3-52 ade2-101 leu2-Δ1 trp1-Δ63 his3-Δ200 lys2-Δ1 swi6::TRP1 |
| YPH-lsr2 | MATa ura3-52 leu2-Δ1 trp1-Δ63 his3-Δ200 lys2-Δ1 swi6::TRP1 ade2-101- GAL1 res2+ -ADE2 |
| YPH-ts | MATa ura3-52 ade2-101 leu2-Δ1 trp1-Δ63 his3-Δ20 swi6::TRP1 lys2-Δ1-TMP-HIS3-LYS2 |
| YPH-tsr2 | MATa ura3-52 ade2-101 leu2-Δ1 trp1-Δ63 swi6::TRP1 ade2-101- GAL1 res2+-ADE2 lys2-Δ1-TMP-HIS3-LYS2 |
| N3-141S | h− leu1-32 ura4-D18 rep2::ura4+ |
| K156-D1 | h− leu1-32 ura4-D18 res1::ura4+ |
| M222 | h− leu1-32 ura4-D18 res2::ura4+ |
Construction of Assay System in S. cerevisiae
A LacZ transcriptional unit driven by the triple MluI sequence containing the core promoter of the S. cerevisiae cytochrome c gene (CYC1 −1 ∼ −178) was excised from pSPΔ178.3 M (Lowndes et al., 1991) and subcloned into the single-copy plasmid pRS313 (Sikorski and Hieter, 1989) with the ADH transcriptional terminator. This plasmid was used as a LacZ reporter for monitoring the activation of MCB by Cdc10–Res2. The S. cerevisiae wild-type strain YPH499 (Sikorski and Hieter, 1989) was disrupted for the SWI6 gene by a one-step gene replacement (YPH-ls). The res2+-coding region fused with the GAL1 promoter (Tanaka et al., 1990) was subcloned into the YIP vector containing the ADE2 gene as a selective marker and integrated at the ade2 locus in the cells disrupted for the SWI6 gene (YPH-lsr2).
The HIS3 reporter gene was constructed as follows. The coding region of the HIS3 gene and the 166 bp promoter of the S. cerevisiae thymidine synthase gene (TMP1 −1 to −166 bp) containing two MCB elements (McIntosh et al., 1991) were ligated, subcloned into a GAPDH terminator-containing YIP vector with the LYS2 gene as a selective marker, and integrated into the lys2 locus of YPH499 followed by disruption of the SWI6 gene (YPH-ts). The res2+ gene driven by the GAL1 promoter was integrated at the ade2 locus in YPH-ts to obtain a final host strain YPH-tsr2. Disruption and integration were confirmed by genomic Southern blotting. The vp16-fused cdc10+ gene was constructed by ligating the VP16 activation domain (78 amino acids from 413 to 490 aa) (Sadowski et al., 1988) to the initiation codon of cdc10+, joined to the GAL1 promoter, and inserted into a LEU2 marker-containing single-copy plasmid (Sikorski and Hieter, 1989). The rep2+ coding region was cloned into the multicopy plasmid pKT10-GAPDH driven by the GAPDH promoter (Tanaka et al., 1990).
Assay for Transcriptional Activity of Res2–Cdc10–Rep2 in the Reconstitution System
The assay host strains transfected with the indicated expression constructs and the LacZ reporter plasmid were inoculated in synthetic minimal medium (SD) containing 3% galactose and 0.2% sucrose at 30°C and grown to log phase. The cells were harvested and ruptured by freeze and thaw, and then β-galactosidase activity was measured as described (Clontech, Cambridge, United Kingdom).
The HIS3 selection host cells were transfected with the indicated constructs and selected on SD plate containing 2% glucose at 30°C for 3 d. The transfectants were spotted on 3% galactose/0.2% sucrose SD plate containing 4 mM 3-aminotriazole (3AT) and incubated at 30°C for 10 d. 3AT was used to inhibit the basal activity of the HIS3 gene product in this strain.
Yeast One- and Two-Hybrid Systems
The yeast one- and two-hybrid assay systems were performed using the commercial Matchmaker two-hybrid system (Clontech). To construct the full-length and deletion mutants of rep2+, fragments from the indicated amino acid to the end of ORF were excised from pcL-rep2+ by PCR. To destroy the zinc-finger motif in Rep2, 177 Cys (tgc) was changed to Gly (ggt) and 180 Cys (tgc) to Ser (tcc). These constructs were inserted into pGBT9. The res2+ gene fused with the GAL4 transactivation domain (pGAD424) was constructed as described previously (Nakashima et al., 1995)
The S. cerevisiae reporter strain SFY526 was transfected with pGAD424-X and pGBT9-rep2+ for the one-hybrid system and with pGAD424-res2+ and pGBT9-rep2+ for the two-hybrid system. Transformants were cultured to log phase in SD medium at 30°C and then harvested and ruptured by freeze and thaw, and β-galactosidase activity was measured as described (Clontech).
Antibodies
The anti-Res1/2 antibody was described previously (Nakashima et al., 1995). The anti-FLAG M2 affinity gel was purchased from IBI (New Haven CT), and the anti-FLAG D8 polyclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Protein Extraction and Immunoprecipitation
The rep2 (N3–141S), res1 (K156-D1), and res2 (M222) disruptants were transfected with FLAG-tagged deletion mutants of rep2+, which were induced by culturing in thiamine-free pombe minimal medium (PM) at 30°C for 13–17 h depending on the constructs to produce similar levels of protein. The crude extract was prepared as described (Nakashima et al., 1995), and protein concentration was determined by the Bradford method. Crude cell extracts (4.5 mg/ml) were incubated at 4°C for 1 h with 20 μl of anti-FLAG M2 affinity gel containing 0.15 M NaCl followed by centrifugal sedimentation of immunoaffinity gel-bound proteins. Affinity gel-bound proteins were washed five times with 500 μl of buffer H (Booher et al., 1989) containing 0.15 M NaCl, 1 mM PMSF, 20 μg/ml pepstatin A and leupeptin, and 10 μg/ml aprotinin, separated by SDS-PAGE (7.5% gel for anti- Res1/2 and 15% for anti-FLAG), and analyzed by Western blotting using anti-Res1/2 and anti-FLAG D8 antibodies as described previously (Jinno et al., 1994) and in the IBI protocol.
Suppression of rep2 Disruptant Cells by Deletion Mutants of the rep2+ Gene
For assay in S. pombe, all of the constructs were expressed from the SV40 promoter (Okazaki et al., 1990). The deletion mutants of rep2+ were constructed by PCR amplification followed by insertion into the pcL vector.
The rep2 disruptant was transformed with the indicated constructs as described (Okazaki et al., 1990). One-half of the transfectant was selected for Leu+ at 30°C, and the other half was selected first at 30°C for 16 h and then at 18°C for 2 wk. The ratios of colonies formed at the nonpermissive temperature to those formed at the permissive temperature were expressed as percentage suppression.
Cell Cycle Distribution and Expression of cdc18+ mRNA
The rep2 disruptant (N3–141S) was transfected with the indicated plasmids and selected for Leu+ at 30°C. Transformants were cultured at 30°C to midlog phase in PM medium and shifted to 18°C. Flow cytometry and Northern blotting of cdc18+ mRNA were performed for the cells before and after a 53 h incubation at 18°C. The probe was the 32P-labeled BamHI fragment of cdc18+. Flow cytometry was performed as described previously (Nakashima et al., 1995) using the FACScan system and CellFIT cell cycle analysis program and the software LYSISII (Becton Dickinson, San Jose, CA).
RESULTS
Reconstruction of Transcriptional Activation by Res2–Cdc10 in Budding Yeast
One clear demonstration of the requirement for Rep2 in the Res2–Cdc10 activity is the reconstitution of this transcriptional system in an evolutionary distant organism. S. cerevisiae is a suitable organism for such an experiment because Swi6 cannot functionally be substituted with its fission yeast homologue Cdc10 (Lowndes et al., 1992a). Figure 1 schematically represents the reconstitution system we constructed. In this system, two reporter genes were used to confirm the dependence of Res–Cdc10-activated transcription on MCB and its independence from the core promoter used. One is the LacZ coding sequence ligated to a core sequence (−1 ∼ −178) of the CYC (cytochrome c) gene promoter fused with artificially synthesized three repeats of the MluI sequence as a Res–Cdc10-responsive MCB cis-element. The other is the His3 coding sequence ligated to the TMP promoter (−1 ∼ −166) containing the two endogenous MCB motifs. The resulting reporter genes were expressed in appropriate host cells from a single-copy plasmid or integrant. Accordingly, promoter activation was assayed by determining induced β-galactosidase activity or the cell’s ability to grow without histidine.
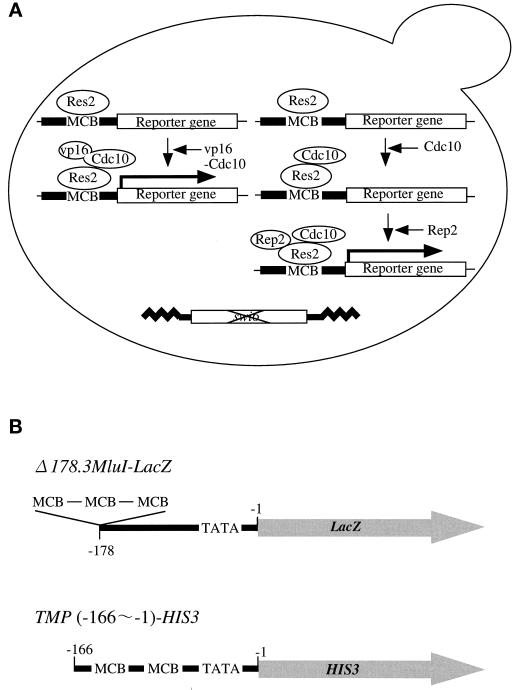
Figure 1.
Schematic representation of the assay systems for in vivo transcriptional activation by Res2–Cdc10–Rep2. (A) Assays for transcription activation by Res2–Cdc10–Rep2 were carried out with a Δswi6 strain of S. cerevisiae and a single copy of a reporter gene whose promoter contains two or three MCB elements. Res2 by itself would not activate reporter gene, whereas the Res2–vp16–Cdc10 complex would. Res2–Cdc10 would have no or very low transcriptional activity, but coexpression of Rep2 with Res2–Cdc10 is expected to activate the reporter gene. (B) Structure of reporter genes in Δ178.3 MluI-LacZ, the promoter of S. cerevisiae CYC1 (−1 ∼ −178) containing exogenously inserted triple MluI sequences (MCB element) is fused with the LacZ coding sequence. In TMP-HIS3, the S. cerevisiae TMP promoter (−1 ∼ −166) containing two endogenous MCB motifs at −159 and −122 is ligated to the S. cerevisiae His3 coding sequence.
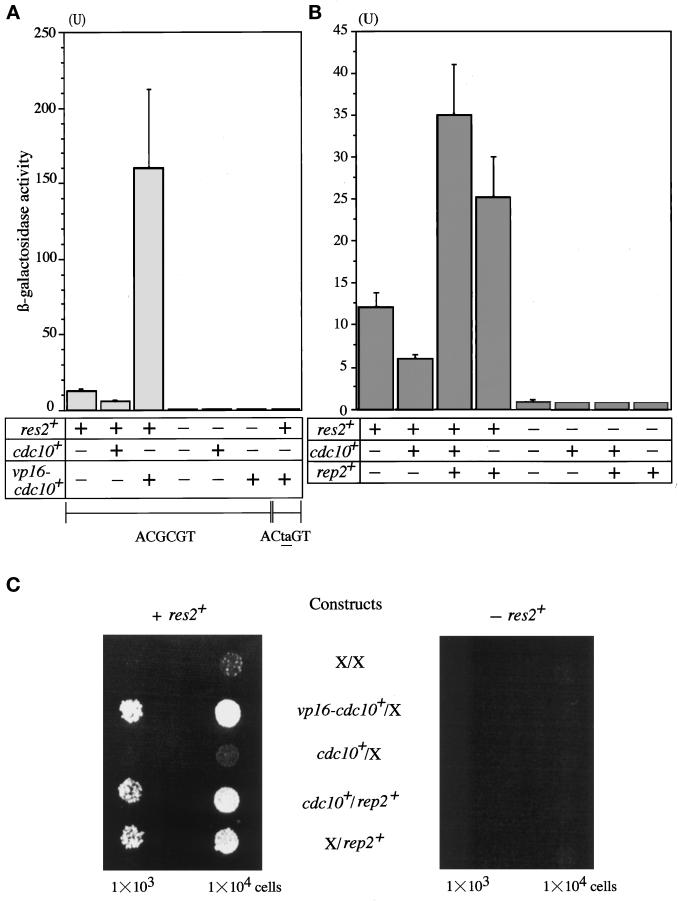
As already noted, the budding yeast contains Cdc10–Res homologues that can activate the MluI sequence used as MCB. To reduce background levels caused by this homologous system, the SWI6 gene was deleted from the host cells. In addition, the res2+ gene under the control of the GAL1 promoter was integrated into the ade2 locus of host cells. The proper function of the enhancer and the inducibility of the artificial promoter were confirmed by β-galactosidase activity strongly induced by coexpression of Res2 and vp16 (transactivator activity)-fused Cdc10. This induction was absolutely dependent on the MCB cis-element. When the MluI sequence was mutated to ACtaGT, induction of β-galactosidase was completely failed (Figure 2A). In this control experiment, expression of Res2 alone slightly induced β-galactosidase activity.
Figure 2.
(A and B) Reconstruction of transcriptional activation by Res2–Cdc10 in budding yeast. Rep2–Res2–Cdc10 complex activates MCB (triple MluI sequences). The LacZ reporter host cells were transfected with the indicated plasmids. The transformants were then induced for the expression of res2+ and cdc10+ by incubating in SD medium containing 3% galactose and 0.2% sucrose at 30°C for 16 h. Cells were harvested and ruptured by freeze and thaw followed by colorimetric assay of β-galactosidase. For the experiment shown in the far right column of A, the LacZ reporter gene driven by the CYC minimal promoter containing triple repeats of the mutated MluI sequences (ACtaGT) was used. Experiments were repeated three times, and values are expressed as means ± SD. (C) Rep2–Res2–Cdc10 complex activates the TMP promoter. The HIS3 reporter host cells with or without expressing res2+ were transfected with the indicated constructs. The transfectants were spotted on 3% galactose/0.2% sucrose SD plate containing 4 mM 3-AT and incubated at 30°C for 10 d.
However, coexpression of Cdc10 did not elevate β-galactosidase activity but rather repressed its level. Coexpression of vp16-fused Cdc10 induced β-galactosidase activity >10-fold. This induction depended on the coexpression of Res2. Similar results were obtained with the His3 selection host (Figure 2C). These results show that Res2–Cdc10 alone can recognize, but cannot activate, MCB.
Rep2 Activates the Res2–Cdc10 Complex
We next examined whether Rep2 could activate Res2–Cdc10 in the reconstitution system. In this experiment, the rep2+ coding sequence was inserted into a GAPDH promoter-driven multicopy expression vector and expressed in the host cells with or without expression of Res2 and Cdc10. As shown in Figure 2B, in the presence of Res2 and Cdc10, Rep2 coexpression activated the MCB-containing CYC promoter six- to sevenfold as measured by β-galactosidase activity. Interestingly, coexpression of only Res2 and Rep2 induced β-galactosidase activity, but to a slightly low level. Because the basal level of β-galactosidase activity induced by Res2 alone was a bit higher, the induction by Rep2 coexpression was only twofold. In fission yeast, overexpression of Res1 completely and overexpression of Res2 at least largely dispense Cdc10 (Tanaka et al., 1992; Caligiuri and Beach, 1993; Miyamoto et al., 1994; Zhu et al., 1994). This situation thus appears to be reproduced with the reconstitution system in a heterogeneous organism. As expected, the activation of the promoter by Rep2 absolutely depended on Res2. These results were confirmed with the reconstitution system using the HIS3 selection host. Virtually identical or even clearer results were obtained by using this system (Figure 2C). These results support our previous assignment of Rep2 to a transcriptional activator subunit for the Res2–Cdc10 complex.
Transactivation Domain of Rep2 Is Located at the C Terminus
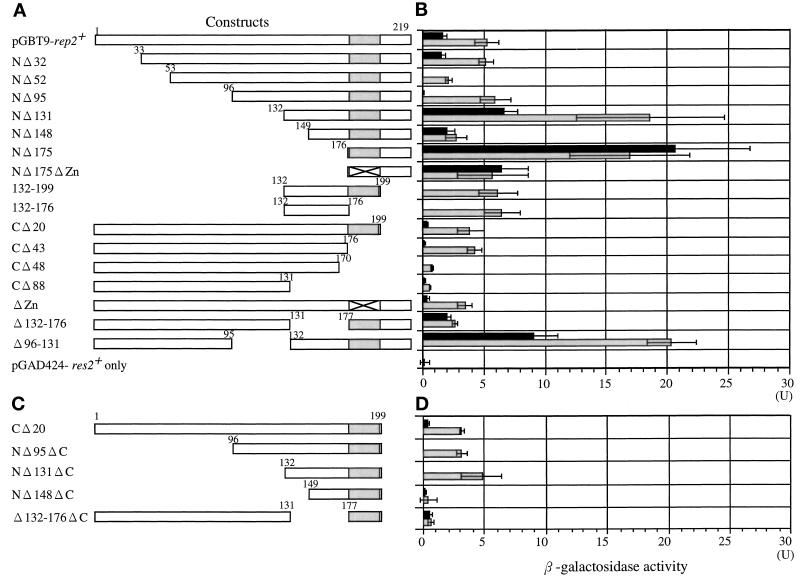
If Rep2 is indeed a transcriptional activator subunit for Res2–Cdc10, it must contain a domain that is responsible for transcriptional activation. We examined this question by using a budding yeast one-hybrid system. A series of N- and C-terminal and internal deletion mutants of rep2+ were constructed, fused with the Gal4 DNA binding domain at their N terminus, and expressed in S. cerevisiae containing a LacZ reporter gene with a Gal4 binding element in the promoter (Figure 3A). The transcriptional activation ability of the deletion mutants was then assayed by measuring induced β-galactosidase activity (Figure 3B, black bar).
Figure 3.
(A and C) Identification of transactivation and Res2 binding domains in Rep2. Schematic representation of deletion mutants analyzed in this study. The intact rep2+ gene is shown at the top. The zinc-finger motif is shaded. The amino acid numbers are shown at each protein coding region. Full-length and deletion mutants of rep2+ were fused to a DNA binding domain of Gal4. (D) One-hybrid and two-hybrid analysis for transcriptional activator and Res2 binding domains in Rep2. The DNA binding domain hybrid was transfected into the yeast reporter strain SFY526 together with (gray bar) or without (black bar) Res2-fused Gal4 activation domain. After growth on selective medium, each transformant was assayed for β-galactosidase activity. Black bars indicate transactivation activity, and the difference between gray and black bars reflects Res2 binding activity. Experiments were repeated four times in B and three times in D; data are presented as means ± SD.
In this hybrid construct, full-length Rep2 showed a relatively low LacZ induction. Progressive N-terminal deletion of Rep2 initially abolished, but further deletion restored, transcriptional activator activities, which reached the maximum (an ∼10-fold greater induction than intact Rep2) when all the molecules but the C-terminal 44 amino acids were deleted. This result suggests that the transcriptional activator activity is localized within this region. Consistently, deletion of the C-terminal 20 amino acids from NΔ131 abolished the transcriptional activator activity. Analysis with progressive C-terminal or internal deletion mutants yielded results consistent with the N-terminal deletion analysis data.
The 44-amino acid region contains a zinc-finger motif (Nakashima et al., 1995). The next question we examined is whether this finger is essential for activity. Two cysteine residues forming the zinc-finger motif were substituted with glycine and serine, respectively. The resulting zinc-fingerless Rep2 molecule showed a reduced ability to activate the reporter gene, suggesting that this motif is important for transcriptional activator function. However, the importance of this motif for Rep2 activity appears to occur only in the budding yeast one-hybrid system. We failed to find any significant requirement for this motif in fission yeast and in the budding yeast reconstitution system (see below). In addition, the apparent requirement for the N-terminal 33–95 amino acid region in Rep2 transcriptional activator function was seen only in budding yeast. In fission yeast, this region was dispensable (see below).
Identification of Res2 Binding Domain
The second key property required for Rep2 function as a transcriptional activator subunit is the ability to physically interact with Res2, because Rep2 is required for the activity of Res2–Cdc10 but not Res1–Cdc10 (Nakashima et al., 1995; Baum et al., 1997). Rep2 forms a complex with Res2–Cdc10 in vivo as well as in vitro (Nakashima et al., 1995). To confirm this and identify the region essential for Res2 binding, we used a two-hybrid binding analysis, in which the same Rep2 deletion constructs fused with the Gal4 DNA binding domain (GD-Rep2 deletion mutant) and Res2 fused with the Gal4 transcriptional activator domain (GT-Res2) were coexpressed. Because at least some GD-Rep2 deletion mutants contain transcriptional activator function, their Res2 binding abilities were initially measured by assaying increases in β-galactosidase activity from the level obtained by expression of GD-Rep2 mutants alone (black bars) to that obtained by coexpression of GD-Rep2 and GT-Res2 (gray bars). Such indirect assays tended to yield quantitatively less accurate results, which would be improved by use of transcriptional activation domainless Rep2; however, we did not initially use such a construct because the Res2 binding domain might overlap with, or be proximal to, the transcriptional activator domain.
Deletion of 131 amino acids from the N terminus was judged to have no marked effect on binding to Res2, but deletion of 148 amino acids or more abolished binding activity as indicated by NΔ148 and NΔ175. On the other hand, deletion of the 43-amino acid transcriptional activator domain (CΔ43) did not affect binding to Res2, but deletion of an additional 5 amino acids (CΔ48) almost completely abolished binding ability. These results suggest that the region of amino acid 132 to 176 is required for Res2 binding. Consistently, internal deletion of this region (Δ132–176) completely abolished binding ability. Because the tentatively assigned Res2 binding region did not overlap with the tentatively assigned transactivator domain, we performed the same assay with transactivator domainless Rep2 constructs shown in Figure 3C, to confirm the results with the original constructs. As shown in Figure 3D, the same results were obtained with the transactivator domainless constructs. These results indicate that the 45-amino acid sequence from amino acid 132 to 176 contains a Res2 binding ability.
Res2 Binding Domain Is Required for Res2 Binding In Vivo
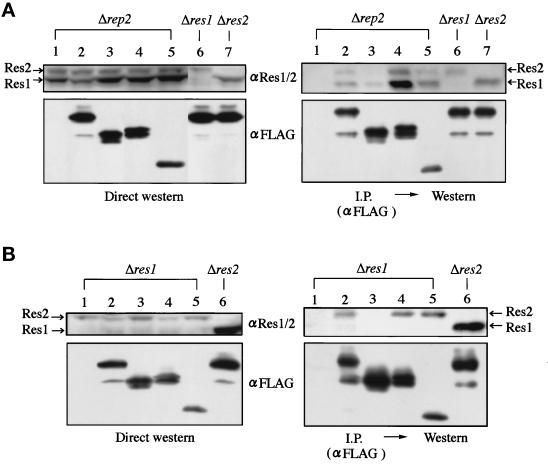
To confirm the assignment of the Res2 binding domain, we examined the ability of Rep2 deletion mutants to associate with Res2 in an in vivo situation. FLAG-tagged, Res2 binding domainless rep2+ was placed under the control of the thiamine-inducible promoter, expressed in the rep2 (Figure 4A) or res1 (Figure 4B) disruptant, immunoprecipitated with anti-FLAG antibody, and assayed for coprecipitation of Res2 by Western blotting with anti-Res1/2 antibody. The anti-Res1/2 antibody detects Res2 (top band) and Res1 (bottom band) (Nakashima et al., 1995) (Figure 4A, lanes 6 and 7). As we showed previously, not only Res2 but also Res1 coprecipitates with Rep2 (Figure 4A) (Nakashima et al., 1995), but the biological significance of the association of Rep2 with Res1 is still unclear. It could be an artifact of an overexpressed situation because we failed to obtain any results suggesting their functional interaction (Nakashima et al., 1995; Sturm and Okayama, 1996).
Figure 4.
The putative Res2 binding domain in Rep2 is required for its Res2 binding in vivo. Full-length and deletion mutants of rep2+ tagged with FLAG epitope were inserted into the pREP1 vector and transfected into the rep2 (N3–141s), res1 (K156-D1), and res2 disruptants (M222). The structures of rep2+ deletion mutants are shown in Figure 3A except for the Δ22–131 aa mutant, which was constructed by fusing the sequence of amino acids 1–21 with NΔ131. Cell extracts (4.5 mg protein) prepared from the transformants grown in thiamine-free medium at 30°C to express the FLAG-tagged Rep2 were immnoprecipitated with the anti-FLAG antibody. Cell extracts (200 μg protein) (left panels) and immunoprecipitates (right panels) were separated in SDS-polyacrylamide gels and analyzed by Western blotting with anti-Res1/2 or anti-FLAG antibody. Lane 1, empty vector; lanes 2, 6, 7, full-length rep2+; lane 3, Δ132–176 aa; lane 4, CΔ43; lane 5, Δ22–131 aa.
In good agreement with the two-hybrid assay results, the same or higher amount of Res2 protein coprecipitated with any of the tested mutant Rep2 proteins that lack the regions N- or C-terminal to the border of the Res2 binding domain but hold the intact Res2 binding domain. Furthermore, no Res2 protein coprecipitated with the Rep2 lacking the putative Res2 binding domain (Δ132–176), despite the presence of the same amount of Res2 in this cell extract as in others.
Recent analysis indicated that Res2 forms a heteroduplex complex with Res1 in S. pombe (Ayte et al., 1997; Baum et al., 1997; Zhu et al., 1997). To further confirm the results in the absence of such a complication, we performed the same assay as above, but with the res1 disruptant as a host, and obtained identical results. Again, Res2 protein coprecipitated with all the Rep2 constructs containing the putative Res2 binding domain but not with the one lacking the binding domain (Δ132–176). In this experiment, even a faint amount of Res2 failed to coprecipitate with the binding domainless Rep2. These results show that the 45-amino acid sequence from 132 to 176 is essential and sufficient for Res2 binding in vivo, leading us to conclude that the Res2 binding domain resides in this region.
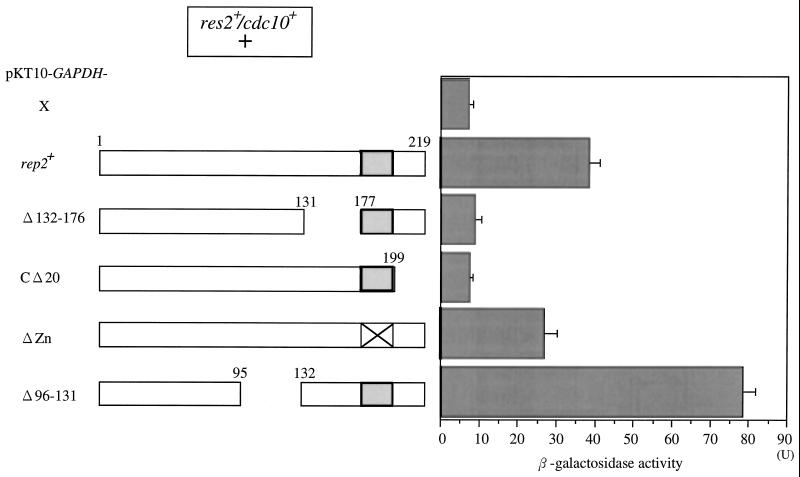
Both Transactivation and Res2 Binding Domains Are Required for Rep2 Function in the Reconstitution System and in Fission Yeast
To corroborate the results obtained by the one- and two-hybrid systems and the in vivo coimmunoprecipitation assay, the functional importance of the identified domains was examined in the reconstitution system (Figure 5). In the reconstitution system, deletion of the C-terminal 20 amino acids (CΔ20), which were essential for transcriptional activator function in the one-hybrid system, completely abrogated the ability of Rep2 to induce β-galactosidase. Similarly, deletion of the Res2 binding domain (Δ132–176) completely abolished Rep2 function. Interestingly, deletion of the zinc-finger motif (ΔZn) only slightly decreased Rep2 activity.
Figure 5.
Transactivation and Res2 binding domains are required for Rep2 function in the reconstitution system. Full-length and deletion mutants of rep2+ were inserted into the pKT10-GAPDH vector and transfected into the LacZ reporter host cells harboring res2+ and cdc10+, which were then induced for res2+ and cdc10+ by incubating at 30°C for 16 h in SD medium containing 3% galactose and 0.2% sucrose followed by β-galactosidase assay. Experiments were repeated three times, and data are presented as means ± SD.
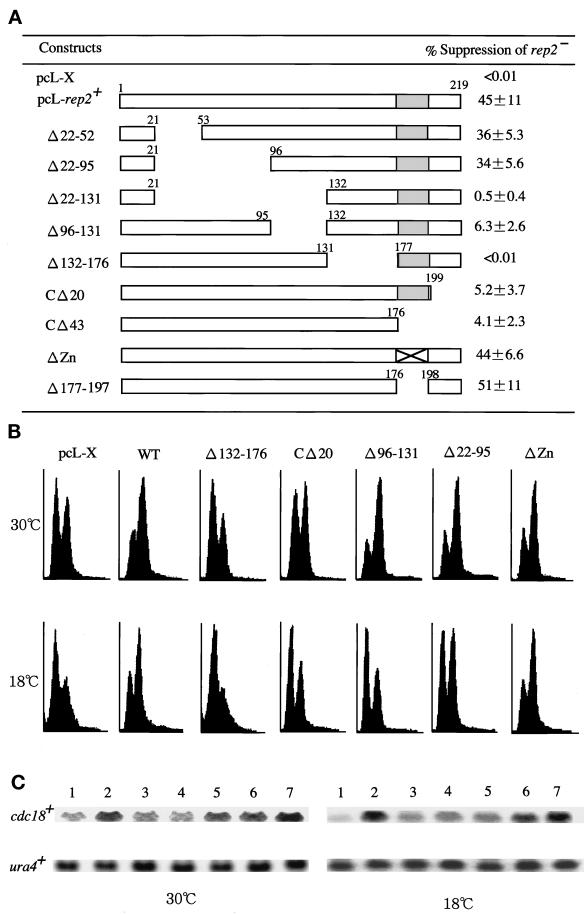
To further corroborate the experimental data, the activity of the rep2+ deletion mutant genes was assayed in fission yeast. The fission yeast host used was a rep2 deletion mutant cell. This mutant cell can grow at regular growth temperatures but is slow in traversing the G1–S transition. Consequently, a significant G1 population is noticeable. In addition, this cell cannot grow at 18°C or lower temperatures (Nakashima et al., 1995). Accordingly, the activity of the rep2+ gene mutants was assayed by two methods. One was rescue of the cold-sensitive growth arrest of the mutant. The other was acceleration of the slow cell cycle start of the mutant and simultaneous induction of cdc18+ mRNA at 30°C. To carry out these assays, various deletion mutants of rep2+ were inserted in the SV40 early promoter-based pcL vector, transfected into the rep2 disruptant, and selected at 18°C. In this assay, the activity of the rep2+ gene deletion mutants was measured as percentage colony formation at 18°C against at 30°C. As shown in Figure 6A, the rep2 genes lacking the putative Res2 binding domain or the C-terminal 20 amino acids completely or nearly completely failed to rescue the mutant, confirming the results obtained with the one- or two-hybrid and reconstitution systems. Similar results were also obtained with the second assay for G1 acceleration as well as elevation of cdc18+ mRNA (Figure 6, B and C). The rep2 gene lacking the putative Res2 binding domain showed no detectable activity in this assay either. On the other hand, the C-terminal 20-amino acid-deleted rep2 gene had only a marginal activity with a slight decrease in G1 population but no apparent changes in cdc18+ mRNA upon its expression.
Figure 6.
Both transactivation and Res2 binding domains are required for Rep2 function in fission yeast. (A) Rescue of low-temperature growth inability of rep2− cells by expression of rep2+ deletion mutants. Full-length and various deletion mutants of rep2+ were inserted into the pcL vector and transfected into the rep2 disruptant (N3–141S) followed by selection at 18°C. The ratio of colonies formed at 18°C to those at 30°C is expressed as percentage suppression in the right column. pcL-X is the pcL vector with no insert and is used as a negative control. Experiments were repeated four times, and data are presented as means ± SD. (B) Flow cytometry of rep2 disruptants expressing each rep2+ deletion mutant. The rep2 disruptant was transfected with the indicated constructs and selected for Leu+ transformants. The transformants were grown to log phase at 30°C in PM medium and then incubated at 18°C for 53 h. Cells were sampled both at 30°C and after a 53 h incubation at 18°C, and analyzed by flow cytometry. (C) The level of cdc18+ mRNA expressed in the rep2 disruptants rescued by rep2+ deletion mutant genes. Total RNA was prepared from the rep2 disruptants at the same time points as described in B. The level of the cdc18+ transcript was determined by Northern blot hybridization. ura4+ was probed as a loading control. Lane 1, empty vector; lane 2, full-length rep2+; lane 3, Δ132–176 aa; lane 4, CΔ20; lane 5, Δ96–131 aa; lane 6, Δ22–95 aa; lane 7, ΔZn finger.
The one-hybrid assay showed that the zinc-finger motif was important for transcriptional activator function, although its role was obscure when assayed in the reconstitution system. In fission yeast, however, this motif was totally dispensable under our assay conditions. Moreover, deletion of the 21-amino acid region containing the zinc-finger motif had no apparent effect on the Rep2 activity in this assay. We therefore tentatively conclude that not only the zinc finger but also the entire 21 amino acids containing this motif are dispensable for Rep2 function as a transcriptional activator subunit for the Res2–Cdc10 complex under the assay conditions used.
As shown already, in the one- and two-hybrid and reconstitution systems, the region (36 amino acids) just upstream of the Res2 binding domain was totally dispensable or rather inhibitory to Rep2 function (Figures 3 and 5). This region, however, was found to be essential for Rep2 function in this rescue assay. Deletion of this region nearly completely eliminated the ability of Rep2 to rescue the low-temperature growth arrest of the rep2 disruptant. The reason for this apparent contradiction became clear in the second assay.
The rep2 disruptant displays a large G1 peak during exponential growth at 30°C, which is due to slow cell cycle start caused by partial sequestering of MCB by the inactive Res2–Cdc10 complex. Expression of wild-type rep2+ suppressed this G1 peak and concurrently increased the level of cdc18+ mRNA (Figure 6, B and C). In this assay, rep2+ lacking the 36-amino acid region was as active as wild-type gene; however, when this assay was carried out at 18°C, the same temperature as for the rep2 rescue assay, the rep2+ mutant gene failed to show any significant activity. These results suggest that this 36-amino acid region is essential for Rep2 at low temperatures but dispensable at the regular growth temperatures.
On the basis of all these results taken together, we conclude that the Rep2 molecule contains a Res2 binding domain within amino acids 132–176 and a main transcriptional activator domain within the C-terminal 22 amino acids.
DISCUSSION
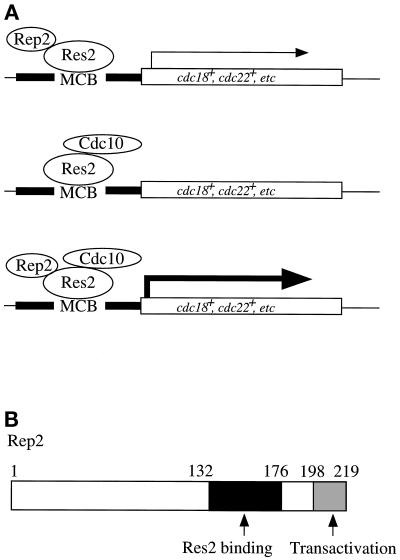
The Res–Cdc10-led transcriptional regulation of genes required for S-phase onset, particularly of cdc18+ that encodes a component of the prereplicative complex, is a key step in regulating the start of the cell cycle. One critical question concerning this transcriptional system is how it is regulated. We recently showed that at least the Res2–Cdc10 complex itself is inactive as transcriptional activator and requires Rep2, a nitrogen starvation-repressible zinc-finger protein, for activity in the mitotic cell cycle (Nakashima et al., 1995). Because Rep2 has a transcriptional activation ability in the one-hybrid system, binds to Res2 in vivo as well as in vitro, and is essential for MCB activation but not for the MCB binding ability of the Res2–Cdc10 complex (Zhu et al., 1994; Nakashima et al., 1995; Sturm and Okayama, 1996), we tentatively assigned Rep2 to a transcriptional activator subunit for Res2–Cdc10 in the mitotic cycle. In this article we show that Rep2 fulfills all the criteria required for a transcriptional activator subunit. First, Rep2 forms a complex with Res2–Cdc10 in vivo. Rep2 has a defined region required for Res2 binding in vitro as well as in vivo and an ability to activate Res2–Cdc10 in the budding yeast two-hybrid and reconstitution systems. Second, Rep2 has a defined sequence capable of transcriptional activation when fused with a DNA binding domain, and this region is also essential for Rep2’s ability to activate Res2–Cdc10. Third, unless provided with Rep2, the Res2–Cdc10 complex itself has no significant ability to activate MCB in fission yeast as well as in the budding yeast reconstitution system. We therefore conclude that Rep2 is a transcriptional activator subunit for Res2–Cdc10 (Figure 7A).
Figure 7.
(A) Activity of the Res2–Cdc10–Rep2 complex suggested by analysis. When overproduced, the Res2 molecule binds and weakly activates MCB in the presence of Rep2. In the absence of Rep2, the Res2–Cdc10 complex can bind but cannot activate MCB. The Res2–Cdc10–Rep2 ternary complex strongly activates MCB. (B) Functional domains in Rep2. The Rep2 molecule contains a Res2 binding domain (black box) at amino acids 132–176 and a transactivator domain (gray box) at amino acids 198–219.
Deletion analysis of rep2+ gene led to identification of a Res2 binding and a transcriptional activator domain locating at amino acids 132–176 and 198–219, respectively (Figure 7B). All of the genetic and biochemical data are consistent with the functional assignment of these two domains. One unexpected outcome is the failure to assign function to the zinc-finger motif that is located between these two regions and that we speculated was important for either Res2 binding or transcriptional activation. In the one-hybrid analysis, this motif was required for the transcriptional activation ability of Rep2, although to a lesser extent in the reconstitution system; however, in fission yeast, not only this motif but also the entire 21-amino acid region containing this motif was dispensable for Rep2 activity under the assay conditions used. Our data do not exclude the possibility that this motif might be involved in some other functions, such as facilitation of Res2 binding and stabilization of the Rep2 protein molecule, which might be detectable only in a low level expression.
One region with an unexpected function locates at amino acids 96–131, just upstream of the Res2 binding domain. This region is either dispensable or inhibitory to Rep2 activity at the regular growth temperature, but absolutely essential at a low temperature of 18°C. The reason for this is unclear at present, but one possibility is that this region might be required for proper protein folding of Rep2 at low temperatures. No matter what the reason, the presence of such a domain in Rep2, however, seems to be reasonable because Res2 appears to preferentially function at low temperatures in the mitotic cycle. Cells deleted for res2+ show cold sensitivity for growth (Zhu et al., 1994).
ACKNOWLEDGMENTS
We thank L. Johnston for the pSPΔ178.3 M and A. Tho-e, M. Nishizawa, Y. Kikuchi, Y. Matsui, and Y. Uezono for the S. cerevisiae strains, plasmids, and critical advice on constructing the assay system in S. cerevisiae. We thank the members of H.O.’s laboratory for their critical advice.
REFERENCES
- Andrews BJ. Gene expression. Dialogue with the cell cycle. Nature. 1992;355:393–394. doi: 10.1038/355393a0. [DOI] [PubMed] [Google Scholar]
- Ayte J, Leis JF, DeCaprio JA. The fission yeast protein p73res2 is a essential component of the mitotic MBF complex and a master regulator of meiosis. Mol Cell Biol. 1997;17:6246–6254. doi: 10.1128/mcb.17.11.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayte J, Leis JF, Herrera A, Tang E, Yang H, DeCaprio JA. The Schizosaccharomyces pombe MBF complex requires hetero dimerization for entry into S phase. Mol Cell Biol. 1995;15:2589–2599. doi: 10.1128/mcb.15.5.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Wuarin J, Nurse P. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 1997;16:4647–4688. doi: 10.1093/emboj/16.15.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Alfa CE, Hyams JS, Beach DH. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Caligiuri M, Beach DH. Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell. 1993;72:607–619. doi: 10.1016/0092-8674(93)90079-6. [DOI] [PubMed] [Google Scholar]
- Dirick L, Moll T, Auer H, Nasmyth K. A central role for SWI6 in modulating cell cycle start-specific transcription in yeast. Nature. 1992;357:508–513. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink RG. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Lowndes NF, Johnson AL, Sugino A. A cell-cycle-regulated transfactor, DSC1, controls expression of DNA synthesis genes in yeast. Cold Spring Harbor Symp Quant Biol. 1991;56:169–176. doi: 10.1101/sqb.1991.056.01.022. [DOI] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Lowndes NF, Johnson AL, Breeden L, Johnston LH. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature. 1992a;357:505–508. doi: 10.1038/357505a0. [DOI] [PubMed] [Google Scholar]
- Lowndes NF, Johnson AL, Johnston LH. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated transfactor. Nature. 1991;350:247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- Lowndes NF, McInerny CJ, Johnson AL, Fantes PA, Johnston LH. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+ Nature. 1992b;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- McInerny CJ, Kersey PJ, Creanor J, Fantes PA. Positive and negative roles for cdc10+ in cell cycle gene expression. Nucleic Acids Res. 1995;23:4761–4768. doi: 10.1093/nar/23.23.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh EM, Atkinson T, Storms RK, Smith M. Characterization of a short, cis-acting DNA sequence which conveys cell cycle stage-dependent transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:329–337. doi: 10.1128/mcb.11.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M, Tanaka K, Okayama H. res2+, a new member of the cdc10+/SWI4 family, controls the “start” of mitotic and meiotic cycles in fission yeast. EMBO J. 1994;13:1873–1380. doi: 10.1002/j.1460-2075.1994.tb06456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T, Schwob E, Koch C, Moore A, Auer H, Nasmyth K. Transcription factors important for starting the cell cycle in yeast. Philos Trans R Soc Lond B Biol Sci. 1993;340:351–360. doi: 10.1098/rstb.1993.0078. [DOI] [PubMed] [Google Scholar]
- Nakashima N, Tanaka K, Sturm S, Okayama H. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle “start” function of Res2-Cdc10. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High frequency transformation method and library transducing vectors for cloning mammalian cDNAs by transcomplementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Marks J, Simanis V. The activity of S. pombe DSC-1-like factor is cell cycle regulated and dependent on the activity of p34cdc2. EMBO J. 1993;12:4325–4334. doi: 10.1002/j.1460-2075.1993.tb06117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm S, Okayama H. Domains determining the functional distinction of the fission yeast cell cycle “start” molecules Res1 and Res2. Mol Biol Cell. 1996;7:1967–1976. doi: 10.1091/mbc.7.12.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Tamanoi F, Kaziro Y, Matsumoto K, Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990;10:4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Okazaki K, Okazaki N, Ueda T, Sugiyama A, Nojima H, Okayama H. A new cdc gene required for S phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SWI4 gene products. EMBO J. 1992;11:4923–4932. doi: 10.1002/j.1460-2075.1992.tb05599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Smiley J, Andrews B, Campbell JL. Regulation of the yeast DNA replication genes through the MluI cell cycle box is dependent on SWI6. Proc Natl Acad Sci USA. 1992;9:9479–9483. doi: 10.1073/pnas.89.20.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Takeda T, Nasmyth K, Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Takeda T, Whitehall S, Peat N, Jones N. Functional characterization of the fission yeast Start-specific transcription factor Res2. EMBO J. 1997;16:1023–1034. doi: 10.1093/emboj/16.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]