Abstract
Cardiac ATP-sensitive potassium (KATP) channels are metabolic sensors formed by the association of the inward rectifier potassium channel Kir6.2 and the sulphonylurea receptor SUR2A. SUR2A adjusts channel gating as a function of intracellular ATP and ADP and is the target of pharmaceutical openers and blockers which, respectively, up- and down-regulate Kir6.2. In an effort to understand how effector binding to SUR2A translates into Kir6.2 gating modulation, we examined the role of a 65-residue SUR2A fragment linking transmembrane domain TMD2 and nucleotide-binding domain NBD2 that has been shown to interact with Kir6.2. This fragment of SUR2A was replaced by the equivalent residues of its close homologue, the multidrug resistance protein MRP1. The chimeric construct was expressed in Xenopus oocytes and characterized using the patch-clamp technique. We found that activation by MgADP and synthetic openers was greatly attenuated although apparent affinities were unchanged. Further chimeragenetic and mutagenetic studies showed that mutation of three residues, E1305, I1310 and L1313 (rat numbering), was sufficient to confer this defective phenotype. The same mutations had no effects on channel block by the sulphonylurea glibenclamide or by ATP, suggesting a role for these residues in activatory – but not inhibitory – transduction processes. These results indicate that, within the KATP channel complex, the proximal C-terminal of SUR2A is a critical link between ligand binding to SUR2A and Kir6.2 up-regulation.
ATP-sensitive potassium channels (KATP) are potassium-selective channels widely distributed among most types of excitable cells, where their primary function is to couple membrane excitability to intracellular metabolic levels (Seino & Miki, 2003; Nichols, 2006) by sensing the balance between cytoplasmic ATP, which inhibits them, and MgADP which activates them (Tarasov et al. 2004; Alekseev et al. 2005). This metabolic sensing role is involved in major physiological processes: it plays a role in the cascade linking insulin secretion to glycaemia in the pancreatic β-cell, and participates in ischaemic pre-conditioning and thereby cardio-protection during heart failure (Ashcroft et al. 1984; Jahangir & Terzic, 2005; Yamada et al. 2006). KATP channels possess an unusual architecture made of two subunits: the inward-rectifier potassium channel Kir6.2 (Inagaki et al. 1995) and the sulphonylurea receptor SUR (Aguilar-Bryan et al. 1995) of the ATP-binding cassette (ABC) protein family. Four Kir6.2 subunits assemble to form the K+-selective pore of the channel, inhibited by nucleotides, and four SUR subunits surround this pore and regulate its gating, activating it in the presence of MgADP or KATP channel openers and inhibiting it in the presence of sulphonylureas (Clement et al. 1997; Moreau et al. 2005b). Stoichiometry control is ensured by the presence on both Kir6.2 and SUR of Arginine-based endoplasmic reticulum (ER) retention signals, allowing only correctly assembled octameric channels to reach the plasma membrane (Zerangue et al. 1999).
SUR has strong homologies with other members of subfamily C of ABC proteins. SUR possesses the core domain of ABC proteins (Fig. 1A), composed of two transmembrane domains TMD1 and TMD2 and two cytoplasmic nucleotide binding domains NBD1 and NBD2 (Moreau et al. 2005b). In addition, it has a supplementary N-terminal transmembrane domain TMD0 composed of five transmembrane helices which is linked to the core ABC domain by a cytosolic loop L0. This domain is found in other ABC proteins like the multidrug resistance protein MRP1, where both TMD0 and L0 have been reported to play a non-essential role in transport activity (Bakos et al. 1998, 2000; Westlake et al. 2003) trafficking (Westlake et al. 2005) and dimerization (Yang et al. 2007). For SUR, TMD0 is indispensable as it is involved in attachment to Kir6.2 (Babenko & Bryan, 2003; Chan et al. 2003). Other domains of SUR have also been shown to interact with Kir6.2: the transmembrane domains, which interact with Kir6.2 transmembrane helix M1 and N-terminus (Schwappach et al. 2000), TMD2–NBD2 which co-immunoprecipitates with Kir6.2 (Chan et al. 2003), and a cytoplasmic region at the TMD2–NBD2 junction (Rainbow et al. 2004a).
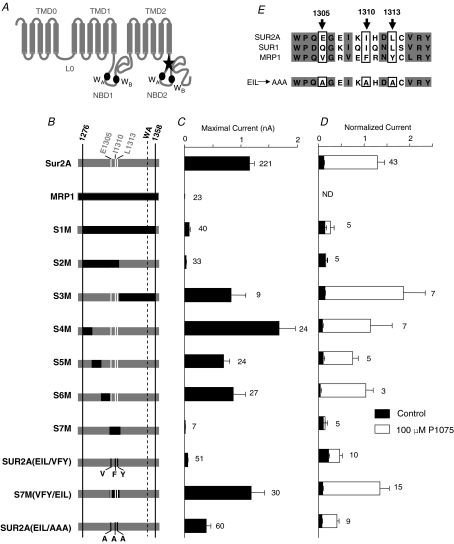
Figure 1. Three C-terminal cytoplasmic residues of SUR2A are essential for KATP channels proper expression and activation by P1075.
A, putative membrane topology of SUR2A (NBD, nucleotide binding domain; WA, Walker A motif; WB, Walker B motif). The star indicates the region identified by Rainbow et al. (2004a) and mutated to obtain the S1M chimera. B, schematic representation of the chimeras. SUR2A and MRP1 elements are drawn in grey and black, respectively. For clarity, residues E1305, I1310 and L1313 are indicated by white stripes. C, maximal current amplitudes in absence of nucleotides, and D, P1075 responses were measured in inside-out patches excised from oocytes co-expressing Kir6.2 and wild-type or chimeric SURs or MRP1 as indicated. P1075 (100 μm) was applied in the presence of 100 μm ATP, and currents were normalized to the current measured in the absence of nucleotides immediately before opener application. Application of 100 μm ATP alone (black bars) was used as a control. Numbers at right of bars indicate the number of patches included in each average. E, alignment of the amino acid sequences of human SUR1, rat SUR2A and human MRP1. Three residues (boxed) are conserved in SURs and not in MRP1: E1305, I1310 and L1313. These residues were replaced in SUR2A by alanines to generate chimera SUR2A(EIL/AAA) at bottom.
If SUR/Kir6.2 physical interactions are well documented, little information is available to date on the mechanisms linking both subunits of the KATP channel, and it remains unclear how nucleotide or drug binding to SUR is converted into Kir6.2 gating modulations. All SUR/Kir6.2 interacting regions identified represent potential transduction pathways to be explored. Solid evidence has already been provided that TMD0 domain and L0 exert a direct influence on Kir6.2 gating (Babenko & Bryan, 2003; Chan et al. 2003; Fang et al. 2006). However, most of these works concluded that there was a major role of TMD0 in the control of the Kir6.2 open probability (Po) but none of them defined it as essential in pharmacological responsiveness of the channel, except for the report of a heterozygous mutation in cytoplasmic loop 2, F132L, responsible for changes in ATP and tolbutamide sensitivity (Proks et al. 2006). Regions other than TMD0 could therefore be essential in transduction processes. A potential candidate is the 65-residue segment described by Rainbow et al. (2004a), located between the last transmembrane helix 17 and NBD2. Indeed, it was described to make a physical interaction with Kir6.2, and co-expression of this fragment with wild-type SUR2A and Kir6.2 prevented proper membrane expression of the channels, suggesting a critical interface disruption in this case (Rainbow et al. 2004a,b). These results raise the hypothesis that this C-terminal cytoplasmic segment of SUR2A may be of crucial importance in connecting functionally the two subunits of KATP channels, and may thereby be a signal transmission pathway linking ligand binding to SUR2A to Kir6.2 gating modulation.
In this study, a chimeric approach between SUR2A and MRP1 was adopted to assess the functional role of an extended 82-residue version of this fragment. We found that responses to KATP channel openers and MgADP were greatly attenuated. Further studies showed that this phenotype could be conferred by only three residues located near the N-terminus of NBD2. Moreover, mutating these residues did not change channel sensitivity to blockers, suggesting a role for these residues in activation – but not inhibition – transduction processes between SUR2A and Kir6.2.
Parts of this work have been published in abstract form (Dupuis et al. 2008).
Methods
Molecular biology
Experimental conditions were essentially as previously described (Moreau et al. 2005a). All constructs were derived from mouse Kir6.2 (GenBank accession no. D50581), rat SUR2A (GenBank accession no. D83598) and human MRP1 (GenBank accession no. L05628) and subcloned in Xenopus oocyte expression vectors derived from pGEMHE (Liman et al. 1992). Mutations were introduced by PCR using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) and the coding sequence of each construct was entirely verified by sequencing. The exact amino acid composition of SUR2A–MRP1 chimeric constructs and mutants were: S1M, SUR2A(M1-V1275) + MRP1(E1262-R1342) + SUR2A(M1358-K1545); S2M, SUR2A(M1-V1275) + MRP1(E1262-L1300) + SUR2A(R1316-K1545); S3M, SUR2A(M1-V1315) + MRP1(R1301-R1342) + SUR2A(M1358-K1545); S4M, SUR2A(M1-V1275) + MRP1(E1262-E1271) + SUR2A(S1286-K1545); S5M, SUR2A(M1-E1285) + MRP1(K1272-P1283) + SUR2A(P1299-K1545); S6M, SUR2A(M1-V1298) + MRP1(P1284-G1291) + SUR2A(E1307-K1545); S7M, SUR2A(M1-Q1304) + MRP1(V1290-L1300) + SUR2A(R1316-K1545); SUR2A(EIL/VFY), SUR2A with mutations E1305V, I1310F and L1313Y; S7M(VFY/EIL), S7M with mutations V1305E, F1310I and Y1313L; SUR2A(EIL/AAA), SUR2A with mutations E1305A, I1310A and L1313A.
After amplification and linearization, plasmid DNAs were transcribed in vitro using the T7 mMessage mMachine Kit (Ambion) to produce cRNAs for subsequent Xenopus oocyte microinjection.
Oocyte preparation and microinjection
Female Xenopus laevis were anaesthetized with 3-aminobenzoic acid ethyl ester (1 g l−1) for ∼20 min. A mini-laparotomy was then performed: a small incision was cut on the side of the abdomen and several ovarian sacs were removed. The incision was then sutured and the animal was allowed to recover. Each animal was used three times and then killed by decapitation. Animal handling and experiments fully conformed to French regulations and were approved by local governmental veterinary services (authorization no. 28-03-15 from the Ministère de l'Agriculture, Direction des Services Vétérinaires to Michel Vivaudou). Stage V or VI Xenopus laevis oocytes were defolliculated by a 60 min incubation at 19°C in a 2 mg ml−1 type A collagenase solution (Sigma-Aldrich). Selected oocytes were injected the next day with cRNAs encoding wild-type Kir6.2 (2 ng) and wild-type or chimeric SURs (6 ng). Injected oocytes were then stored at 19°C in Barth's solution (KCl 1 mm, MgSO4 0.82 mm, NaCl 88 mm, NaHCO3 2.4 mm, CaCl2 0.41 mm, Ca(NO3)2 0.3 mm, Hepes 16 mm, pH 7.4) supplemented with 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 100 μg ml−1 gentamycin.
Electrophysiology
Three to five days after injection, oocytes were manually devitellinized and channels were characterized by the patch-clamp technique in the excised inside-out configuration at room temperature using a RK300 amplifier (BioLogic) and a Digidata 1322A acquisition system with Axoscope 9 software (Axon Instruments). Patch pipettes contained: 154 mm K+, 146 mm Cl−, 5 mm Mg2+ and 10 mm Pipes-KOH (pH 7.1). The cytoplasmic face of the patch was bathed in solutions containing 174 mm K+, 40 mm Cl−, 1 mm EGTA, 1 mm Mg2+, 10 mm Pipes-KOH (pH 7.1) and methanesulphonate as the remaining anion. ATP, ADP, SR47063, P1075 and glibenclamide were added as specified. Membrane potential was held at −50 mV during all experiments. Application of various solutions to the patch was performed using a RSC-100 automated sewer pipes system (Bio-Logic; Vivaudou & Forestier, 1995). Pipe switching time was set at 250 ms. Data acquisition and analysis were performed using in-house software. Baseline fluctuations were removed by interactive fitting with a spline curve and subtraction of this fit with the signal. Non-linear curve fittings were performed with Origin software (OriginLab). The following standard Hill equations were used for fitting.
For activation by openers:
| (1) |
where |X| is the concentration of activator, i0 is the control normalized current in absence of activator, imax is the maximal activator-induced current, K½ the concentration for half-maximal activation and h the Hill coefficient.
For inhibition by nucleotides and glibenclamide:
| (2) |
where |X| is the concentration of inhibitor, i0 is the control normalized current in absence of inhibitor, K½ the concentration for half-maximal inhibition and h the Hill coefficient.
For combined activation and inhibition by ADP:
| (3) |
where i0 is the control normalized current in the absence of ADP, imax is the current that would be obtained if inhibition was absent, Ka½ the concentration for half-maximal activation, ha the Hill coefficient for activation, K½ the concentration for half-maximal inhibition, and hi the Hill coefficient for inhibition.
Results are displayed as mean ± s.e.m. Error bars in figures represent s.e.m. and are only shown if greater than symbols.
Results
Implication of three C-terminal residues of SUR2A in KATP channel expression and activation by P1075
The aim of this work was to assess whether the proximal C-terminal Kir6.2-interacting segment of SUR2A (Fig. 1A), described by Rainbow et al. (2004a) as responsible for disrupting association between KATP channel subunits SUR2A and Kir6.2, could be implicated in the allosteric transmission processes linking effectors binding on SUR2A to Kir6.2 gating. In order to address this question, we pursued an approach based on the construction of chimeras between the cardiac muscle isoform SUR2A and its close homologue the multidrug resistance protein 1 (MRP1). MRP1 was the ideal candidate for the development of this kind of strategy because it is very homologous to SUR isoforms but exerts neither physical nor functional influence on Kir6.2. In contrast, SUR2A is exclusively expressed in association with Kir6.2 and is dedicated to its regulation.
Our strategy, illustrated in Fig. 1B, consisted of co-expressing wild-type or chimeric SUR2A with wild-type Kir6.2 in Xenopus oocytes. Using the patch-clamp technique in the inside-out configuration, the resulting KATP channels were monitored both for maximal currents (Fig. 1C) and change in activity produced by a saturating dose (100 μm) of the pinacidil analogue P1075 (Fig. 1D), as reporters of channel surface expression and functionality, respectively. As demonstrated in the following, these two parameters proved appropriate to discriminate between SUR2A and mutant phenotypes since nearly all of our constructs gave clear responses to both assays. As expected, whereas Kir6.2/SUR2A channels were characterized by nanoamp-amplitude currents (1150 ± 90 pA), no Kir6.2/MRP1 currents could be recorded (Fig. 1C). Moreover, in the presence of 100 μm ATP (sufficient to produce 90% channel inhibition in most cases) a concentration of 100 μm P1075 strongly activated the Kir6.2/SUR2A channels (Fig. 1C). P1075 response was not tested on Kir6.2/MRP1 channels since no currents were recordable. In the same conditions, we next tested the S1M chimera, consisting SUR2A with an 82-residue TMD2–NBD2 region replaced by the homologous part of MRP1. This first chimera was poorly expressed since only very small currents could be recorded (72 ± 20 pA). It was also weakly activated by 100 μm P1075 (Fig. 1C), and therefore displayed what we designate as a MRP1-like phenotype.
We then replaced progressively shorter regions of SUR2A with their corresponding segments in MRP1. The S1M segment was split into two equal parts to generate chimeras S2M and S3M, as illustrated in Fig. 1B. Chimera S2M displayed the same small currents and weak P1075 activation phenotype, whereas the S3M chimera was found to produce nanoamp-size currents and to retain full sensitivity to P1075. The same splitting procedure was applied to the S2M segment to obtain four chimeras: S4M, S5M, S6M and S7M, respectively. Chimeras S4M, S5M and S6M showed a Kir6.2/SUR2A phenotype. This was not the case for chimera S7M which showed the same MRP1-like defective phenotype as observed for chimeras S1M and S2M.
Primary sequence alignments of SUR isoforms and MRP1 within a 16-residue region encompassing the S7M segment revealed a high conservation as shown in Fig. 1E. Three residues within this segment were found to be similar in SURs but different in MRP1: E1305, I1310 and L1313 of SUR2A. Alignment of SUR2A with Sav1866 (Dawson & Locher, 2006) and MsbA (Ward et al. 2007) reveals that the three residues stand before (E1305) and within the first β-sheet in the N-terminus of NBD2 (I1310 and L1313), respectively (see online Supplemental Fig. 1). These three residues were swapped between SUR2A and MRP1 to generate construct SUR2A(EIL/VFY). Figure 1C and D shows that this construct resembled chimera S7M, with low currents (49 ± 11 pA) and weak activation by P1075, leading to the conclusion that mutating these residues is sufficient to confer a MRP1-like deficient phenotype. To confirm this hypothesis, we reintroduced the initial SUR2A residues in the S7M chimera, generating S7M(VFY/EIL). This construct proved to be fully responsive to P1075 and displayed wild-type-like amplitude currents. This phenotype reversion validated the essential role of residues E1305, I1310 and L1313 in current levels and P1075 sensitivity of Kir6.2/SUR2A KATP channels. Mutating fewer residues proved not to be successful since all constructs mutated for less than three residues (for example SUR2A(IL/FY), data not shown) proved to display intermediate phenotypes.
Two main hypotheses can explain altered P1075 sensitivity. The first one is that mutating these three amino acids is sufficient to instigate a disruption in the physical association between SUR2A and Kir6.2 subunits, which would be in good accordance with previous work by Rainbow et al. (2004a). The second possibility is that these three residues stand at the SUR2A–Kir6.2 interface and mediate signal transduction. In order to see whether side-chain sizes could be of importance in physical association – in which case reducing their sizes might restore association and channel activity – we mutated residues E1305, I1310 and L1313 to alanines, generating construct SUR2A(EIL/AAA). This construct yielded much larger currents than Kir6.2/SUR2A(EIL/VFY) (420 ± 90 pA versus 49 ± 11 pA, respectively) but was significantly less activated by P1075 than wild-type channels (Student's t test; P < 0.00001) (Figs 1C, and 2A and B). This result suggests that physical association is near normal in the SUR2A(EIL/AAA) construct, but that P1075 binding or functional coupling is altered by the triple mutation.
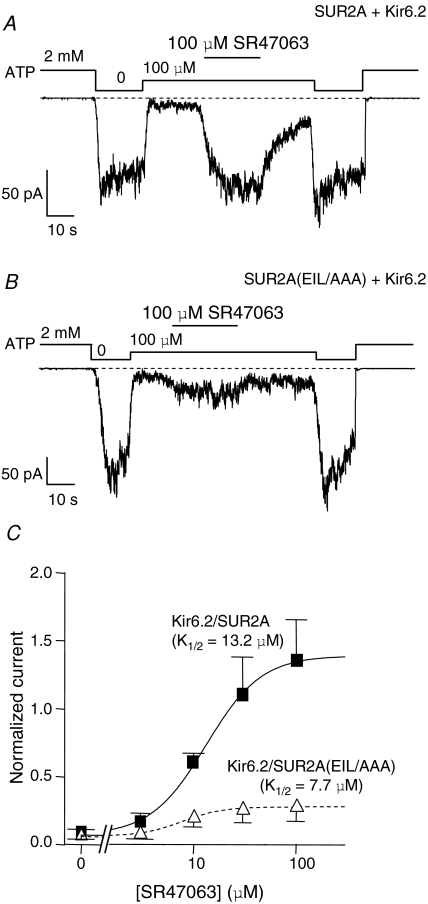
Figure 2. Mutation to alanines of residues E1305, I1310 and L1313 in SUR2A reduces drastically the extent of activation by P1075 without changing affinity.
A and B, representative patch-clamp recordings illustrating the responses of wild-type (A) and SUR2A(EIL/AAA) (B) channels to 10 μm P1075 in the presence of 100 μm ATP. C, mean P1075 dose–response relationships for Kir6.2/SUR2A (▪) and Kir6.2/SUR2A(EIL/AAA) (▵) in the presence of 100 μm ATP. Currents were normalized to the current measured in 0 ATP before application of ATP and opener. Each point represents data from 6 patches for Kir6.2/SUR2A and 8 patches for Kir6.2/SUR2A(EIL/AAA). Fitting using eqn (1) yielded K1/2 = 6.5 ± 1.3 μm (h = 1.1 ± 0.2) for Kir6.2/SUR2A and K1/2 = 4.0 ± 1 μm (h = 0.94 ± 0.2) for Kir6.2/SUR2A(EIL/AAA).
To examine whether mutagenesis altered drug binding, we submitted wild-type and mutant channels to increasing P1075 concentrations to evaluate affinity (Fig. 2C). P1075 apparent affinities appeared to be very similar for Kir6.2/SUR2A (K1/2 = 6.5 μm), Kir6.2/SUR2A(EIL/AAA) (K1/2 = 4.0 μm), and Kir6.2/SUR2A(EIL/VFY) (K1/2 = 8.0 μm, data not shown) channels suggesting that mutations did not alter P1075 binding to SUR2A.
SUR2A(EIL/AAA) channels show severe SR47063 activation defects without change in drug affinity
We also tested the ability of the SUR2A(EIL/AAA) mutant to respond to SR47063, another KATP channel opener from the benzopyran family. In the presence of 100 μm ATP, 100 μm SR47063 robustly activated the Kir6.2/SUR2A channels but activated Kir6.2/SUR2A(EIL/AAA) channels to a much lesser extent (14.3 ± 1.3- versus 4.7 ± 0.6-fold activation, respectively, P < 0.000001, data not shown) as illustrated in Fig. 3A and B. Both mutant and wild-type channels were submitted to increasing concentrations of SR47063, and dose–response curves indicated little difference in SR47063 apparent affinity between Kir6.2/SUR2A(EIL/AAA) (K1/2 = 7.7 μm), Kir6.2/SUR2A(EIL/VFY) (K1/2 = 7 μm, data not shown) and Kir6.2/SUR2A (K1/2 = 13.2 μm), suggesting that mutations did not alter SR47063 binding to SUR2A. Therefore, it seems as if Kir6.2/SUR2A(EIL/AAA) channels are no longer able to convert SR47063 binding at SUR2A into Kir6.2 activation.
Figure 3. SUR2A(EIL/AAA) channels show severe SR47063 activation defects without change in affinity.
A and B, representative patch-clamp recordings illustrating the responses of wild-type and SUR2A(EIL/AAA) channels to 100 μm SR47063 in the presence of 100 μm ATP. C, mean SR47063 dose–response relationships for Kir6.2/SUR2A (▪) and Kir6.2/SUR2A(EIL/AAA) (▵) in the presence of 100 μm ATP. Currents were normalized to the current measured in 0 ATP before application of ATP and opener. Each point represents data from 3 patches for Kir6.2/SUR2A and 5 patches for Kir6.2/SUR2A(EIL/AAA). Fitting using eqn (1) yielded K1/2 = 13.2 ± 1.1 μm (h = 1.61 ± 0.2) for Kir6.2/SUR2A and K1/2 = 7.7 ± 0.7 μm (h = 2.2 ± 0.4) for Kir6.2/SUR2A(EIL/AAA).
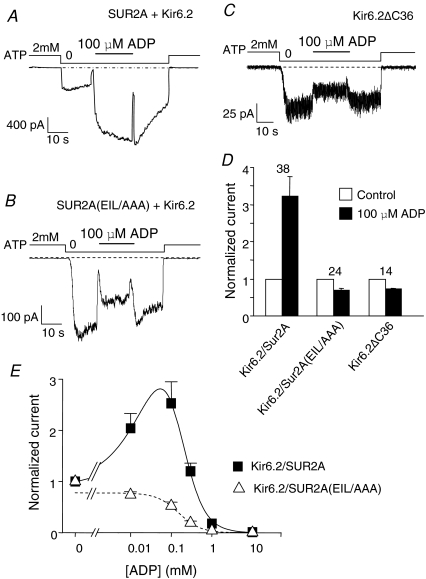
E1305, I1310 and L1313 triple mutations in SUR2A abolish MgADP activation
In the cell environment, KATP channels are physiologically activated by MgADP. Both Kir6.2 and SUR2A possess cytoplasmic nucleotide-binding domains that are inhibitory and activatory, respectively. For submillimolar MgADP, the balance of the two effects leans toward activation. Subunit uncoupling should shift the effect of MgADP toward inhibition. The MgADP response of Kir6.2/SUR2A(EIL/AAA) channels was therefore tested and compared to that of wild-type channels and of Kir6.2 channels truncated of their 36 C-terminal residues (Kir6.2ΔC36) that form SUR-less channels (Tucker et al. 1997). As shown in Fig. 4, wild-type channels were strongly activated by 100 μm MgADP whereas SUR2A(EIL/AAA) channels were clearly inhibited, much like Kir6.2 alone. Over the full range of ADP, wild-type channels are activated at the lower concentrations and inhibited at the higher concentrations reflecting the combination of SUR-mediated activation and Kir6.2-mediated inhibition. For SUR2A(EIL/AAA) channels, activation is abolished and ADP only retains its inhibitory effect. Therefore, our results suggest that SUR2A(EIL/AAA) channels behave towards MgADP as if SUR2A was still physically present but unable to balance MgADP inhibitory action on Kir6.2 by exerting its activating influence anymore.
Figure 4. E1305, I1310 and L1313 triple mutations in SUR2A abolish MgADP activation.
A–C, representative currents recorded in inside-out patches from Xenopus oocytes expressing either Kir6.2/SUR2A (A), Kir6.2/SUR2A(EIL/AAA) (B) or Kir6.2ΔC36 (C). MgADP (100 μm) was applied in the absence of ATP. D, average normalized currents for Kir6.2/SUR2A, Kir6.2/SUR2A(EIL/AAA) and Kir6.2ΔC36 measured before (open bars) and after (filled bars) application of 100 μm MgADP in the absence of nucleotides. Numbers above bars indicate the number of patches included in the averages. E, MgADP dose–response relationships for Kir6.2/SUR2A (▪) and Kir6.2/SUR2A(EIL/AAA) (▵) in the absence of ATP. Currents were normalized to the current measured in 0 ATP. Each point represents data from 5 to 15 patches for Kir6.2/SUR2A and 5–14 patches for Kir6.2/SUR2A(EIL/AAA). Fitting of eqn (3) to the Kir6.2/SUR2A data yielded Ka1/2 = 124 ± 9 μm (ha = 0.94 ± 0.03) and Ki1/2 = 230 ± 6 μm (hi = 1.93 ± 0.05). Fitting of eqn (2) to the Kir6.2/SUR2A(EIL/AAA) data yielded K1/2 = 152 ± 16 μm (h = 1.6 ± 0.2).
Inhibitory mechanisms are not affected by E1305/I1310/L1313 triple mutations
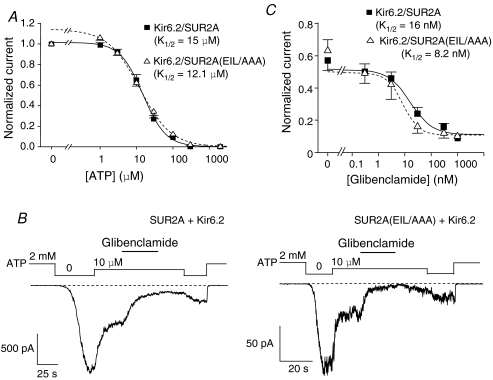
Having tested a range of channel activators, which were all inefficient at activating Kir6.2/SUR2A(EIL/AAA) channels, we examined their sensitivities to the inhibitory actions of ATP and glibenclamide. As illustrated in Fig. 5A, patches were first exposed to increasing concentrations of ATP. Kir6.2/SUR2A channels were blocked by ATP with a Ki of 15 μm, and Kir6.2/SUR2A(EIL/AAA) channels proved to have a very similar Ki of 12.1 μm, showing no real difference in ATP inhibition sensitivity between both channel populations. Moreover, application of 300 nm glibenclamide, a dose sufficient to produce 90% inhibition of Kir6.2/SUR2A channels (Fig. 5B), induced a clear wild-type-like inhibition of Kir6.2/SUR2A(EIL/AAA) channels both in the absence (data not shown) and the presence of ATP (10 μm). As illustrated in Fig. 5C, glibenclamide sensitivity was similar for both Kir6.2/SUR2A (Ki = 16 nm) and Kir6.2/SUR2A(EIL/AAA) (Ki = 8.2 nm) channels. Unchanged sensitivities both for ATP and glibenclamide raise the conclusion that inhibitory mechanisms are probably unaffected by mutations E1305/I1310/L1313 to alanines.
Figure 5. SUR2A(EIL/AAA) channels respond like wild-type to ATP and glibenclamide.
A, ATP dose–response relationships for Kir6.2/SUR2A (▪) and Kir6.2/SUR2A(EIL/AAA) (▵). Currents were normalized to the current measured in 0 ATP. Each point represents data from 21–44 patches for Kir6.2/SUR2A and 8–28 patches for Kir6.2/SUR2A(EIL/AAA). Fitting of eqn (2) yielded K1/2 = 15 ± 0.7 μm (h = 1.39 ± 0.07) for Kir6.2/SUR2A and K1/2 = 12.1 ± 0.9 μm (h = 1.01 ± 0.05) for Kir6.2/SUR2A(EIL/AAA). B, representative currents recorded in inside-out patches from Xenopus oocytes expressing either Kir6.2/SUR2A or Kir6.2/SUR2A(EIL/AAA). Glibenclamide (300 nm) was applied in the presence of 10 μm ATP. C, glibenclamide dose–response relationships for Kir6.2/SUR2A (▪) and Kir6.2/SUR2A(EIL/AAA) (▵) in the presence of 10 μm ATP. Currents were normalized to the current measured in 0 ATP. Each point represents data from 5–12 patches for Kir6.2/SUR2A and 4–7 patches for Kir6.2/SUR2A(EIL/AAA). Fitting of eqn (2) yielded K1/2 = 16 ± 9.5 μm (h = 1.04 ± 0.6) for Kir6.2/SUR2A and K1/2 = 8.2 ± 1.4 μm (h = 1.43 ± 0.19) for Kir6.2/SUR2A(EIL/AAA).
Discussion
Within the KATP channel complex, little information is available on the molecular mechanisms involved in cross-regulation of both subunits, and it remains unclear how nucleotide or drug binding to SUR is converted into Kir6.2 gating modulations. In this study, we identified three SUR2A residues, E1305 I1310 and L1313, that are intimately involved in KATP channel activation both by K+ channel openers (P1075, SR47063) and MgADP. These residues could possibly form part of a transduction pathway functionally coupling SUR2A and Kir6.2 subunits through the cytosolic segment of SUR2A interconnecting transmembrane helix 17 to NBD2.
Residues E1305, I1310 and L1313 are critical for SUR2A–Kir6.2 association
The critical role of residues E1305, I1310 and L1313 was discovered by following a systematic chimeric strategy designed to address the question whether a TMD2–NBD2 interconnecting fragment of SUR2A previously described to be in interaction with Kir6.2 could play a role in channel expression and regulation. Chimeras incorporating progressively shorter fragments of MRP1 in a SUR2A background led to the finding that these three residues are associated with a reduction in channel expression. This expression defect agrees with the observations of Rainbow et al. (2004a) and argues in favour of these residues being part of a critical Kir6.2-interacting region. Nevertheless, we observed that replacing E1305, I1310 and L1313 by AAA rather than VFY had less drastic consequences on surface expression as shown by the larger SUR2A(EIL/AAA) currents. It is noteworthy that mutation of SUR1 l1350 – cognate to SUR2A L1313 highlighted here – to a glutamine (L1350Q) is found in patients suffering from congenital hyperinsulinism and seems to induce the disease by preventing surface expression of KATP channels in pancreatic β-cells (Yan et al. 2007). Taken together, these results show that residues E1305, I1310 and L1313 are not absolutely essential for physical association, since mutating them to alanines only partly alters surface expression, but suggest that they are likely to stand at the interface between SUR2A and Kir6.2.
Proximal C-terminus of SUR2A is involved in the activation processes linking ligand binding to SUR2A and Kir6.2 gating modifications
Since all SUR–Kir6.2 interfacing regions represent potential transduction pathways to be explored, as illustrated by the involvement of Kir6.2-interacting TMD0/L0 in channel gating (Fang et al. 2006), the question arose of the role of the three previously identified residues in channel regulation. Characterization of Kir6.2/SUR2A(EIL/AAA) channels revealed that responses to the pinacidil analogue P1075, to the cromakalim analogue SR47063 and to the physiological KATP channel activator MgADP were drastically reduced compared to wild-type. Since the experiments performed were purely functional, we asked whether these residues act at the binding step or at the transducing step between binding and channel activation. The former hypothesis is not improbable since the mutated residues lie in proximity to the presumed KATP channel openers (KCOs) and ADP binding sites, located at transmembrane helix 17 (Moreau et al. 2000) and NBD2 (Matsuo et al. 2005), respectively. For synthetic openers, this hypothesis is contradicted by the experimental evidence showing that triple mutations induced no significant changes in affinity. For ADP, activation being abolished, it is not possible to compare affinities and it is conceivable that ADP unresponsiveness could arise from an altered interaction with the NBDs. However, the fact that both KCOs and MgADP were similarly affected by the triple mutations even though they probably bind to distinct sites on SUR argues in favour of a perturbation of a common pathway downstream of the binding sites. We therefore feel that the most appropriate interpretation of our results is that residues E1305, I1310 and L1313 are part of the transducing machinery linking effectors binding to SUR and Kir6.2 activation.
One may point out that, if the S7M chimera remained weakly responsive to KCOs (Fig. 1D), mutation of these three residues did not completely abolish the responses to P1075 and SR47063, suggesting that other unknown determinants exist in the proximal region investigated. Anyhow, the residual effects were very weak, implying that these other determinants are of minor importance compared with residues E1305, I1310 and L1313. Conversely, restoration of these residues in the S7M MRP1-like phenotype chimera was sufficient to reverse the unresponsiveness to activators and restored both KCOs and MgADP activation as shown for P1075 (data not shown for SR47063 and MgADP). While we are aware of the pitfalls of overinterpreting mutagenesis results, the combination of experiments showing both loss-of-function in the SUR2A context and gain-of-function in the S7M chimera context argues strongly in favour of the implication of residues E1305, I1310 and L1313 in the activation processes instigated by KCOs and MgADP.
KATP channel activation and inhibition processes may rely on distinct transduction pathways
To date, KATP channel activation and inhibition processes are poorly understood, and it is hard to define whether they rely on a single or multiple pathways linking SUR2A and Kir6.2. Recently, an aspartate/glutamate-rich stretch located after NBD1 in SUR2A (residues 948–962) was described as critical both for activation (MgADP, KCOs) and inhibition (glibenclamide) processes (Karger et al. 2008). This stretch could form part of a common transduction pathway in association with either the TMD0 domain or/and the C-terminal region investigated in our work, which were both reported to interact with Kir6.2. However, we surprisingly observed that mutating the proximal C-terminus of SUR2A resulted in impaired KATP channel activation but not inhibition. Indeed, contrary to activators (MgADP, KCOs), both ATP and glibenclamide inhibition were unaffected by our mutations. These observations definitely invalidate a single-pathway model and are much more in accordance with the idea that multiple transduction pathways functionally linking SUR2A to Kir6.2 may exist: one for activation processes, which we believe act through the SUR2A TMD2–NBD2 linker (Fig. 6), and distinct pathways for inhibition processes which could implicate the TMD0 domain. Only a representative sample of effectors was tested here, and whether other functional processes act through yet unknown pathways remains to be determined and will require further investigations. However, these results suggest that, within the KATP channel complex, the proximal C-terminus of SUR2A is a critical link between ligand binding to SUR and Kir6.2 up-regulation.
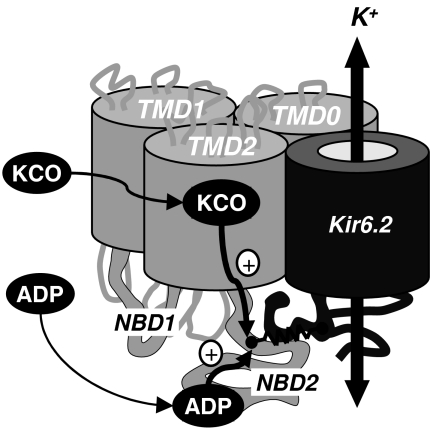
Figure 6. TMD2–NBD2 linker as a common pathway for activator between SUR2A and Kir62.
In this highly schematic model of the Kir6.2–SUR2A complex, both TMD0 and TMD2 interact with Kir6.2 and contribute to signal transmission. Binding of KCOs (P1075, SR47063) at transmembrane helix 17 of TMD2 or MgADP at NBD2 triggers Kir6.2 activation through a cytoplasmic link between SUR2A C-terminus and Kir6.2 cytosolic regions.
Acknowledgments
We are grateful to Dr S. Seino (Chiba, Japan) for mouse Kir6.2 and rat SUR2A, and Dr S. P. Cole (Kingston, ON, Canada) for human MRP1. J.P.D. was supported by a PhD studentship from C.E.A. (Commissariat à l'Energie Atomique).
Supplemental material
Online supplemental material for this paper can be accessed at: http://jp.physoc.org/cgi/content/full/jphysiol.2008.152744/DC1 and http://www.blackwell-synergy.com/doi/suppl/10.1113/jphysiol.2008.152744
References
- Aguilar-Bryan L, Nichols CG, Wechsler SW, Clement JP, Boyd AE, Gonzalez G, et al. Cloning of the b cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- Alekseev AE, Hodgson DM, Karger AB, Park S, Zingman LV, Terzic A. ATP-sensitive K+ channel channel/enzyme multimer: metabolic gating in the heart. J Mol Cell Cardiol. 2005;38:895–905. doi: 10.1016/j.yjmcc.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft FM, Harrison DE, Ashcroft SJ. Glucose induces closure of single potassium channels in isolated rat pancreatic β-cells. Nature. 1984;312:446–448. doi: 10.1038/312446a0. [DOI] [PubMed] [Google Scholar]
- Babenko AP, Bryan J. SUR domains that associate with and gate KATP pores define a novel gatekeeper. J Biol Chem. 2003;278:41577–41580. doi: 10.1074/jbc.C300363200. [DOI] [PubMed] [Google Scholar]
- Bakos E, Evers R, Calenda G, Tusnady GE, Szakacs G, Varadi A, et al. Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1) J Cell Sci. 2000;113:4451–4461. doi: 10.1242/jcs.113.24.4451. [DOI] [PubMed] [Google Scholar]
- Bakos E, Evers R, Szakacs G, Tusnady GE, Welker E, Szabo K, et al. Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J Biol Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- Chan KW, Zhang H, Logothetis DE. N-terminal transmembrane domain of the SUR controls trafficking and gating of Kir6 channel subunits. EMBO J. 2003;22:3833–3843. doi: 10.1093/emboj/cdg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement JP, Kunjilwar K, Gonzalez G, Schwanstecher M, Panten U, Aguilar-Bryan L, et al. Association and stoichiometry of KATP channel subunits. Neuron. 1997;18:827–838. doi: 10.1016/s0896-6273(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Revilloud J, Moreau C, Vivaudou M. Three C-terminal residues from the sulphonylurea receptor contribute to the functional coupling between the KATP channel subunits SUR2A and Kir6.2. Biophysical J. 2008;94:435a. doi: 10.1113/jphysiol.2008.152744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K, Csanady L, Chan KW. The N-terminal transmembrane domain (TMD0) and a cytosolic linker (L0) of sulphonylurea receptor define the unique intrinsic gating of KATP channels. J Physiol. 2006;576:379–389. doi: 10.1113/jphysiol.2006.112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Gonoi T, Clement JP, Namba N, Inazawa J, Gonzalez G, et al. Reconstitution of I-KATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- Jahangir A, Terzic A. KATP channel therapeutics at the bedside. J Mol Cell Cardiol. 2005;39:99–112. doi: 10.1016/j.yjmcc.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger AB, Park S, Reyes S, Bienengraeber M, Dyer RB, Terzic A, et al. Role for SUR2A ED domain in allosteric coupling within the KATP channel complex. J Gen Physiol. 2008;131:185–196. doi: 10.1085/jgp.200709852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Kimura Y, Ueda K. KATP channel interaction with adenine nucleotides. J Mol Cell Cardiol. 2005;38:907–916. doi: 10.1016/j.yjmcc.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Moreau C, Gally F, Jacquet-Bouix H, Vivaudou M. The size of a single residue of the sulfonylurea receptor dictates the effectiveness of KATP channel openers. Mol Pharmacol. 2005a;67:1026–1033. doi: 10.1124/mol.104.008698. [DOI] [PubMed] [Google Scholar]
- Moreau C, Jacquet H, Prost AL, D'Hahan N, Vivaudou M. The molecular basis of the specificity of action of KATP channel openers. EMBO J. 2000;19:6644–6651. doi: 10.1093/emboj/19.24.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau C, Prost AL, Derand R, Vivaudou M. SUR, ABC proteins targeted by KATP channel openers. J Mol Cell Cardiol. 2005b;38:951–963. doi: 10.1016/j.yjmcc.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Nichols CG. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–476. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- Proks P, Arnold AL, Bruining J, Girard C, Flanagan SE, Larkin B, et al. A heterozygous activating mutation in the sulphonylurea receptor SUR1 (ABCC8) causes neonatal diabetes. Hum Mol Genet. 2006;15:1793–1800. doi: 10.1093/hmg/ddl101. [DOI] [PubMed] [Google Scholar]
- Rainbow RD, James M, Hudman D, Al-Johi M, Singh H, Watson PJ, et al. Proximal C-terminal domain of sulphonylurea receptor 2A interacts with pore-forming Kir6 subunits in KATP channels. Biochem J. 2004a;379:173–181. doi: 10.1042/BJ20031087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbow RD, Lodwick D, Hudman D, Davies NW, Norman RI, Standen NB. SUR2A C-terminal fragments reduce KATP currents and ischaemic tolerance of rat cardiac myocytes. J Physiol. 2004b;557:785–794. doi: 10.1113/jphysiol.2004.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwappach B, Zerangue N, Jan YN, Jan LY. Molecular basis for KATP assembly: transmembrane interactions mediate association of a K+ channel with an ABC transporter. Neuron. 2000;26:155–167. doi: 10.1016/s0896-6273(00)81146-0. [DOI] [PubMed] [Google Scholar]
- Seino S, Miki T. Physiological and pathophysiological roles of ATP-sensitive K+ channels. Prog Biophys Mol Biol. 2003;81:133–176. doi: 10.1016/s0079-6107(02)00053-6. [DOI] [PubMed] [Google Scholar]
- Tarasov A, Dusonchet J, Ashcroft F. Metabolic regulation of the pancreatic β-cell ATP-sensitive K+ channel – a pas de deux. Diabetes. 2004;53:S113–S122. doi: 10.2337/diabetes.53.suppl_3.s113. [DOI] [PubMed] [Google Scholar]
- Tucker SJ, Gribble FM, Zhao C, Trapp S, Ashcroft FM. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- Vivaudou M, Forestier C. Modification by protons of frog skeletal muscle KATP channels: effects on ion conduction and nucleotide inhibition. J Physiol. 1995;486:629–645. doi: 10.1113/jphysiol.1995.sp020840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Cole SP, Deeley RG. Role of the NH2-terminal membrane spanning domain of multidrug resistance protein 1/ABCC1 in protein processing and trafficking. Mol Biol Cell. 2005;16:2483–2492. doi: 10.1091/mbc.E04-12-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Qian YM, Gao M, Vasa M, Cole SP, Deeley RG. Identification of the structural and functional boundaries of the multidrug resistance protein 1 cytoplasmic loop 3. Biochemistry. 2003;42:14099–14113. doi: 10.1021/bi035333y. [DOI] [PubMed] [Google Scholar]
- Yamada S, Kane GC, Behfar A, Liu XK, Dyer RB, Faustino RS, et al. Protection conferred by myocardial ATP-sensitive K+ channels in pressure overload-induced congestive heart failure revealed in KCNJ11 Kir6.2-null mutant. J Physiol. 2006;577:1053–1065. doi: 10.1113/jphysiol.2006.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Liu Y, Dong ZZ, Xu JK, Peng H, Liu ZQ, et al. Regulation of function by dimerization through the amino-terminal membrane-spanning domain of human ABCC1/MRP1. J Biol Chem. 2007;282:8821–8830. doi: 10.1074/jbc.M700152200. [DOI] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane KATP channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.