Abstract
Proprotein convertase subtilisin/kexin type 9 (PCSK9) posttranslationally regulates hepatic low-density lipoprotein receptors (LDLRs) by binding to LDLRs on the cell surface, leading to their degradation. The binding site of PCSK9 has been localized to the epidermal growth factor-like repeat A (EGF-A) domain of the LDLR. Here, we describe the crystal structure of a complex between PCSK9 and the EGF-A domain of the LDLR. The binding site for the LDLR EGF-A domain resides on the surface of PCSK9's subtilisin-like catalytic domain containing Asp-374, a residue for which a gain-of-function mutation (Asp-374–Tyr) increases the affinity of PCSK9 toward LDLR and increases plasma LDL-cholesterol (LDL-C) levels in humans. The binding surface on PCSK9 is distant from its catalytic site, and the EGF-A domain makes no contact with either the C-terminal domain or the prodomain. Point mutations in PCSK9 that altered key residues contributing to EGF-A binding (Arg-194 and Phe-379) greatly diminished binding to the LDLR's extracellular domain. The structure of PCSK9 in complex with the LDLR EGF-A domain defines potential therapeutic target sites for blocking agents that could interfere with this interaction in vivo, thereby increasing LDLR function and reducing plasma LDL-C levels.
Keywords: hypercholesterolemia, proprotein convertase, crystal structure
Proprotein convertase (PC) subtilisin/kexin type 9 (PCSK9) is a serine protease of the PC family that has profound effects on plasma low-density lipoprotein (LDL)-cholesterol (LDL-C) levels through its ability to mediate LDL receptor (LDLR) protein degradation (1, 2). The link between PCSK9 and plasma LDL-C levels was first established by the discovery of missense mutations in PCSK9 that were present in patients with an autosomal dominant form of familial hypercholesterolemia (FH) (3). These mutations were speculated to result in a gain-of-function of PCSK9 owing to their mode of inheritance. Subsequently, loss-of-function mutations in PCSK9 were discovered that are associated with low plasma LDL-C levels and significantly reduced coronary heart disease (CHD) (4, 5).
The overall domain structure of PCSK9 is similar to other PC family members. It includes a signal peptide, followed by a prodomain, a subtilisin-like catalytic domain, and a variable C-terminal domain (6). The prodomain serves a dual role as a chaperone for folding and as an inhibitor of catalytic activity (7–9). Autocatalysis between Gln-152 and Ser-153 (VFAQ:SIP) separates the prodomain from the catalytic domain, but the prodomain remains bound, occluding the catalytic site (10, 11). For other PC family members, a second catalytic cleavage is required to release the prodomain, which unmasks the catalytic site, resulting in an active protease (7). No site of secondary cleavage has been identified that activates PCSK9.
The crystal structure of apo-PCSK9 revealed a tightly bound prodomain that is predicted to render the active site inaccessible to exogenous substrates (12–14). The structure of the PCSK9 prodomain and catalytic domain is similar to that of other subtilisin-like serine proteases. The C-terminal domain of PCSK9 contains three six-stranded β-sheet subdomains arranged with quasi-threefold symmetry. This domain shares structural homology to the adipokine resistin and has been speculated to mediate protein–protein interactions (12–14).
Recent studies have provided insights into the site and mode of PCSK9's action. Addition of recombinant PCSK9 to the medium of cultured hepatocytes results in a dramatic reduction in LDLR number. PCSK9 binds directly to the LDLR on the cell surface and PCSK9-stimulated degradation of the LDLR requires autosomal recessive hypercholesterolemia (ARH), an adaptor protein needed for internalization of LDLRs (11). The affinity of PCSK9 binding to the LDLR is enhanced at acidic pH, suggesting that PCSK9 binds more avidly to LDLRs in the lysosomal/endosomal compartments (12, 15, 16). One gain-of-function mutant, Asp-374–Tyr, was ≈10-fold more active than wild-type PCSK9 in mediating degradation of LDLRs, owing to an ≈5- to 30-fold increased affinity of PCSK9 for the LDLR (11, 12, 15). Although autocatalysis of PCSK9 is required for proper folding and secretion, catalytic activity was not required for PCSK9-mediated LDLR degradation when added to cultured cells (17, 18).
The LDLR is a multidomain protein whose extracellular domain (ECD) consists of an N-terminal ligand binding domain [seven cysteine-rich repeats (R1–R7) that mediate binding to LDL and β-VLDL], followed by the epidermal growth factor (EGF)-precursor homology domain [a pair of EGF-like repeats (EGF-A and EGF-B) separated from a third EGF-like repeat (EGF-C) by a β-propeller domain], and an “O-linked sugar” domain (19, 20). After endocytosis of the receptor/ligand complex, bound lipoproteins are released in the acidic environment of the endosome, and the LDLR recycles to the cell surface. Ligand release has been proposed to result from a pH-dependent conformational change in the LDLR-ECD because of an intramolecular interaction between ligand binding modules R4 and R5 with the β-propeller domain (21).
Zhang et al. (16) recently mapped the residues of the LDLR that are required for binding to PCSK9. PCSK9 binds in a calcium-dependent manner to the first EGF-like repeat (EGF-A) of the EGF-precursor homology domain of the LDLR. The integrity of the EGF-precursor homology domain is essential for normal LDLR turnover. Deletion of this region results in a failure of internalized LDLRs to release bound ligand and prevents recycling to the cell surface (22). Several natural mutations in the EGF-precursor homology domain of LDLR that hinder receptor recycling are present in patients with FH (23, 24). The EGF-A domain itself forms several intramolecular interactions proposed to be important for LDLR stability. The EGF-A and EGF-B tandem pair form an extended rod-like conformation stabilized by calcium binding and interdomain hydrophobic packing interactions (25, 26). The interaction of EGF-A with ligand binding module R7 confers a rigid conformation on this region of the LDLR across a wide pH range (27). This rigidity has been proposed to facilitate the acid-dependent closed conformation of the LDLR (28).
To determine the residues of PCSK9 that interact with the LDLR, we have determined the crystal structure of PCSK9 in complex with the EGF-A domain of the LDLR. The structure defines the mode of binding of PCSK9 to the EGF-A domain and demonstrates that LDLR binding does not require dissociation of the prodomain from PCSK9. This information may facilitate the development of therapeutic antibodies or peptides that could be used to disrupt the PCSK9–LDLR interaction and thus function as an inhibitor of PCSK9's action.
Results and Discussion
For structural studies, we used a recombinant PCSK9 protein lacking the N-terminal 21 aa of the prodomain region (Δ53-PCSK9). Δ53-PCSK9 underwent normal autocatalysis and secretion when expressed in HEK 293S cells (data not shown). To test the relative binding affinity of Δ53-PCSK9 compared with full-length PCSK9, competition binding assays were performed at pH 7.4 by using a fluorophore-labeled PCSK9 and purified LDLR-ECD. Unexpectedly, Δ53-PCSK9 displayed a >7-fold greater affinity for the LDLR-ECD compared with the full-length PCSK9 protein (EC50 of 27.8 nM for Δ53-PCSK9 versus 203 nM for wild-type PCSK9) [supporting information (SI) Table 2 and SI Fig. 4A]. The affinity of binding of both PCSK9 and Δ53-PCSK9 increased ≈3-fold when the pH was lowered from 7.0 to 6.0 (SI Table 2 and SI Fig. 4B).
We repeated the binding studies by using a purified LDLR fragment consisting of the EGF-AB tandem pair that was found to be more stable in solution than the EGF-A domain alone. Compared with the ECD, EGF-AB was ≈3-fold less efficient at competing for binding of fluorophore-labeled LDLR-ECD to PCSK9 (EC50 of 119 nM for EGF-AB versus 35.3 nM for LDLR-ECD). As previously reported for the LDLR-ECD, Δ53-PCSK9 had an ≈3- to 4-fold higher affinity for the EGF-AB at pH 6.0 compared with pH 7.0 (SI Table 2 and SI Fig. 4 C and D).
Structure of the Complex.
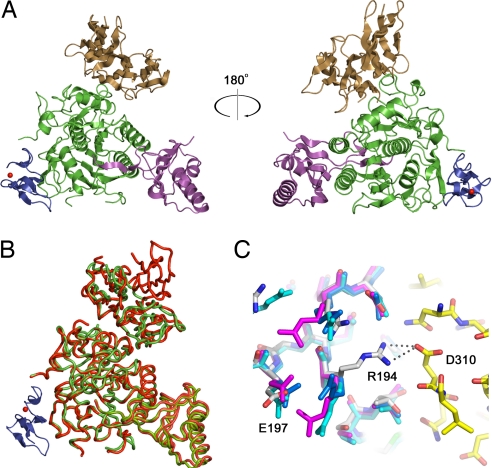
Crystals of Δ53-PCSK9 in complex with the EGF-AB pair of the LDLR were grown at pH 4.8, and the structure was refined to 2.4 Å (Table 1). Electron density for the EGF-B domain was poor, so EGF-B was not included in the final model. Electron density is visible for the entire EGF-A domain of the LDLR (residues 293–332) and for a calcium ion within the EGF-A domain. In the structure of the PCSK9:EGF-A complex (Fig. 1A), PCSK9 adopts a fold that is identical to apo-PCSK9, except for a subdomain within the C-terminal variable domain (residues 532–601) that is visible in the apo-PCSK9 structures (12–14) but is disordered in the PCSK9:EGF-A complex (Fig. 1B). Aside from this domain, the structures of apo-PCSK9 and PCSK9 bound to EGF-A are virtually identical, with a rmsd of 1.5 Å across Cα atoms. The first ordered residue of the prodomain is Thr-61, similar to the apo-PCSK9 structures (12–14). The lack of an ordered N terminus in the prodomain of the PCSK9:EGF-A complex and the increased affinity of Δ53-PCSK9 for the LDLR-ECD (SI Table 2) suggest that residues 31–60 do not directly interact with LDLR.
Table 1.
Data collection and refinement statistics
| Space group | P4(1)2(1)2 |
| a, b, Å | 116.952 |
| c, Å | 134.878 |
| Resolution, Å (final shell) | 40.0 – 2.41 (2.47–2.41) |
| Reflections | |
| Total | 162,854 |
| Unique | 36,583 |
| Completeness, % | 98.9 (90.0) |
| Rsym, % | 6.2 |
| Rcryst, % | 20.3 (25.9) |
| Rfree, % | 24.0 (31.4) |
| rmsd | |
| Bond length, Å | 0.014 |
| Bond angle, ° | 1.483 |
Fig. 1.
The PCSK9:EGF-A complex. (A) PCSK9, with the prodomain (magenta), the subtilisin-like catalytic domain (green), and the C-terminal domain (brown), and the EGF-A domain of LDLR (blue) is represented as a ribbon diagram. The bound calcium ion within the EGF-A domain is shown as a red sphere. (B) Superposition of the PCSK9:EGF-A complex and apo-PCSK9. The PCSK9:EGF-A complex is shown with PCSK9 in green, EGF-A in blue, and bound calcium as a red sphere. Apo-PCSK9 is shown in red. (C) The apo-PCSK9 (Protein Data Bank ID code 2P4E, blue; Protein Data Bank ID code 2PMW, cyan; Protein Data Bank ID code 2QTW, magenta) structures superimposed onto the PCSK9:EGF-A complex [carbon, gray (PCSK9) or yellow (EGF-A); nitrogen, blue; oxygen, red]. Arg-194 from PCSK9 forms a salt bridge with Asp-310 of EGF-A, breaking an intramolecular salt bridge with Glu-197.
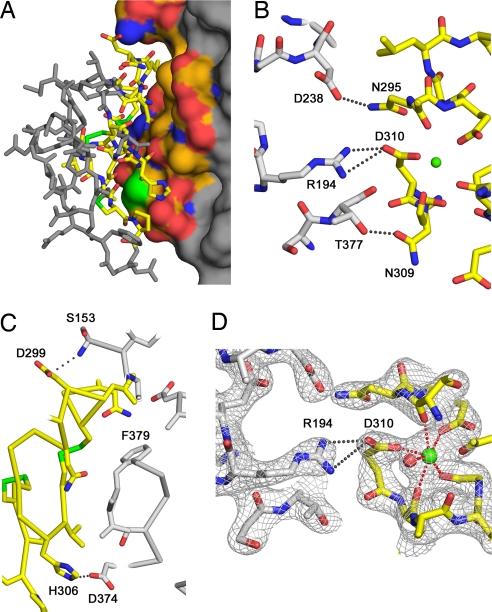
EGF-A binds a surface of PCSK9 that is formed primarily by residues 367–381. This region is >20 Å from the catalytic site of PCSK9, and ≈500 Å2 of solvent-accessible surface are buried on each molecule (Fig. 2A). The prodomain and the C-terminal domain of PCSK9 do not contact the EGF-A domain. PCSK9 primarily contacts the N-terminal region of EGF-A, making no contacts with the C terminus of EGF-A. An antiparallel β-sheet is formed between residues 377–379 of PCSK9 and 308–310 of EGF-A. In addition, key interactions with EGF-A are made by Arg-194 and Asp-238 of PCSK9 (Fig. 2B). The N terminus of the catalytic domain of PCSK9, residues 153–155, also contributes to the interface (Fig. 2C). The interface between PCSK9 and EGF-A is primarily hydrophobic with a number of specific polar interactions surrounding the interface. Phe-379 is at the center of this hydrophobic surface and makes a number of contacts to EGF-A. Other residues that contribute to this hydrophobic face include Ile-369 and Cys-378. Cys-378 contacts Leu-318 of EGF-A, a residue that was previously determined to be important for specificity (16).
Fig. 2.
Interface between PCSK9 and EGF-A. (A) The surface of PCSK9 buried upon binding of EGF-A. The surface of PCSK9 buried upon binding to EGF-A is colored according to element type: carbon, orange; nitrogen, blue; oxygen, red; sulfur, green. Areas of PCSK9 not involved in binding are colored gray. EGF-A is represented as a stick model. Residues within EGF-A involved in binding are colored according to element type: carbon, yellow; nitrogen, blue; oxygen, red. Residues not involved in binding are colored gray. (B) Interactions between PCSK9 (gray) and EGF-A (yellow) near the calcium (green sphere) binding site of EGF-A. (C) Autocatalysis of PCSK9 is required for binding to EGF-A. Autocatalysis of PCSK9 (gray) between residues 152–153 results in a free N-terminal amine that interacts with EGF-A (yellow). The site of the gain-of-function mutation, Asp-374, on PCSK9 is positioned to interact with His-306 of EGF-A. (D) Calcium coordination within the EGF-A domain after binding to PCSK9. The sigmaA weighted, 2Fo − Fc electron density map contoured at 1σ shows the calcium ligands arranged as a pentagonal bipyramid. Asp-310 coordinates calcium (green sphere) and forms a salt bridge with Arg-194 of PCSK9. A water molecule (red sphere) acts as an additional ligand.
Numerous salt bridges and hydrogen bonds formed between PCSK9 and EGF-A likely contribute to the specificity of PCSK9 binding to the EGF-A domain of LDLR over other EGF-like domains. Hydrogen bonds between PCSK9–Asp-238 and EGF-A–Asn-295, PCSK9–Thr-377 and EGF-A–Asn-309, and a salt bridge between the N-terminal amine of PCSK9–Ser-153 to EGF-A–Asp-299 contribute to this specificity. The corresponding residues in EGF-B (Asp-335, Asp-339, Val-348) or the EGF-A domain of the closely related very-LDLR (VLDLR: Asn-331, Val-335, Lys-345) or apolipoprotein E receptor-2 (apoER2: Asn-306, His-310, Thr-320) cannot participate in all of these interactions (SI Fig. 5). Mutation of Asn-295 to Ala in the LDLR decreases binding to PCSK9, demonstrating the importance of this interaction (16). The repositioning of PCSK9–Ser-153 after autocatalysis [>25 Å removed from Gln-152 (12–14)] is required for forming the observed salt bridge to EGF-A–Asp-299, consistent with the inability of secreted, uncleaved PCSK9 to bind to the LDLR-ECD (17). Nassoury et al. (29) reported that the uncleaved proform of PCSK9 binds to the LDLR in the endoplasmic reticulum when both proteins are overexpressed in cells. In this study, uncleaved PCSK9 was also shown to bind to an LDLR chimera protein lacking the EGF-precursor homology domain. Therefore, PCSK9 may form weak contacts with other regions of the LDLR that are revealed by high expression levels, explaining a recent report of PCSK9 binding to the VLDLR and apoER2 (30).
A comparison of the EGF-A binding surface in apo-PCSK9 to the PCSK9:EGF-A complex shows that Arg-194 is the only residue of PCSK9 that makes a significant conformational change upon binding to EGF-A (Fig. 1C). In apo-PCSK9, Arg-194 forms an intramolecular salt bridge with Glu-197. In the structure of the complex, Arg-194 is bent away from Glu-197 and forms a salt bridge with EGF-A–Asp-310, a calcium-coordinating residue in the EGF-A domain (21, 25–27). Mutations of Asp-310 of EGF-A to Glu severely decrease binding of LDLR to PCSK9, demonstrating the specificity of this interaction (16). The equivalent aspartate residue (Asp-64) from an EGF-like domain of factor IX uses both of its side-chain oxygens to coordinate calcium (31); however, in the structure of the PCSK9:EGF-A complex, Asp-310 coordinates calcium through only one of its side-chain oxygen atoms, whereas the other is involved in the salt bridge with Arg-194 of PCSK9 (Fig. 2D and SI Fig. 6). The calcium coordination seen in PCSK9-bound EGF-A is similar to the coordination geometry seen in the EGF-like domain of C1s where an Asn residue replaces Asp-310 and contributes only one ligand (32) (SI Fig. 6). The other calcium ligands in EGF-A include: a side-chain oxygen from Glu-296; the carbonyl oxygens of Thr-294, Leu-311, and Gly-314; and a water molecule arranged as a pentagonal bipyramid. The carbonyl oxygen of Cys-292 (Met in the crystal) is 2.9 Å from the calcium and is positioned to act as a seventh ligand. Removal of calcium coordinating residues within the EGF-A domain has been shown to impair LDL release and uncouple the fixed orientation between ligand binding module R7 and EGF-A (27). The observed calcium coordination in the PCSK9:EGF-A complex could signify a conformational change in the EGF-A domain upon PCSK9 binding.
Mutagenesis of PCSK9.
The crystal structure of the PCSK9:EGF-A complex identified Arg-194 and Phe-379 of PCSK9 as critical residues for EGF-A binding, with Arg-194 forming a salt bridge with EGF-A–Asp-310 and Phe-379 making several contacts, through both its main chain and side chain, to EGF-A (Fig. 2). To confirm that these residues are critical for PCSK9 binding to full-length LDLR, purified proteins were prepared that harbor amino acid changes in these key residues (Arg-194–Gln or Phe-379–Ala), and the mutant proteins were tested for their ability to bind to the LDLR-ECD. Neither mutation affected PCSK9 autocatalysis or secretion rate (SI Fig. 7A). As with wild-type PCSK9, the catalytic fragment of the mutant proteins copurified with bound prodomain as revealed by SDS/PAGE and Coomassie staining of the purified mutant proteins (SI Fig. 7B).
Ligand blotting was performed to assess the ability of immobilized mutant PCSK9 proteins to associate with fluorophore-labeled LDLR-ECD. To ensure equivalent amounts of PCSK9 were present in each lane, blots were coincubated with a fluorophore-labeled anti-PCSK9 mAb that does not interfere with LDLR binding (data not shown). The gain-of-function mutant Asp-374–Tyr bound LDLR-ECD with greater affinity than wild-type PCSK9 (SI Fig. 7C), in agreement with previous reports (11, 12, 15). Mutation of either Arg-194 or Phe-379 decreased PCSK9 binding to LDLR-ECD by >90%. Thus, Arg-194 and Phe-379 in the catalytic domain of PCSK9 represent critical surface residues for the binding of PCSK9 to the LDLR.
Natural Mutations in PCSK9.
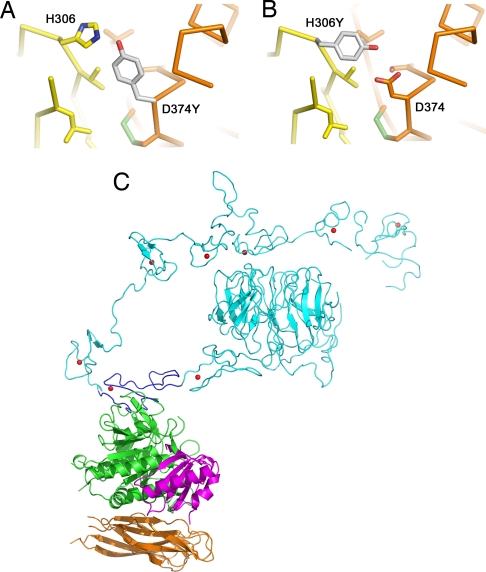
The gain-of-function mutation Asp-374–Tyr increases the affinity of PCSK9 for LDLR ≈5- to 30-fold (11, 12, 15). PCSK9–Asp-374 is positioned 4 Å from EGF-A–His-306. In the crystal structure at pH 4.8, EGF-A–His-306 is presumably protonated and forming a salt bridge with PCSK9–Asp-374. A mutation of Asp-374 to Tyr would place the hydroxyl group of tyrosine ≈3 Å from EGF-A–His-306, allowing for a favorable hydrogen bond (Fig. 3A). Of note, the FH-associated mutation His-306–Tyr would create the reciprocal change, suggesting that this mutation in LDLR would increase its affinity toward PCSK9 (Fig. 3B), thereby causing enhanced degradation of the LDLR.
Fig. 3.
Mutations in PCSK9 and LDLR. (A) The gain-of-function mutation Asp-374–Tyr in PCSK9 increases binding to LDLR. Asp-374 in PCSK9 modeled as tyrosine (gray) is in position to hydrogen bond to His-306 of LDLR. (B) The FH mutation His-306–Tyr in LDLR. His-306 in LDLR modeled as tyrosine (gray) is in position to hydrogen bond to Asp-374 of PCSK9. (C) Model for full-length LDLR-ECD bound to PCSK9. The EGF-A domain (blue) of the LDLR-ECD (cyan) at acidic pH and the PCKSK9:EGF-A complex were superimposed. PCSK9 (prodomain, magenta; subtilisin-like catalytic domain, green; C-terminal domain, brown) binds on the outside surface of LDLR and would not interfere with the interaction of ligand binding modules R4 and R5 with the β-propeller domain.
Other gain-of-function mutations, Ser-127–Arg and Phe-216–Leu, display modest changes in affinity for the LDLR (12, 15), suggesting that the hypercholesterolemic phenotype associated with these mutations is not directly a result of an increase in affinity for LDLR. PCSK9 containing the Ser-127–Arg mutation is internalized at approximately the same rate as wild-type PCSK9 (15), but processing and secretion are less efficient for this mutant (10, 33). Ser-127 is located in the prodomain and is >40 Å from the EGF-A binding site. Although it is possible that other domains of LDLR interact with PCSK9, these domains are not absolutely required for binding (16), making it unlikely that Ser-127 or neighboring residues contribute significantly to the binding affinity of PCSK9 for the LDLR through other direct contacts. The distance of Ser-127 from the EGF-A binding surface suggests that the phenotype of this mutation is likely a result of another mode of PCSK9 regulation in vivo.
Phe-216 of PCSK9 is located within a disordered loop in the structure of the PCSK9:EGF-A complex and apo-PCSK9 (12–14). PCSK9 carrying the Phe-216–Leu gain-of-function mutation does not have increased affinity for LDLR (12) and Phe-216 does not interact with the EGF-A domain in the structure of the PCSK9:EGF-A complex. The Phe-216–Leu mutation has been proposed to reduce a Furin/PC5/6A-dependent cleavage of PCSK9 after residue Arg-218 (34). This cleavage event would remove two residues (Ser-153 and Arg-194) that form key salt bridges in the PCSK9:EGF-A complex, presumably weakening or destroying the interaction. Indeed, alteration of Arg-194 in PCSK9 by site-directed mutagenesis decreased binding to the LDLR-ECD by 90% (SI Fig. 7C). Thus, the structure of the PCSK9:EGF-A complex provides a molecular basis for the FH phenotype associated with the Phe-216–Leu mutation.
Loss-of-function mutations in PCSK9 resulting in missense or nonsense mutations generally result in lowered levels of secreted PCSK9 (35). Among the loss-of-function mutations that are processed and secreted normally, Ala-443–Thr is ≈20 Å from the EGF-A binding site and does not play a direct role in binding to EGF-A. It has been suggested that the Ala-443–Thr mutation results in a higher susceptibility to Furin/PC5/6A-dependent cleavage (34) and may result in lower levels of functional PCSK9. PCSK9 carrying the Arg-46–Leu mutation has been shown to have a modest decrease in affinity toward the LDLR (15). Arg-46 is not present in our structure. Because Thr-61 is ≈55 Å from EGF-A, it is unlikely that Arg-46 could interact with EGF-A without a significant conformational change occurring in the prodomain. It should be noted that Δ53-PCSK9 binds with increased affinity to the LDLR-ECD at physiological pH (SI Table 2), suggesting that the region of the prodomain containing Arg-46 could affect binding of PCSK9 to the LDLR in an indirect manner.
pH Dependence of Binding.
The LDLR is proposed to undergo a conformational change at acidic pH from an opened to a closed conformation, thereby facilitating ligand release (21, 28). A model of the low pH form of LDLR bound to PCSK9 was generated by superimposing the EGF-A domain from the LDLR-ECD structure (21) onto the corresponding residues in the PCSK9:EGF-A structure (Fig. 3C). Based on this model, PCSK9 interacts only with the EGF-A domain and binds on the outside of the closed, low pH form of the LDLR. As such, PCSK9 would not be predicted to sterically interfere with the intramolecular interaction of ligand binding modules R4 and R5 to the β-propeller domain or with lipoprotein binding. However, an effect imposed by PCSK9 on either lipoprotein binding or adoption of the closed conformation by the LDLR cannot be excluded.
The crystal structure of the LDLR-ECD at pH 5.3 shows an alternative orientation of EGF-A relative to EGF-B from that observed in the isolated EGF-AB domain NMR structure obtained at pH 6.5 (25, 26). Given the lack of change in apo-PCSK9 domain organization and structure at pH values ranging from 4.6 to 10.5 (12–14) and in PCSK9 in complex with EGF-A, it is possible that the enhanced affinity of PCSK9 toward LDLR at acidic pH is caused by a conformational change in the LDLR. Consistent with this model, in vitro binding studies comparing association and dissociation rates between PCSK9 and LDLR reveal that the major difference at neutral versus acidic pH is a change in the association rate. At low pH, the association rate increases ≈100-fold, whereas the dissociation rate is virtually unchanged (15). Our binding studies indicate that at acidic pH, PCSK9 has increased affinity toward the EGF-AB fragment used in our structural studies (SI Table 2). The in vitro binding studies with the EGF-AB fragment and the crystal structure of the PCSK9:EGF-A complex suggest that the pH-dependent increase in affinity resides, at least partially, within the EGF-AB fragment. It has been suggested that a region rich in histidine residues on the surface of the C-terminal domain of PCSK9 might be responsible for the pH-dependent increase in affinity toward LDLR (12–14). This region of the C-terminal domain is disordered in our structure; therefore, we cannot rule out a contribution of the C-terminal domain of PCSK9 to the pH-dependent changes in binding affinity, either through interacting with domains other than EGF-A or influencing the conformation of PCSK9. In the PCSK9:EGF-A structure, only a single histidine contributes to the binding interface, His-306 of EGF-A. Based on our model, it is unlikely that PCSK9 interacts with other domains of the LDLR without significant conformational changes to either PCSK9 and/or the LDLR. Our structural and biochemical data are consistent with a model where LDLR, through a low pH-induced conformational change and/or protonation of His-306, is responsible for the pH-dependent increase in affinity toward PCSK9.
PCSK9-Dependent LDLR Degradation.
The cellular mechanism by which PCSK9 increases LDLR degradation remains unresolved. The structure of the PCSK9:EGF-A complex is consistent with a model where enhanced affinity of PCSK9 for a low-pH conformation of the LDLR prevents receptor recycling. PCSK9 binding to the LDLR may alter the conformation of the low-pH form of the LDLR. For instance, binding of PCSK9 near the N-terminal region of EGF-A might affect known interdomain interactions involving the EGF-A domain, either through steric hindrance by the PCSK9 molecule itself or by affecting the EGF-A conformation, calcium coordination, and/or stability. Known intramolecular EGF-A interactions include the interdomain packing of EGF-A with EGF-B, which is important for LDLR stability (25), and the interaction of EGF-A with ligand binding module R7, which confers a rigid conformation on this region of the LDLR across a wide pH range (27). This rigidity is proposed to facilitate the acid-dependent closed conformation that allows ligand release and LDLR recycling from the endosomal compartment (28). Alternatively, PCSK9 may bind to an as-yet-unknown factor that directly promotes degradation of LDLR.
Conclusions
PCSK9 has emerged as one of the most important determinants of plasma LDL-C levels as illustrated by a recent report that described a compound heterozygote with two inactivating mutations in PCSK9 (35). That healthy individual had no circulating PCSK9 and a strikingly low plasma LDL-C concentration of 14 mg/dl. The effects of heterozygous loss-of-function mutations in PCSK9 (Tyr-142X and Cys-679X in African-Americans and Arg-46–Leu in Caucasians) on CHD have been reported in a large biracial 15-year prospective study (5). The Tyr-142X and Cys-679X nonsense mutations in PCSK9 reduced LDL-C levels by 28% and decreased the frequency of CHD by 88%. Caucasians with an Arg-46–Leu allele had a mean reduction in LDL-C levels of 15% and manifest a 47% reduction in CHD (5). Thus, PCSK9 represents a validated therapeutic target for the treatment of hypercholesterolemia; however, the demonstration that catalytic activity is required for PCSK9 maturation and secretion but not for degradation of LDLRs suggests that small-molecule inhibitors of PCSK9 catalytic activity will need to function in the endoplasmic reticulum (17, 18). The elucidation of the structure of the PCSK9:EGF-A complex now provides the structural foundation for the development of alternative approaches to inhibit PCSK9-mediated LDLR degradation. If PCSK9 functions primarily as a secreted protein in plasma, agents that interfere with the PCSK9:LDLR interaction at the cell surface also have the potential to lower plasma LDL-C levels in individuals with hypercholesterolemia.
Materials and Methods
Expression Constructs for Truncated and Mutant Forms of PCSK9.
A full-length human PCSK9 cDNA followed by a FLAG-epitope tag (DYKDDDDK) under control of the cytomegalovirus promoter-enhancer (pCMV-PCSK9-FLAG) was used for generation of truncated (Δ53-PCSK9) and mutant forms of PCSK9 (33). Δ53-PCSK9, Arg-194–Gln, and Phe-379–Ala mutant forms of PCSK9 were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Purification of Recombinant Proteins.
Δ53-PCSK9 was stably expressed in HEK-293S cells (36). PCSK9(Arg-194–Gln) and PCSK9(Phe-379–Ala) were expressed in HEK-293 cells after transient transfection by using FuGene 6 transfection reagent (Roche) according to the manufacturer's instructions. FLAG-tagged proteins were purified from conditioned medium as described (11). The EGF-AB fragment (residues 293–372) was expressed as a GST fusion in Rosetta-gamiB cells (Novagen) and purified by glutathione chromatography. The GST tag was removed by tobacco etch virus protease cleavage, and the EGF-AB fragment was further purified by ion exchange and size exclusion chromatography. Reverse-phase chromatography and mass spectrometry were used to monitor proper folding and disulfide bond formation. The LDLR-ECD was expressed and purified as described (21).
Pulse–Chase Analysis.
HEK 293A cells (Q-Biogene) were cultured in DMEM (Cellgro) supplemented with 10% (vol/vol) FCS, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, and 1 g/liter glucose, and the pulse–chase was carried out as described (11). Immunoprecipitation of cell and medium extracts was performed as described (11).
In Vitro Binding Measurements.
The pH dependence of 125I-labeled PCSK9 binding to LDLR-ECD or EGF-AB fragment was analyzed by ligand blotting as described (16). Other binding assays were performed by using purified PCSK9 and LDLR-ECD proteins labeled through amine-linkage with DyLight800 fluorophore dye with the DyLight antibody labeling kit per the manufacturer's instructions (Pierce). Blots were scanned by using the LI-COR Odyssey Infrared Imaging System, and band intensity was quantified with Odyssey v2.0 software.
Crystallization and Structure Determination.
The complex of Δ53-PCSK9 and EGF-AB was formed by adding a 4-fold molar excess of EGF-AB to Δ53-PCSK9 and further purified by size exclusion chromatography. Initial crystals were obtained with the Fluidigm TOPAZ system. Crystals of PCSK9:EGF-A were formed by mixing equal volumes of the complex (6 mg/ml in 10 mM Tris, pH 7.5, 50 mM NaCl, 0.25 mM CaCl2, 0.01% NaN3) with 0.3 M (NH4)H2PO4 and equilibrating the mixture under Al's oil against H2O. Crystals were transferred stepwise into a solution of 0.05 M (NH4)H2PO4, 5 mM CaCl2, and 35% glycerol and flash-frozen in a −160°C nitrogen stream. Crystals belong to space group P4(1)2(1)2 (a = b = 117.0 Å, c = 134.9 Å). Diffraction data were collected at the Advanced Photon Source (Argonne, IL) beam line 19-ID and processed with HKL2000 (37) and the CCP4 suite (38). The structure was determined by molecular replacement using the program PHASER (39). The search model included residues 61–449 of PCSK9 (Protein Data Bank ID code 2P4E). The model was built with the program COOT (40). Initial refinement was performed with CNS (41), and the final cycles of refinement were performed with REFMAC (42). Figures were generated with PYMOL (www.pymol.org).
Other Materials and Methods.
Detailed descriptions of experimental protocols and reagents are described in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Maya Palnitkar, Lisa Henry, Y. K. Ho, Linda Donnelly, Lisa Beatty, and members of the laboratories of J.D. and J.D.H. for assistance and comments during the course of this project; Helen Hobbs for critical reading of the manuscript; Norma Duke and Jack Lazarz [Argonne National Laboratory (Argonne, IL)], Chad Brautigam, and Diana Tomchick for assistance with data collection. This work was supported in part by a grant from the Welch Foundation (to J.D.), the Perot Family Foundation, and National Institutes of Health Grants HL-38049 and HL-20948 (to J.D.H.). M.C.M. is supported by Medical Scientist Training Grant GM08014. T.A.L. was supported by a fellowship from the Canadian Institutes of Health Research. J.D. is an Investigator in the Howard Hughes Medical Institute. Results shown in this article are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BPS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712064105/DC1.
References
- 1.Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: Its role in LDL metabolism. Trends Biochem Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert G. Unraveling the functional significance of PCSK9. Curr Opin Lipidol. 2007;18:304–309. doi: 10.1097/MOL.0b013e3281338531. [DOI] [PubMed] [Google Scholar]
- 3.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 6.Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: Explanation of their specificity. J Mol Biol. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 7.Anderson ED, et al. The ordered and compartment-specific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J Biol Chem. 2002;277:12879–12890. doi: 10.1074/jbc.M108740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker D, Shiau AK, Agard DA. The role of pro regions in protein folding. Curr Opin Cell Biol. 1993;5:966–970. doi: 10.1016/0955-0674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama K. Furin: A mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjannet S, et al. NARC-1/PCSK9 and its natural mutants: Zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 11.Lagace TA, et al. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham D, et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14:413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- 13.Hampton EN, et al. The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain. Proc Natl Acad Sci USA. 2007;104:14604–14609. doi: 10.1073/pnas.0703402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piper DE, et al. The crystal structure of PCSK9: A regulator of plasma LDL-cholesterol. Structure (London) 2007;15:545–552. doi: 10.1016/j.str.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Fisher TS, et al. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J Biol Chem. 2007;282:20502–20512. doi: 10.1074/jbc.M701634200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang DW, et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 17.McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J Biol Chem. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- 18.Li J, et al. Secreted PCSK9 promotes LDL receptor degradation independently of proteolytic activity. Biochem J. 2007;406:203–207. doi: 10.1042/BJ20070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell DW, et al. Domain map of the LDL receptor: Sequence homology with the epidermal growth factor precursor. Cell. 1984;37:577–585. doi: 10.1016/0092-8674(84)90388-x. [DOI] [PubMed] [Google Scholar]
- 20.Rudenko G, Deisenhofer J. The low-density lipoprotein receptor: Ligands, debates, and lore. Curr Opin Struct Biol. 2003;13:683–689. doi: 10.1016/j.sbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Rudenko G, et al. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 22.Davis CG, et al. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 23.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 24.van der Westhuyzen DR, et al. Deletion of two growth-factor repeats from the low-density-lipoprotein receptor accelerates its degradation. Biochem J. 1991;277:677–682. doi: 10.1042/bj2770677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha S, et al. Solution structure of the LDL receptor EGF-AB pair: A paradigm for the assembly of tandem calcium binding EGF domains. Structure (London) 2001;9:451–456. doi: 10.1016/s0969-2126(01)00606-2. [DOI] [PubMed] [Google Scholar]
- 26.Kurniawan ND, Aliabadizadeh K, Brereton IM, Kroon PA, Smith R. NMR structure and backbone dynamics of a concatemer of epidermal growth factor homology modules of the human low-density lipoprotein receptor. J Mol Biol. 2001;311:341–356. doi: 10.1006/jmbi.2001.4867. [DOI] [PubMed] [Google Scholar]
- 27.Beglova N, Jeon H, Fisher C, Blacklow SC. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol Cell. 2004;16:281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 28.Beglova N, Blacklow SC. The LDL receptor: How acid pulls the trigger. Trends Biochem Sci. 2005;30:309–317. doi: 10.1016/j.tibs.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Nassoury N, et al. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR. Traffic. 2007;8:718–732. doi: 10.1111/j.1600-0854.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 30.Poirier S, et al. The proprotein convertase PCSK9 induces the degradation of LDLR and its closest family members VLDLR and APOER2. J Biol Chem. 2008;283:2363–2372. doi: 10.1074/jbc.M708098200. [DOI] [PubMed] [Google Scholar]
- 31.Rao Z, et al. The structure of a Ca2+-binding epidermal growth factor-like domain: Its role in protein–protein interactions. Cell. 1995;82:131–141. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 32.Gregory LA, Thielens NM, Arlaud GJ, Fontecilla-Camps JC, Gaboriaud C. X-ray structure of the Ca2+-binding interaction domain of C1s: Insights into the assembly of the C1 complex of complement. J Biol Chem. 2003;278:32157–32164. doi: 10.1074/jbc.M305175200. [DOI] [PubMed] [Google Scholar]
- 33.Park SW, Moon YA, Horton JD. Posttranscriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J Biol Chem. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 34.Benjannet S, Rhainds D, Hamelin J, Nassoury N, Seidah NG. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: Functional consequences of natural mutations and post-translational modifications. J Biol Chem. 2006;281:30561–30572. doi: 10.1074/jbc.M606495200. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Z, et al. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am J Hum Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves PJ, Thurmond RL, Khorana HG. Structure and function in rhodopsin: High-level expression of a synthetic bovine opsin gene and its mutants in stable mammalian cell lines. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.