Abstract
Allostery is a common mechanism of regulation of enzyme activity and specificity, and its signatures are readily identified from functional studies. For many allosteric systems, structural evidence exists of long-range communication among protein domains, but rarely has this communication been traced to a detailed pathway. The thrombin mutant D102N is stabilized in a self-inhibited conformation where access to the active site is occluded by a collapse of the entire 215–219 β-strand. Binding of a fragment of the protease activated receptor PAR1 to exosite I, 30-Å away from the active site region, causes a large conformational change that corrects the position of the 215–219 β-strand and restores access to the active site. The crystal structure of the thrombin-PAR1 complex, solved at 2.2-Å resolution, reveals the details of this long-range allosteric communication in terms of a network of polar interactions.
Keywords: protease activated receptor, thrombin, x-ray crystallography
Ever since its original formulation (1, 2), the allosteric concept of enzyme regulation has captivated the interest of structural biologists. The idea that events occurring at a given site of the protein can be transmitted long-range to affect affinity or catalytic efficiency at a distant site offers an elegant explanation for linkage and cooperativity (3) but poses a challenge to the crystallographer who seeks to identify the communication pathway underlying the functional effects. Perutz (4) was the first to offer a molecular explanation for the allosteric mechanism of hemoglobin cooperativity. For multimeric proteins like hemoglobin, conformational transitions tend to affect the quaternary structure and signatures of allostery have been detected crystallographically (5–9). Allostery is not limited to multimeric assemblies and, in fact, many monomeric proteins feature conformational plasticity that translate into allosteric behavior at equilibrium and steady state (6, 10, 11). For such systems, the structural signatures of long-range communication tend to manifest themselves as small changes in tertiary structure or H-bonding connectivity (12, 13) and lack the amplification seen in large quaternary structural reorganization. Identification of the pathway of communication underlying an allosteric mechanism remains a difficult task in general and continues to receive utmost attention (11, 13). Alternative approaches have been proposed to identify such pathways based on the statistical analysis of evolutionary records of protein sequences (14) or anisotropic thermal diffusion (15). Although promising and insightful, such approaches must ultimately find validation by structural investigation.
Among Na+-activated allosteric enzymes (6), the serine protease thrombin has received much attention in view of its critical roles in hemostasis and thrombosis (12, 16, 17). Thrombin features considerable structural plasticity and exists predominantly in two forms at equilibrium, the Na+-free slow form E (≈40% of the molecules in vivo) and the Na+-bound fast form E:Na+ (≈60% of the molecules in vivo) that are responsible, respectively, for the anticoagulant and procoagulant functions of the enzyme (18). A third form, E* (≈1% of the molecules in vivo), is in equilibrium with E and is unable to bind Na+ (19). Na+ binds 15-Å away from residues of the active site (20, 21) and enhances activity toward fibrinogen and the protease activated receptors (PAR) PAR1 and PAR4 (18, 22). Structural details on how Na+ binding influences allosterically the active site have emerged recently (12, 21), but mutagenesis and spectroscopic studies vouch for more extensive, global effects of Na+ binding on the conformation of the enzyme (12, 19, 23). In addition to the allosteric effect of Na+, thrombin activity and specificity is influenced allosterically by binding of ligands to exosite I (24–28), a domain located 25-Å away from the active site and on the opposite pole of the molecule relative to the Na+ site (16, 29). The exact mechanism of this long-range communication remains controversial (30, 31). A recent structure of murine thrombin bound to a fragment of PAR3 at exosite I has revealed a significant conformational change of the 60-loop that opens the active site fully for optimal substrate diffusion (32). Such structural transition is relevant to the cofactor function of PAR3 on PAR4 cleavage (33–35) and to the cofactor function of thrombomodulin on protein C activation that is at the basis of the anticoagulant activity of thrombin (36, 37). We now provide crystallographic evidence of a much larger conformational change experienced by thrombin when exosite I is bound to the extracellular fragment of PAR1, which is the primary receptor responsible for aggregation of human platelets (33, 34, 38). The findings reveal the full extent of structural flexibility accessible to this important allosteric enzyme and a precise pathway of long-range communication.
Results
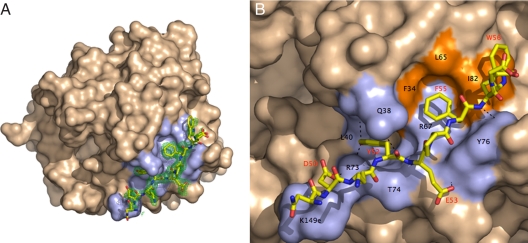
Human thrombin inactivated with the single-site mutation D102N was crystallized in complex with the extracellular fragment of human PAR1, 42SFLLRNPNDKYEPFWEDEEKN62, corresponding to the sequence downstream from the cleavage site at Arg-41 (38) and containing the hirudin-like motif 52YEPFWE57 predicted to bind to exosite I (33, 38). Although the structure was solved at a resolution of 2.2 Å, only the sequence 49NDKYEPFWE57 of the extracellular fragment of PAR1 could be traced in the electron density map, leaving the tethered ligand domain 42SFLLRN47 and the acidic tail of the fragment unresolved (Fig. 1). A similar problem recently was encountered for the cleaved fragment of murine PAR3 bound to exosite I of murine thrombin (32). The tethered ligand domain presumably folds away from the thrombin surface after cleavage at Arg-41, and its conformation becomes disordered, as documented by NMR studies (39). As recently seen for the binding epitope of PAR3 (32), the PAR1 fragment engages exosite I of thrombin in a number of polar and hydrophobic interactions starting with Asp-50 whose Oδ1 atom H bonds to the NH1 and NH2 atoms of Arg-73 and the Nζ atom of Lys-149e of thrombin (Table 1 and Fig. 1). Other polar interactions involve the O and OH atoms of Tyr-52 of PAR1 with the Nε2 atom of Gln-38, the NH1 atom of Arg-73, and the N atom of Leu-40 of thrombin. The Oε1 and N atoms of Glu-53 H bond to the N atom of Tyr-76 and the O atom of Thr-74 of thrombin, respectively. Finally, a weak H bond couples the N atom of Trp-56 to the OH atom of Tyr-76 of thrombin. The hydrophobic interactions are substantial and involve Phe-34, Leu-65, and Ile-82 that accommodate in a cluster Tyr-52, Phe-55, and Trp-56 of PAR1. Phe-34 is edge-to-face with respect to Tyr-52 on one side and Phe-55 on the other side. Phe-55 is in van der Waals' contact with the entire hydrophobic cluster in exosite I and is also engaged in a cation–π interaction by Arg-67 of thrombin. Trp-56 interacts with Ile-82 of thrombin at the boundary of the binding epitope. The contacts documented by the crystal structure are in agreement with the results of Ala scanning mutagenesis of thrombin (40). In particular, mutagenesis studies have pointed out the peculiar contribution of Arg-67 to PAR1 recognition. The guanidinium group of Arg-67 is within 4.2 Å from the benzene ring of Phe-55 of PAR1, in optimal cation–π interaction, but not so relative to Phe-50 of PAR3 (32), which could explain why the R67A mutation is far more deleterious for PAR1 binding relative to PAR3 (40). Ala scanning mutagenesis of PAR1 have singled out Tyr-52, Glu-53, and Phe-55 as important determinants of recognition by exosite I (38), in agreement with the results of the crystal structure. However, the important role of Asp-50 of PAR1 in the interaction with thrombin has been missed by previous mutagenesis studies (38, 40).
Fig. 1.
Structure of the human thrombin mutant D102N in complex with the extracellular fragment of human PAR1. (A) Thrombin is rendered in surface representation (wheat) with residues <4 Å from the bound fragment of PAR1 (stick model) colored in light blue. The orientation is centered on the 30-loop that separates exosite I on the right from the active site cleft on the left. The 60-loop occupies the upper rim of the active site. The electron density 2Fo − Fc map (green mesh) is contoured at 1.0σ. (B) Details of the molecular contacts at the thrombin–PAR1 interface, with hydrophobic regions of the thrombin epitope colored in orange and polar regions colored in light blue. H bonds are depicted as broken lines. Residues involved in contacts <4 Å are listed in Table 1 and are labeled in black for thrombin and red for PAR1. The extracellular fragment of PAR1 engages exosite I through polar and hydrophobic interactions.
Table 1.
Interatomic contacts (<4 Å) between thrombin and PAR1
| Thrombin | PAR1 | Distance, Å |
|---|---|---|
| Arg-73 NH1 | Asp-50 Oδ1 | 2.94 |
| Arg-73 NH2 | Asp-50 Oδ1 | 3.06 |
| Lys-149e Nζ | Asp-50 Oδ1 | 3.07 |
| Gln-38 Nε2 | Tyr-52 O | 3.34 |
| Leu-40 N | Tyr-52 OH | 3.88 |
| Arg-73 NH1 | Tyr-52 OH | 3.13 |
| Phe-34 | Tyr-52 | vdW |
| Thr-74 O | Glu-53 N | 2.95 |
| Tyr-76 N | Glu-53 Oε1 | 2.67 |
| Phe-34, Leu-65, Ile-82 | Phe-55 | vdW |
| Tyr-76 OH | Trp-56 N | 3.86 |
| Ile-82 | Trp-56 | vdW |
vdW, van der Waals' contact.
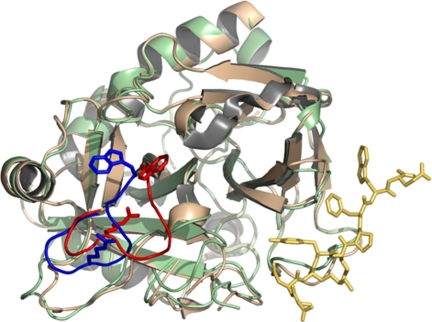
A previous structure of thrombin bound to a fragment of PAR1 revealed a nonproductive binding mode bridging two thrombin molecules in the crystal lattice without any significant conformational change in the enzyme (41). The structure of bound D102N reported here shows the fragment of PAR1 recognizing exosite I basically in the same conformation as reported previously (41). However, the structure documents a large conformational change that propagates from Phe-34 and Arg-73 in exosite I to Trp-215 in the aryl binding site and Arg-221a in the 220-loop located up to 28-Å away on the opposite side of the active site relative to exosite I. The conformational change is revealed by comparison of the structure of D102N bound to the fragment of PAR1 at exosite I with that of the free mutant solved recently (42). The free mutant D102N folds in a self-inhibited conformation with the active site occluded (Fig. 2). The 215–219 β-strand collapses into the active site, with the indole ring of Trp-215 losing its contact with Phe-227 and making hydrophobic contact with the catalytic His-57. Further downstream from Trp-215, Arg-221a relinquishes its ion–pair interaction with Glu-146 in the autolysis loop and penetrates the 220-loop to engage the carboxylate of Asp-189 in the primary specificity pocket with its guanidinium group. The drastic rearrangement of the 215–219 β-strand and the 220-loop shift the orientation of the Cys-191:Cys-220 disulfide bond and propagate the perturbation up to the oxyanion hole where the backbone N atom of Gly-193 is flipped relative to wild type (42). Binding of PAR1 to exosite I reverses all of these drastic changes and shifts the conformation of D102N into that of wild-type thrombin (21, 41) using a long-range allosteric communication between exosite I and the active site.
Fig. 2.
Allosteric effect induced by binding of the extracellular fragment of PAR1 (stick model in gold) to exosite I of thrombin (ribbon model in light green) on the conformation of the 215–219 β-strand and the 220-loop (blue). The position of Trp-215 and Arg-221a is indicated as a stick model. Thrombin is shown in the standard Bode orientation (29) with the active site cleft in the middle and exosite I to the right. Comparison with the free structure of thrombin (ribbon model in wheat, with the 215–219 β-strand and the 220-loop, Trp-215, and Arg-221a in red) shows a drastic rearrangement that pushes the 215–219 β-strand back >6 Å. Trp-215 and Arg-221a relocate >9 Å to restore access to the active site and primary specificity pocket that was obliterated in the free form. The allosteric communication between exosite I and the 215–219 β-strand and 220-loop spans almost 30 Å across the thrombin molecule (see also Fig. 3) and reveals a possible mechanism for the conversion of thrombin from its inactive form E* into the active form E.
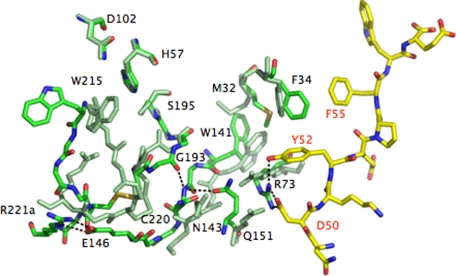
The crystal structure of D102N bound to PAR1 at exosite I details a plausible, yet necessarily hypothetical pathway underlying this long-range communication. In what follows, we propose a possible sequence of events consistent with the changes documented in the electron density maps. The allosteric communication is triggered by a change in the conformation of Phe-34 and Arg-73 in exosite I. The Cε2 atom of Phe-34 is 4.3-Å away from the S atom of Met-32 in the free form of D102N. Upon binding of PAR1 to exosite I, the phenyl ring of Phe-34 rotates to optimize its interaction with Phe-55 and Tyr-52 of PAR1 and pulls the Cε atom of Met-32 along causing a 5.6-Å shift (Fig. 3). The indole ring of Trp-141 switches places with Met-32 and causes a >1-Å concerted translation of the entire 141–146 β-strand. Contributing to this shift is the repositioning of Arg-73 for engagement of Asp-50 and Tyr-52 of PAR1 that pushes Gln-151 toward Asn-143, causing electrostatic clash between the side chain O atoms coming within 2.96 Å. Pro-152 pushes Gly-142 back, contributing to the realignment of the 141–146 and 191–193 β-strands that become stabilized by H bonds between the backbone N atom of Asn-143 with the Oε1 atom of Gln-151 on one side and the backbone O atom of Glu-192 on the other side (Fig. 3). The latter H bond flips back the peptide bond with Gly-193 into the position seen in the wild type and reconstitutes the oxyanion hole with the correct orientation of the backbone N atom. As the 141–146 β-strand is pushed back, the backbone O atom of Asn-143 collides with the S atom of Cys-220. As a result, the Cys-191:Cys-220 disulfide bond twists and relocates 4.29-Å deeper inside the protein, causing a major rearrangement in the backbone of the 215–219 β-strand and the 220-loop. The shift in the 141–146 β-strand initiated at Trp-141 and amplified by Arg-73 propagates to the hinge region of the flexible autolysis loop and orders the side chain of Glu-146 for a strong ion–pair with Arg-221a in the 220-loop, whose guanidinium group moves >9 Å from the interior of the primary specificity pocket to the surface of the protein. The entire 215–219 β-strand moves back >6 Å and Trp-215 reestablishes its stacking interaction with Phe-227 moving >10-Å away from the catalytic His-57, thereby restoring access to the active site. Asp-221 restores its important ion–pair interaction with Arg-187 whose guanidinium group moves away from the Na+ binding pocket.
Fig. 3.
Molecular basis of the allosteric communication between exosite I and the 215–219 β-strand and 220-loop spanning almost 30 Å across the thrombin molecule (see also Fig. 2). The extracellular fragment of PAR1 is rendered as a stick model with C atoms in yellow and the thrombin residues in the PAR1 bound form are rendered as stick models with C atoms in green. Residues of the free form of thrombin are rendered as stick models uniformly colored in light green. Relevant H bonds are indicated as broken lines. The allosteric communication initiates with a rotation of the benzene ring of Phe-34 and a shift in the side chain of Arg-73 (labeled in black, as all other thrombin residues) in exosite I because of binding of PAR1 via Phe-55, Tyr-52, and Asp-50 (labeled in red). The changes propagate to the 141–146 β-strand via Met-32 and Gln-151. In turn, that reestablishes H bond connections with the 191–193 β-strand and restores the oxyanion hole and the orientation/location of the Cys-191:Cys-220 disulfide bond, which relocates the entire 215–219 β-strand and 220-loop in their canonical positions to free access to the active site and the primary specificity pocket. Trp-215 folds back almost 10 Å into the aryl binding site, relinquishing its hydrophobic interaction with the catalytic His-57 (the catalytic Asp-102 and Ser-195 are also shown for completeness). Arg-221a leaves the interior of the primary specificity pocket where it binds to Asp-189 (data not shown) and moves >9 Å to the surface to engage Glu-146 in a strong bidentate ion–pair. Glu-146 is disordered in the free structure. Additional changes involving the 186-loop restoring access to the Na+ binding site are omitted for clarity. The allosteric communication documented in the structure of the thrombin–PAR1 complex relative to the free form of the enzyme is testimony to the flexibility of the thrombin fold and proves that the various forms of the enzyme (E*, E, and E:Na+) interconvert under the influence of ligand binding to distinct domains.
Binding of PAR1 to exosite I converts the self-inhibited conformation of the mutant D102N into one that is catalytically competent, with the oxyanion hole correctly structured and access to the active site and primary specificity pocket open to substrate docking. The peculiar self-inhibited conformation documented originally for D102N (42) therefore can convert into a catalytically active conformation through a structural transition that can be traced to a set of residues organized in four layers (Fig. 3). A first layer directly in contact with ligands recognizing exosite I (Phe-34 and Arg-73), a second layer of “transducing” residues connecting to the 141–146 β-strand (Met-32 and Gln-151), a third layer comprising the interactions between the 141–146 and 191–193 β-strands (Trp-141, Asn-143, and Glu-192) and a final layer where such interactions are transmitted to the 215–219 β-strand and the 220-loop via the Cys-191:Cys-220 disulfide bond and Glu-146.
Discussion
The allosteric nature of thrombin was established more than a decade ago from functional studies (43). Two well documented pathways of allosteric regulation exist in the thrombin molecule: one involving the Na+ site and the other involving exosite I. Binding of Na+ to thrombin enhances activity toward procoagulant and prothrombotic substrates like fibrinogen and PARs (12, 44), whereas binding of thrombomodulin to exosite I enhances activity toward the anticoagulant protein C (37, 45). A significant linkage also exists between the two allosteric sites (28, 46, 47). The structural mechanism underlying these physiologically important mechanisms of allosteric regulation has been subject to intense investigation. The structure of thrombin bound to a fragment of thrombomodulin at exosite I failed to reveal significant conformational changes in the active site (30). Such changes might have been obliterated by the presence of the active site inhibitor used in the crystallization. A number of peptides targeting exosite I influence allosterically the active site of thrombin and bring about significant changes in activity and even substrate specificity (24, 27, 28, 47, 48). One of these peptides, hirugen, is derived from the C-terminal fragment of hirudin. The structure of thrombin bound to hirugen was solved with the active site free (31) but again failed to reveal any significant conformational changes as for the thrombomodulin-bound structure (30). In contrast, the recent structure of murine thrombin bound to a fragment of PAR3 at exosite I reveals a snapshot of the mechanism for the allosteric communication in terms of a shift in the indole ring of Trp-60d and upward movement of the entire 60-loop that open up the active site cleft (32). The resulting facilitated diffusion of substrate into the active site produces an enhancement of kcat/Km as found experimentally (28). Thrombomodulin binding to exosite I may open the active site fully as shown in the thrombin–PAR3 structure and produce the large change in the rate of diffusion of protein C into the active site (49) in addition to enhanced kcat (50). The conformational change documented in the structure of the thrombin mutant D102N bound to a fragment of PAR1 at exosite I differs markedly from that uncovered for PAR3 binding, which opens the question as to the true extent of conformational perturbation induced by binding to exosite I. Consideration of the distinct conformational states that thrombin assumes when free and bound to Na+ rationalizes these structural findings.
A recent kinetic analysis of the mechanism of Na+ binding to thrombin has revealed the existence of three major states of the enzyme (19, 51). The Na+-bound form E:Na+ is the high-activity conformation corresponding to the procoagulant fast form (18, 43), and its structural signatures are well established (21, 29). The Na+-free form is partitioned into two conformations, E and E*, of which only E can interact with Na+ and corresponds to the low-activity, anticoagulant slow form (18, 43). The structure of E reveals several differences with that of E:Na+ (21), but recent mutagenesis and spectroscopic studies suggest that the E→E:Na+ transition is more global because it affects the environment of all nine Trp residues of thrombin located up to 35-Å away from the bound Na+ (12, 19, 23). Similarly, the E*→E transition affects the structure of the enzyme as a whole (12, 19). If E* is indeed an inactive conformation of thrombin (47, 52), then the recent structure of the mutant D102N in the free form offers a possible representation (42). Given the current structural assignments of E*, E, and E:Na+, one can conclude that the structural changes in the 60-loop documented upon PAR3 binding to exosite I of murine thrombin (32) pertain to a conformation of the enzyme that is stabilized in the highly active E:Na+ form (53, 54). In fact, murine thrombin is constitutively stabilized in the E:Na+ form by the D222K replacement in the Na+ site, where Lys-222 provides molecular mimicry of the bound Na+ (54). However, the larger structural effects caused by binding of PAR1 to exosite I in the D102N mutant of human thrombin pertain to the E* conformation of the enzyme that is severely compromised for both substrate and Na+ binding. The consequences of exosite I ligation therefore would differ for the various conformations, E*, E, and E:Na+, accessible to thrombin under conditions relevant to physiological function (19). Binding to exosite I in the E:Na+ form would cause a shift in the 60-loop to open the active site fully. Binding to exosite I in the E* form would cause a larger conformational transition with a long-range communication that can be traced from exosite I to the opposite side of the molecule almost 30-Å away. As a result of this drastic conformational change induced allosterically by binding to exosite I, access to the active site is fully restored and the self-inhibited E* conformation of D102N is converted into a catalytically competent form similar to E or E:Na+. Therefore, it is not surprising that previous structures of thrombin crystallized in the highly active E:Na+ form have failed to reveal large-scale conformational changes upon binding of ligands to exosite I (31, 41), as documented here for the E* form.
Recent studies have stressed the importance of allostery as an intrinsic property of all dynamic proteins (10), encompassing numerous examples of monomeric proteins such as thrombin. Proteins that exist in multiple states in dynamic equilibrium tend to show large conformational transitions linked to ligand binding or substrate catalysis. In the classical example of hemoglobin allostery, the initial shift in the F8 His near the heme upon O2 binding triggers a cascade of structural changes that alter the interaction within and between the α and β chains leading to the T to R transition (4). Similarly, large-scale allosteric changes are observed in multimeric proteins like aspartate transcarbamylase (7), the nicotinic receptor (5), or GroEL (9). Evidence of long-range communication in smaller monomeric proteins is more difficult to obtain (10), but notable successes have been documented recently (8, 11) especially by NMR (55). The results reported here add an important example of how allosteric communication is channeled within a protein in the absence of quaternary structure by using the shift in a preexisting equilibrium between two distinct conformations.
Another relevant aspect of the results reported in this study is the possibility of exploiting allostery for rational drug design. One embodiment of this exciting strategy is to screen the protein surface for potential allosteric sites (56). The equilibrium between inactive and active forms could be used to switch the enzyme on and off upon binding of suitable molecules working as activators or inhibitors. The thrombin mutant W215A/E217A features a remarkable anticoagulant and antithrombotic profile in vitro (57) and in vivo (58, 59) by expressing catalytic activity only in the presence of the cofactor thrombomodulin. The structure of W215A/E217A in the free form shows a collapse of the 215–219 β-strand (60) similar to that seen in the D102N mutant (42), indicating that it may be stabilized in the inactive E* form. Upon binding of thrombomodulin to exosite I, the W215A/E217A mutant acquires full catalytic activity toward protein C (57). Most likely, the W215A/E217A mutant bound to thrombomdulin experiences a E*→E conformational transition similar to that seen for the D102N mutant upon binding of PAR1. Hence, the structural basis of the E*→E transition reported here document how thrombin can be engineered as an allosteric switch to turn on optimal anticoagulant activity in vivo upon interaction with an effector molecule.
Materials and Methods
The human thrombin mutant D102N was constructed, expressed, and purified to homogeneity to enable cocrystallization with relevant substrates (42). The isosteric D102N mutation effectively inactivates the enzyme at pH <8.0 and avoids drastic changes in the polarity of the active site such as those observed with the S195A mutation. A soluble fragment of human PAR1, 42SFLLRNPNDKYEPFWEDEEKN62, corresponding to the sequence downstream from the cleavage site at Arg-41 and containing the hirudin-like motif 52YEPFWE57 predicted to bind to exosite I (33, 38), was synthesized by solid phase, purified to homogeneity by HPLC, and tested for purity by mass spectrometry.
Crystals of human thrombin D102N in complex with human PAR1 were obtained by using the hanging drop vapor-diffusion method. D102N and PAR1 were mixed in 1:11 molar ratio at 4°C for 2 h before crystallization. A solution of D102N (5 mg/ml in 2 μl) in 50 mM choline chloride and 20 mM Mes (pH 6.0) was mixed with an equal volume reservoir solution containing 30% PEG 4000 and 100 mM Mes (pH 6.5) and left to equilibrate at 12°C. Crystals were triclinic, space group P1, with unit cell parameters a = 46.1 Å, b = 50.3 Å, c = 85.1 Å, α = 76.9°, β = 84.3°, and γ = 73.7°, and contained two molecules in the asymmetric unit. A structure of D102N in the free form also was obtained at 1.55-Å resolution (PDB ID code 3BEI) and reproduced the self-inhibited conformation of the mutant reported previously at 1.87 Å (42). Crystals were cryoprotected in the solution containing 15% glycerol and mother liquor for 3 min and frozen in liquid nitrogen to 100 K. X-ray data were collected to 2.2-Å resolution on an ADSC Quantum-315 CCD detector at the Biocars Beamline 14-BM-C of the Advanced Photon Source, Argonne National Laboratories (Argonne, IL). Data processing, indexing, integration, and scaling were performed with the HKL2000 package (61). The structure was solved by molecular replacement with MOLREP from the CCP4 package (62) using the coordinates of the PPACK-inhibited form of human thrombin (PDB ID code 1SHH) (21) as a starting model. Refinement and electron density generation were performed with Crystallography and NMR System software package (63), and 5% of the reflections were randomly selected as a test set for cross-validation. Ramachandran plots were calculated with PROCHECK (64). Results of data collection, processing, and refinement are listed in Table 2. Coordinates of the structure of the human D102N–PAR1 complex have been deposited in the Protein Data Bank (PDB ID code 3BEF).
Table 2.
Crystallographic data for the human thrombin mutant D102N bound to the extracellular fragment of human PAR1 (PDB ID 3BEF)
| Data Collection | |
| Wavelength, Å | 0.9 |
| Space Group | P1 |
| Unit Cell Dimension, Å | a = 46.1, b = 50.3, c = 85.1, α = 76.9°, β = 84.3°, γ = 73.7° |
| Molecules/Asymmetric Unit | 2 |
| Resolution Range, Å | 40.0–2.2 |
| Observations | 81,492 |
| Unique Observations | 34,055 |
| Completeness, % | 94.0 (87.7) |
| Rsym, % | 6.1 (22.1) |
| I/σ(I) | 12.3 (3.1) |
| Refinement | |
| Resolution, Å | 40.0–2.2 |
| ∣F∣/σ(∣F∣) | >0 |
| Rcryst, Rfree | 0.207, 0.248 |
| Reflections (Working/Test) | 31,620/1,657 |
| Protein Atoms | 4,963 |
| Solvent Molecules | 229 |
| rmsd Bond Lengths,* Å | 0.006 |
| rmsd angles,* ° | 1.3 |
| rmsd ΔB, Å2 (mm/ms/ss)† | 3.12/3.58/4.64 |
| <B> protein, Å2 | 37.9 |
| <B> solvent, Å2 | 36.0 |
| Ramachandran Plot | |
| Most favored, % | 98.5 |
| Generously allowed, % | 1.5 |
| Disallowed, % | 0.0 |
*Root-mean-squared deviation (rmsd) from ideal bond lengths and angles and rmsd in B factors of bonded atoms.
†mm, main chain–main chain; ms, main chain–side chain; ss, side chain–side chain.
ACKNOWLEDGMENTS.
We are grateful to Leslie Bush-Pelc and Alaji Bah for their valuable contribution and suggestions. This work was supported in part by the National Institutes of Health Research Grants HL49413, HL58141, and HL73813 (to E.D.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BEF).
References
- 1.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 2.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 3.Wyman J, Gill SJ. Binding and Linkage. Mill Valley, CA: Univ Science Books; 1990. [Google Scholar]
- 4.Perutz MF. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 5.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2006;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 6.Di Cera E. A structural perspective on enzymes activated by monovalent cations. J Biol Chem. 2006;281:1305–1308. doi: 10.1074/jbc.R500023200. [DOI] [PubMed] [Google Scholar]
- 7.Kantrowicz ER, Lipscomb WM. Escherichia coli aspartate transcarbamylase: the molecular basis for a concerted allosteric transition. Trends Biochem Sci. 1990;15:53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- 8.Pellicena P, Kuriyan J. Protein–protein interactions in the allosteric regulation of protein kinases. Curr Opin Struct Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 10.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Prot Struct Funct Genet. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 11.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Di Cera E, Page MJ, Bah A, Bush-Pelc LA, Garvey LC. Thrombin allostery. Phys Chem Chem Phys. 2007;9:1292–1306. doi: 10.1039/b616819a. [DOI] [PubMed] [Google Scholar]
- 13.Yu EW, Koshland DE., Jr Propagating conformational changes over long (and short) distances in proteins. Proc Natl Acad Sci USA. 2001;98:9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Süel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 15.Ota N, Agard DA. Intramolecular signaling pathways revealed by modeling anisotropic thermal diffusion. J Mol Biol. 2007;351:345–354. doi: 10.1016/j.jmb.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 16.Bode W. Structure and interaction modes of thrombin. Blood Cells Mol Dis. 2006;36:122–130. doi: 10.1016/j.bcmd.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Davie EW, Kulman JD. An overview of the structure and function of thrombin. Semin Thromb Hemost. 2006;32(Suppl 1):3–15. doi: 10.1055/s-2006-939550. [DOI] [PubMed] [Google Scholar]
- 18.Dang OD, Vindigni A, Di Cera E. An allosteric switch controls the procoagulant and anticoagulant activities of thrombin. Proc Natl Acad Sci USA. 1995;92:5977–5981. doi: 10.1073/pnas.92.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bah A, Garvey LC, Ge J, Di Cera E. Rapid kinetics of Na+ binding to thrombin. J Biol Chem. 2006;281:40049–40056. doi: 10.1074/jbc.M608600200. [DOI] [PubMed] [Google Scholar]
- 20.Di Cera E, et al. The Na+ binding site of thrombin. J Biol Chem. 1995;270:22089–22092. doi: 10.1074/jbc.270.38.22089. [DOI] [PubMed] [Google Scholar]
- 21.Pineda AO, et al. Molecular dissection of Na+ binding to thrombin. J Biol Chem. 2004;279:31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 22.Dang QD, Guinto ER, Di Cera E. Rational engineering of activity and specificity in a serine protease. Nat Biotechnol. 1997;15:146–149. doi: 10.1038/nbt0297-146. [DOI] [PubMed] [Google Scholar]
- 23.Mengwasser KE, Bush LA, Shih P, Cantwell AM, Di Cera E. Hirudin binding reveals key determinants of thrombin allostery. J Biol Chem. 2005;280:23997–27003. doi: 10.1074/jbc.M502678200. [DOI] [PubMed] [Google Scholar]
- 24.Liu LW, Vu TK, Esmon CT, Coughlin SR. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991;266:16977–16980. [PubMed] [Google Scholar]
- 25.Rezaie AR, He X, Esmon CT. Thrombomodulin increases the rate of thrombin inhibition by BPTI. Biochemistry. 1998;37:693–699. doi: 10.1021/bi971271y. [DOI] [PubMed] [Google Scholar]
- 26.Rezaie AR, Yang L. Thrombomodulin allosterically modulates the activity of the anticoagulant thrombin. Proc Natl Acad Sci USA. 2003;100:12051–12056. doi: 10.1073/pnas.2135346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vindigni A, White CE, Komives EA, Di Cera E. Energetics of thrombin-thrombomodulin interaction. Biochemistry. 1997;36:6674–6681. doi: 10.1021/bi962766a. [DOI] [PubMed] [Google Scholar]
- 28.Ayala Y, Di Cera E. Molecular recognition by thrombin. Role of the slow→fast transition, site-specific ion binding energetics and thermodynamic mapping of structural components. J Mol Biol. 1994;235:733–746. doi: 10.1006/jmbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- 29.Bode W, Turk D, Karshikov A. The refined 1.9-A X-ray crystal structure of d-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: Structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuentes-Prior P, et al. Structural basis for the anticoagulant activity of the thrombin-thrombomodulin complex. Nature. 2000;404:518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 31.Vijayalakshmi J, Padmanabhan KP, Mann KG, Tulinsky A. The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3:2254–2271. doi: 10.1002/pro.5560031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bah A, Chen Z, Bush-Pelc LA, Mathews FS, Di Cera E. Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4. Proc Natl Acad Sci USA. 2007;104:11603–11608. doi: 10.1073/pnas.0704409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 34.Coughlin SR. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost. 2005;3:1800–1814. doi: 10.1111/j.1538-7836.2005.01377.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi-Matsui M, et al. PAR3 is a cofactor for PAR4 activation by thrombin. Nature. 2000;404:609–613. doi: 10.1038/35007085. [DOI] [PubMed] [Google Scholar]
- 36.Esmon CT. Thrombomodulin as a model of molecular mechanisms that modulate protease specificity and function at the vessel surface. FASEB J. 1995;9:946–955. doi: 10.1096/fasebj.9.10.7615164. [DOI] [PubMed] [Google Scholar]
- 37.Esmon CT. The protein C pathway. Chest. 2003;124:26S–32S. doi: 10.1378/chest.124.3_suppl.26s. [DOI] [PubMed] [Google Scholar]
- 38.Vu TK, Wheaton VI, Hung DT, Charo I, Coughlin SR. Domains specifying thrombin-receptor interaction. Nature. 1991;353:674–677. doi: 10.1038/353674a0. [DOI] [PubMed] [Google Scholar]
- 39.Seeley S, Covic L, Jacques SL, Sudmeier J, Baleja JD, Kuliopulos A. Structural basis for thrombin activation of a protease-activated receptor: Inhibition of intramolecular liganding. Chem Biol. 2003;10:1033–1041. doi: 10.1016/j.chembiol.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Ayala YM, Cantwell AM, Rose T, Bush LA, Arosio D, Di Cera E. Molecular mapping of thrombin-receptor interactions. Proteins. 2001;45:107–116. doi: 10.1002/prot.1130. [DOI] [PubMed] [Google Scholar]
- 41.Mathews II, et al. Crystallographic structures of thrombin complexed with thrombin receptor peptides: Existence of expected and novel binding modes. Biochemistry. 1994;33:3266–3279. doi: 10.1021/bi00177a018. [DOI] [PubMed] [Google Scholar]
- 42.Pineda AO, Chen ZW, Bah A, Garvey LC, Mathews FS, Di Cera E. Crystal structure of thrombin in a self-inhibited conformation. J Biol Chem. 2006;281:32922–32928. doi: 10.1074/jbc.M605530200. [DOI] [PubMed] [Google Scholar]
- 43.Wells CM, Di Cera E. Thrombin is a Na(+)-activated enzyme. Biochemistry. 1992;31:11721–11730. doi: 10.1021/bi00162a008. [DOI] [PubMed] [Google Scholar]
- 44.Di Cera E. Thrombin interactions. Chest. 2003;124:11S–17S. doi: 10.1378/chest.124.3_suppl.11s. [DOI] [PubMed] [Google Scholar]
- 45.Esmon CT, Mather T. Switching serine protease specificity. Nat Struct Biol. 1998;5:933–937. doi: 10.1038/2906. [DOI] [PubMed] [Google Scholar]
- 46.Kroh HK, Tans G, Nicolaes GAF, Rosing J, Bock PE. Expression of allosteric linkage between the sodium ion binding site and exosite I of thrombin during prothrombin activation. J Biol Chem. 2007;282:16095–16104. doi: 10.1074/jbc.M610577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai MT, Di Cera E, Shafer JA. Kinetic pathway for the slow to fast transition of thrombin: Evidence of linked ligand binding at structurally distinct domains. J Biol Chem. 1997;272:30275–30282. doi: 10.1074/jbc.272.48.30275. [DOI] [PubMed] [Google Scholar]
- 48.Parry MA, Stone SR, Hofsteenge J, Jackman MP. Evidence for common structural changes in thrombin induced by active-site or exosite binding. Biochem J. 1993;290(Pt 3):665–670. doi: 10.1042/bj2900665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H, Bush LA, Pineda AO, Caccia S, Di Cera E. Thrombomodulin changes the molecular surface of interaction and the rate of complex formation between thrombin and protein C. J Biol Chem. 2005;280:7956–7961. doi: 10.1074/jbc.M412869200. [DOI] [PubMed] [Google Scholar]
- 50.Esmon NL, DeBault LE, Esmon CT. Proteolytic formation and properties of gamma-carboxyglutamic acid-domainless protein C. J Biol Chem. 1983;258:5548–5553. [PubMed] [Google Scholar]
- 51.Gianni S, Ivarsson Y, Bah A, Bush-Pelc LA, Di Cera E. Mechanism of Na+ binding to thrombin resolved by ultra-rapid kinetics. Biophys Chem. 2007;131:111–114. doi: 10.1016/j.bpc.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrell CJ, Bush LA, Mathews FS, Di Cera E. High resolution crystal structures of free thrombin in the presence of K+ reveal the basis of monovalent cation selectivity and an inactive slow form. Biophys Chem. 2006;121:177–184. doi: 10.1016/j.bpc.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Bush LA, Nelson RW, Di Cera E. Murine thrombin lacks Na+ activation but retains high catalytic activity. J Biol Chem. 2006;281:7183–7188. doi: 10.1074/jbc.M512082200. [DOI] [PubMed] [Google Scholar]
- 54.Marino F, Chen ZW, Ergenekan C, Bush-Pelc LA, Mathews FS, Di Cera E. Structural basis of Na+ activation mimicry in murine thrombin. J Biol Chem. 2007;282:16355–16361. doi: 10.1074/jbc.M701323200. [DOI] [PubMed] [Google Scholar]
- 55.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 56.Hardy JA, Wells JA. Searching for new allosteric sites in enzymes. Curr Opin Struct Biol. 2004;14:706–715. doi: 10.1016/j.sbi.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 57.Cantwell AM, Di Cera E. Rational design of a potent anticoagulant thrombin. J Biol Chem. 2000;275:39827–39830. doi: 10.1074/jbc.C000751200. [DOI] [PubMed] [Google Scholar]
- 58.Gruber A, Cantwell AM, Di Cera E, Hanson SR. The thrombin mutant W215A/E217A shows safe and potent anticoagulant and antithrombotic effects in vivo. J Biol Chem. 2002;277:27581–27584. doi: 10.1074/jbc.C200237200. [DOI] [PubMed] [Google Scholar]
- 59.Gruber A, et al. Relative antithrombotic and antihemostatic effects of protein C activator versus low molecular weight heparin in primates. Blood. 2007;109:3733–3740. doi: 10.1182/blood-2006-07-035147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pineda AO, et al. The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket. J Biol Chem. 2004;279:39824–39828. doi: 10.1074/jbc.M407272200. [DOI] [PubMed] [Google Scholar]
- 61.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected by oscillation methods. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 62.Bailey S. The CCP4 suite. Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 63.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 64.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]