Abstract
We present results of combined studies of the enzyme human aldose reductase (h-AR, 36 kDa) using single-crystal x-ray data (0.66 Å, 100K; 0.80 Å, 15K; 1.75 Å, 293K), neutron Laue data (2.2 Å, 293K), and quantum mechanical modeling. These complementary techniques unveil the internal organization and mobility of the hydrogen bond network that defines the properties of the catalytic engine, explaining how this promiscuous enzyme overcomes the simultaneous requirements of efficiency and promiscuity offering a general mechanistic view for this class of enzymes.
Keywords: enzymatic mechanism, helium cooling, subatomic resolution crystallography, x-ray plus neutrons joint refinement
Enzymatic catalysis frequently involves proton translocation (PT). PT can occur on very short time scales (nanoseconds) and over long distances (tens of Angstroms), often involving multiple proton relay sites and proton wires (Grotthus-like mechanisms) (1). The nature of PT processes combined with the weak x-ray scattering signal from hydrogen atoms, which excludes their observation at resolutions <1.2 Å, make the structural characterization of PT processes very difficult. Recent advances in synchrotron and neutron sources, detectors, cryocooling, and software, coupled with the ability to obtain high-quality crystals, have led to an improved level of detail in protein structures, as exemplified in our recent structure of human aldose reductase (h-AR) determined at subatomic resolution (2).
h-AR (Enzyme Commission 1.1.1.21) is a NADPH-dependent enzyme that reduces a wide range of substrates, such as aldehydes, aldoses, and corticosteroids. Because it reduces d-glucose into d-sorbitol, it is believed to cause severe degenerative complications of diabetes (3). The catalytic reaction involves a hydride transfer from carbon C4 of NADPH, which becomes NADP+, and a proton donation from the enzyme (4). To visualize h-AR interactions with the NADPH cofactor, substrates, and inhibitors, and to determine the details of the catalytic mechanism, several x-ray analysis (5–7), site-directed mutagenesis (8–11), and modeling studies (12) have been performed. The extraordinary quality of h-AR crystals allows structural studies at a level of detail not available before for an enzyme of this size.
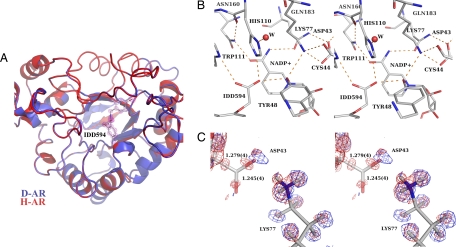
h-AR folds as a (β/α)8 triosephosphate isomerase barrel with the nicotinamide ring of the NADP+ buried at the bottom of a deep cleft (5) (Fig. 1A). The catalytic carbon C4 of the nicotinamide ring of NADP+ is accessible through this cleft, thus defining the catalytic zone of the active site. Residues Asp-43, Tyr-48, Lys-77, His-110, Asn-160, and Gln-183, highly conserved in all enzymes of the aldo-ketoreductase superfamily, form a tight network of hydrogen bonds linked to the nicotinamide ring (Fig. 1B).
Fig. 1.
X-ray results. (A) Ribbon drawing of hydrogenated (S1) (red) and deuterated (S2) (blue) structures (rmsd = 0.7 Å), superposed with x-ray 2Fo−Fc density map (S2) structure, 1.5 rms magenta contours for the inhibitor IDD594. (B) Active-site conformation showing the residues around NADP+ and inhibitor IDD594 for the deuterated structure (S2). (C) Closeup of Lys-77 and Asp-43 in deuterated h-AR superposed with the two difference maps [deuterated (S2), 2 rms blue contours; and hydrogenated (S1), 2 rms red contours]. These maps show the hydrogen atom in the H-bond Asp-43–Lys-77 in two alternative conformations. This shared partial protonation is indicated also by the bond lengths (corresponding to structure S2) in the carboxylate of Asp-43. The H-atom is placed on the Lys-77 side.
Various aspects of the h-AR catalytic mechanism have been studied, and several models have been proposed, with the identity of the residue donating the proton proving controversial. Based on previous low-resolution results of site-directed mutagenesis studies of key active-site residues (Tyr-48, His-110, and Lys-77), Bohren et al. (11) concluded that Tyr-48 is the most likely proton donor, and that His-110 plays a role in orienting the substrate. They also suggested that Lys-77 plays an important role in catalysis. After several papers with alternative propositions (9–14), the Tyr-48-mediated pathway, for which the proton is transferred from Tyr-48 (which has a lowered pKa), while His-110 is singly protonated at Nε2, gained significance (14). This hypothesis was confirmed by the subatomic-resolution (0.66-Å) structure of the complex of h-AR with the brominated inhibitor IDD594 (Fig. 1A) (2), which revealed most of the hydrogen atoms for the well ordered catalytic region, including the single protonation state of His-110. However, although this h-AR structure provided details rarely observed before (e.g., a direct Br-O electrostatic contact and unexpected h-AR main chain conformations), information was still required for a full explanation of the catalytic mechanism about the protonation state of other key catalytic residues, such as Asp-43, Lys-77, and Tyr-48.
Results
Experimental Work.
To better visualize the active-site residue protonation states, we produced fully deuterated samples (15) and collected both x-ray and neutron diffraction data at room temperature. Neutron diffraction is particularly efficient at locating Deuterium (D) atoms, because the scattering length of D is similar to that of C, N, and O atoms. Three different structures of the complex of either the hydrogenated or fully deuterated h-AR with the cofactor NADP+ and the inhibitor IDD594 were analyzed in this work [see Table 1 and supporting information (SI) Text for data collection and refinement statistics], as follows: structure 1 (S1), the hydrogenated enzyme (x-ray, 0.66 Å, 100 K) (2); S2, the fully deuterated enzyme (x-ray, 0.80 Å, 15 K, this work); and S3, the fully deuterated enzyme (neutron, 2.2 Å and x-ray, 1.75 Å, both at 293K, this work). Determination of partially occupied H+/D atoms was done by a combination of information from electron density maps, neutron scattering maps, and bond length analysis.
Table 1.
Summary of data collection and refinement statistics for the three different structures analyzed in this work
| H-AR100K x-rays (S1) | Full D-AR15K x-rays (S2) | Full D-AR293K neutrons (S3) | Full D-AR293K x-rays (S3) | |
|---|---|---|---|---|
| Diffraction data | ||||
| Resolution range, Å | 20.0–0.66 | 50.0–0.8 | 50.0–2.2 | 50–1.75 |
| R(I)merge, % | 2.9 | 2.3 | 22.8 | 2.9 |
| Completeness, % | 89.1 | 99.8 | 73.5 | 98.4 |
| I/σ(I) (all) | 14.5 | 28.1 | 5.1 | 26.8 |
| Refinement | ||||
| Reflections (work/test) | 485,662/25,585 | 306,026/16,125 | 11,884/992 | 31,524/2,952 |
| Rcryst/Rfree with H (D), % | 9.36/10.32 | 9.6/11.3 | 25.65/29.08 | 12.88/16.53 |
| Rcryst/Rfree without H (D), % | 10.56/10.94 | 11.6/12.5 | 34.67/34.76 | 14.20/17.30 |
| PDB ID code | 1US0 | 2QXW | 2R24 | 2R24 |
Combined Structure Analysis at the Catalytic Region.
Analysis of the deuterated structure (S2) showed no overall significant difference with the structure (S1) (Fig. 1). The Cα rmsd was 0.7 Å overall and 0.15 Å in the well ordered active site, implying that the information from both structures can be combined. The electron-density maps for structure (S2) showed clear density for active site amino acid, cofactor, and inhibitor atoms (Fig. 1A) (see SI Text for statistics).
The active site of the 0.66-Å structure (S1) of hydrogenated h-AR (2) was analyzed for partially occupied hydrogen atoms of key catalytic residues. This revealed a significant difference in the C-O bond lengths of Asp-43 carboxylate [1.243(3) vs. 1.271(3) Å], indicating a partial protonation of the carboxylate O2 oxygen in its H-bond with Lys-77. However, this observation was not confirmed by the difference electron density maps, which showed a peak corresponding to a hydrogen atom at the Lys side but not at the Asp side (Fig. 1C).
Although the overall structures (S1 and S2) were seen to be similar, analysis of the active site of structure (S2) provided new fine detail. The Nζ atom of Lys-77 makes H-bonds with the O atom of Cys-44 (as proton donor) and the Oδ2 atom of Asp-43 (as proton acceptor) (Fig. 1B). The S2 structure shows also significant differences in the Asp-43 C-O bond lengths [1.245(4) vs. 1.279(4) Å, Fig. 1C, very similar to those observed for structure S1] and indicates a partial protonation of the carboxylate O2 oxygen in its H-bond with Lys-77.
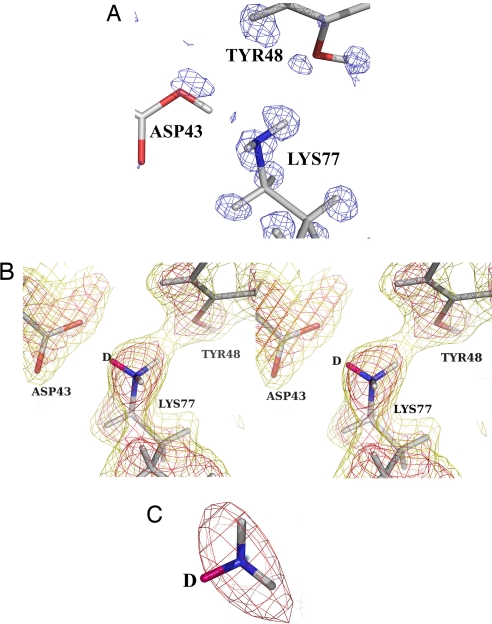
To locate the H-atom in this hydrogen bond, the difference maps for structures S1 and S2 were further compared (Fig. 1C). They suggest indeed that this H-atom is present in two conformations, because a peak is observed near Lys-77 in structure S1 and near Asp-43 in structure S2 (Fig. 2A), and in both structures, the C-O bond lengths indicate partial protonation of Asp-43. The neutron map confirms the neutral state of Lys-77 (Fig. 2 B and C). Furthermore, the room temperature x-ray 2Fo−Fc map shows a diminished density of Cε in Lys-77 (SI Text and SI Fig. 5), which indicates some mobility of the side chain in the neutral state.
Fig. 2.
Comparison of x-ray and neutron results. (A) X-ray model of fully deuterated h-AR-IDD594 complex [0.80-Å data collected at 15K, S2; treatment with HKL2000 (16); refinement with SHELX (17)] superposed with electron density difference map (Fo−Fc, 2 rms blue contours, phases calculated from model without deuteriums). The map suggests partial deuteration of the Asp-43. The model shows the neutral state for the Asp-43-Lys-77 pair. (B) Model from joint x-ray/neutron refinement of fully deuterated h-AR-IDD594 complex (S3) (both data collected at room temperature) superposed with neutron scattering-density map calculated with phases from the model with all deuteriums (2Fo−Fc, 2 rms red contours, 1 rms gold contours, S3). Note that the deuterium atom D (marked in magenta) was included in the model but is only weakly present in the map, confirming the partial protonation of the Asp-43-Lys-77 pair. (C) Closeup of B centered on the Lys-77 head. The map (2 rms red contours) shows strong density for only the two deuteriums marked in light gray.
These interpretations about both related motilities of the proton and the lysine residue were included in a new model of the catalytic mechanism, as described below.
Quantum mechanics/molecular mechanics (QM/MM) modeling of the catalytic mechanism was used to help explain and analyze the different protonation states of the Asp-43–Lys-77 pair, the geometry of the Asp-Lys-Tyr triad, and the proton-transfer energetics after the initial hydride transfer step. Hybrid QM/MM, using density functional theory (DFT) calculations for the bond-breaking and forming processes, provided a satisfactory but perhaps not unique explanation of the observed facts.
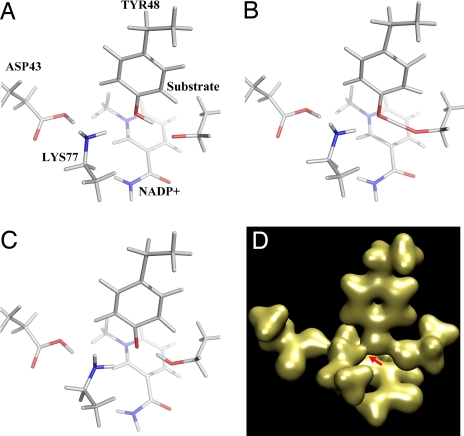
MM/MD calculations were in general agreement with the crystal structures. In the catalytic region, this agreement strongly depends on the charge state of the Asp-43–Lys-77 pair. The PT process itself was studied by using QM/MM simulations. Productive trajectories (with complete PT from Tyr to substrate or inhibitor head; see Fig. 3 and SI Movies 1 and 2) were obtained for the neutral/inhibitor and neutral/substrate models in all five cases simulated for each model. This analysis using the QM/MM neutral pair/substrate model results in the following distances: Tyr-48(O)/IDD594(O) = 3.0 Å; Tyr(O)/Lys(N) = 3.1 Å, and Lys(N)/Asp(O) = 3.2 Å, in excellent agreement with experiment. On the other hand, QM/MM simulations using a charged pair/substrate model results in the recoil of the Tyr head, hindering its ability to donate the proton to the substrate [Tyr-48(O)/IDD594(O) = 4.0 Å; Tyr(O)/Lys(N) = 3.2 Å, and Lys(N)/Asp(O) = 3.0 Å]. Thus, the charged form appears to be a kinetic trap that must be avoided. This is achieved through the destabilization of the charged Lys by the positive nicotinamide head, causing the back donation of the proton from Lys(+) to Asp.
Fig. 3.
QM/MD results. (A–C) Three steps of the QM/MD calculations before (A), during (B), and after (C) PT. The calculations suggest that the presence of the charged nicotinamide head has a marked influence in the behavior of the reaction centre region. The initial hydride transfer step provokes a driving force for PT, because of both the charge on the substrate and the effect on the Lys component of the salt bridge, which donates back a proton to Asp to reduce its positive charge, thus triggering the movement of Tyr and the consequent PT. Note the shortening of the distance between Lys 77 and Tyr-48 between frames A and B. (D) Map corresponding to PT step (B) computed using quantum chemical methods. The image shows the important residues in the pathway, especially Asp-43, Lys-77, and Tyr-48, as well as the substrate. Note the density (red arrow) around the oxygen atom of Tyr-48 extending over the Lys-77 nitrogen, in good agreement with the experiment.
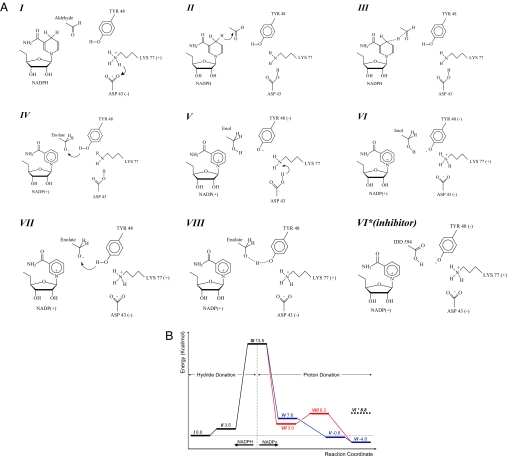
Before every productive PT event, there is a shortening of the Tyr-Lys distance (Fig. 3 SI Text, and SI Appendix). This event is largely reversible in the neutral/inhibitor model. The close values of the energies of the contributing protonation forms in the neutral/substrate model result in oscillatory events with the proton bouncing back and forth from the inhibitor carboxylate to the Tyr. This process seems induced by the presence of the Tyr− after the Tyr proton is transported to the inhibitor. The reaction pathway deduced from this analysis shows rapid proton transport after the hydride donation by the nicotinamide. As shown in Fig. 4A, the pathway follows steps I to VI in numerical order. The previously suggested pathway (14) is obtained by joining steps I, III, VII, VIII, and VI. The energy diagram (Fig. 4B) shows our calculations for all these states, including those corresponding to the previous pathway (VII and VIII), which are in good agreement with the published ones (14), as shown in the SI Fig. 7. Step IV shows a similar density distribution to the experimental one across the Lys-Tyr region (Fig. 3D). Transits IV to V and V to VI are barrierless (Fig. 4B, energy diagram, blue lines) in the free energy hypersurface, resulting from a concerted mechanism (18) (for further details, see SI Text and SI Appendix). Furthermore, step VI* (IDD594 model), showing a similar density along the Lys-Tyr pair than step IV, has the closest geometry to the experimental one from the modeled ones, with Tyr-48(O)/IDD594(O) = 3.2 Å [Experiment (Exp.): 2.92 Å]; Tyr(O)/Lys(N) = 3.4 Å (Exp.: 3.05 Å), and Lys(N)/Asp(O) = 3.2 Å (Exp.: 2.72 Å). By comparison, the distances in step VII, which is part of a previouslysuggested reaction pathway (14), were Tyr-48(O)/IDD594(O) = 4.6 Å; Tyr(O)/Lys(N) = 3.0 Å, and Lys(N)/Asp(O) = 3.1 Å.
Fig. 4.
Proposed reaction pathway. (A) I–VI show the proposed reaction pathway. VII and VIII show the proton donation for a “charged” model, in which both Lys-77 and Asp-43 are charged. VI* shows the complex with the inhibitor IDD 594. (B) Energies calculated for each reaction step. The proposed pathway I–VI is shown in black (before hydride donation) and blue (after hydride donation) lines. The “charged model” pathway is included (red lines) after hydride donation (VII–VIII–VI). Note that for the proposed model, the proton donation is barrierless (blue lines), whereas for the “charged model,” there is an energy barrier (red lines).
In summary, after the hydride donation step, the positive charge in the nicotinamide head stabilizes the neutral bridge between Asp-43 and Lys-77, allowing the relaxation of the Tyr geometry toward the substrate head and the subsequent proton donation. After this last step, Tyr-48 recoils to a locked geometry where the Asp-43–Lys-77 salt bridge is formed again and completes a tight four-charge sandwich involving the Asp(−), Lys(+), Tyr(−), and nicotinamide(+) head (step VI).
Discussion
A detailed structural analysis of a medium-size protein (36 kDa, h-AR in ternary complex with NADP+ and IDD594 inhibitor) is presented here. The x-ray (293K, 100K, and 15K) and neutron (293K) data describe the structure of h-AR corresponding to the state after hydride transfer but just before proton transfer. The experimental data were supplemented with quantum mechanics and molecular dynamics calculations for the entire reaction. These calculations are fully consistent with the state observed in the x-ray and neutron structures. The maps reveal details that cannot be extrapolated from single-method structural studies, including nonconventional protonation states of critical side chains and direct evidence of geometries and densities compatible with rapid proton relocation along an extended network of hydrogen bonds. Our data underscore the importance of Tyr-48 and substantiate this residue in proton transfer, as suggested (14). However, our data show also the role of the Asp-Lys pair operating on the Tyr donor as a crucial motif responsible for the catalytic properties of the enzyme. This proposition, which implies that Tyr-48 is negatively charged after the reaction (step VI), has been independently confirmed by microcalorimetric studies showing that a proton is accepted by Tyr-48 during ligand binding to the h-AR-NADP+ complex (corresponding to step VI) (19).
The ability of Tyr-48 to donate a proton may help enable adoption of multiple productive geometries and allow the processing of different substrates, a key feature of a promiscuous enzyme. Under this assumption, the catalytic center can be conceptually understood as having its catalytic zone placed one step away from the substrate, with Tyr-48 acting as a flexible final proton carrier. This mode of operation of the catalytic site is an extension of the interpretation of enzymes as transition-state (TS) binders, with the TS defined by the geometric arrangement of atoms of the enzyme and substrate in direct contact. The results presented here suggest this enzyme may overcome the difficulty of simultaneously satisfying the requirements of being an effective catalyst and a promiscuous one by using a distal proton donor (Asp-43–Lys-77 pair) acting through the flexible side chain of the final proton donor (Tyr-48), capable of accommodating different substrates.
Although this mechanistic suggestion should be further verified by other enzymology approaches, its application to other promiscuous enzymes is conceptually straightforward and might provide a cross-validation. The direct observation of the properties of the proton network, extending from Asp-43 to IDD594, was possible only by using state-of-the-art x-ray and neutron diffraction data. This work highlights the possibilities provided by the combination of these complementary techniques in structural biology.
Materials and Methods
Experimental.
The x-ray data (S2) were collected at ANL/APS/SBC sector 19ID at 15K. Refinement resulted in a model with 315 residues, NADP+, IDD594, two citrate molecules, and 555 water molecules. The neutron data (S3) were collected from a single crystal of volume 0.15 mm3 (unusually small for neutron diffraction) on the Laue diffractometer, LADI, at the Institut Laue Langevin (ILL) (15, 20, 21). The x-ray data (S3) were collected at Swiss Light Source [(SLS) Villigen, Switzerland], to 1.75-Å resolution and were used simultaneously with the neutron data in a joint refinement using PHENIX (22). The protonation states of amino acid residues were determined, and 301 water molecules, in which both deuterium and oxygen atoms were visualized, were assigned as full D2O molecules with the characteristic “boomerang” shape density (23). Resulting R factor/Rfree were 12.9%/16.6% for x-ray data and 25.6%/29.1% for neutron data. For comparison, the final R factor of a refinement with the neutron data only was 25.6% and Rfree 31.4%. The joint refinement decreased the neutron Rfree factors and improved significantly the neutron maps (see SI Text and SI Fig. 6). The main difference between the two cases was in the B factors, which reflected better the disordered regions in the joint refinement. On the other hand, the mean deviation of Cα atoms between the neutron-only and combined refinements was only 0.1 Å.
Modeling.
Molecular dynamics (MD) was performed by using both a conventional MM/MD methodology and a hybrid QM/MM approach, linking a quantum chemistry code to our own MD driver program Dynga (24). Details are given in SI Text and SI Appendix.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Clemens Schulze-Briesse and Takashi Tomizaki for help during data collection of the RT x-ray dataset at SLS. We thank the Institute of Diabetes Discovery for providing the inhibitor IDD594. This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, and the Hôpital Universitaire de Strasbourg. We acknowledge the use of the 19ID beamline at the Structural Biology Center/Advanced Photon Source supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. P.A. gratefully acknowledges the financial support of the National Institutes of Health-funded Macromolecular Neutron Crystallography Consortium [(NIGMS) Grant R01GM071939]. This work was supported in part by the U.S. Department of Energy under contract no. DE-AC02-05CH11231. This work benefited from the activities of the DLAB (Deuteration Laboratory) consortium funded by the European Union under contract HPRI-2001-50065 and from United Kingdom Engineering and Physical Sciences Research Council (EPSRC)-funded activity within the ILL-EMBL Deuteration Laboratory (DLAB) under Grant GR/R99393/01. This work has been funded in part by the National Cancer Institute–National Institutes of Health (contract no. NO1-CO-12400). This research was sponsored in part by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory (ORNL), managed by UT–Battelle, LLC, for the U.S. Department of Energy under Contract DE-AC05-00OR22725.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2QXW (RX 15K structure) and 2R24 (Neutron/RX 293K structure)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711659105/DC1.
References
- 1.Bernal JD, Fowler RH. A theory of water and ionic solution, with particular reference to hydrogen and hydroxyl ions. J Chem Phys. 1933;1:515–548. [Google Scholar]
- 2.Howard EI, et al. Ultra-high resolution drug design I: Human aldose reductase-inhibitor complex at 0.66 Å shows experimentally protonation states and atomic interactions which have implications for the inhibition mechanism. Proteins Struct Funct Genet. 2004;55:792–804. doi: 10.1002/prot.20015. [DOI] [PubMed] [Google Scholar]
- 3.Yabe-Nishimura C. Aldose reductase in glucose toxicity: A potential target for the prevention of diabetic complications. Pharmacol Rev. 1998;50:21–33. [PubMed] [Google Scholar]
- 4.Wermuth B. In: Enzymology of Carbonyl Metabolism 2: Aldehyde Dehydrogenase, Aldo-Keto Reductase, and Alcohol Dehydrogenase. Flynn TG, Weiner H, editors. New York: Liss; 1985. pp. 209–230. [Google Scholar]
- 5.Rondeau J-M, et al. Novel NADPH-binding domain revealed by the crystal structure of aldose reductase. Nature. 1992;355:469–472. doi: 10.1038/355469a0. [DOI] [PubMed] [Google Scholar]
- 6.Wilson DK, Bohren KM, Gabbay KH, Quiocho FA. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. Science. 1992;257:81–84. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- 7.Tete-Favier F, et al. Aldose reductase from pig lens. Eur J Med Chem. 1995;30:589s–603s. [Google Scholar]
- 8.Tarle I, Borhani DW, Wilson DK, Quiocho FA, Petrash JM. Probing the active site of human aldose reductase. Site-directed mutagenesis of Asp-43, Tyr-48, Lys-77, and His-110. J Biol Chem. 1993;268:25687–25693. [PubMed] [Google Scholar]
- 9.Schlegel BP, Jez JM, Penning TM. Mutagenesis of 3 alpha-hydroxysteroid dehydrogenase reveals a “push-pull” mechanism for proton transfer in aldo-keto reductases. Biochemistry. 1998;37:3538–3548. doi: 10.1021/bi9723055. [DOI] [PubMed] [Google Scholar]
- 10.Schlegel BP, Ratnam K, Penning TM. Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine. Biochemistry. 1998;37:11003–11011. doi: 10.1021/bi980475r. [DOI] [PubMed] [Google Scholar]
- 11.Bohren KM, et al. Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme. Biochemistry. 1994;33:2021–2032. doi: 10.1021/bi00174a007. [DOI] [PubMed] [Google Scholar]
- 12.Lee YS, Chen Z, Kador PF. Molecular modeling studies of the binding modes of aldose reductase inhibitors at the active site of human aldose reductase. Bioorg Med Chem. 1998;6:1811–1819. doi: 10.1016/s0968-0896(98)00139-4. [DOI] [PubMed] [Google Scholar]
- 13.Varnai P, Richards W, Lyne PD. Modelling the catalytic reaction in human aldose reductase. Proteins. 1999;37:218–227. doi: 10.1002/(sici)1097-0134(19991101)37:2<218::aid-prot7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 14.Varnai P, Warshel A. Computer Simulation Studies of the Catalytic Mechanism of Human Aldose Reductase. J Am Chem Soc. 2000;122:3849–3860. [Google Scholar]
- 15.Hazemann I, et al. High-resolution neutron protein crystallography with radically small crystal volumes; application of perdeuteration to human Aldose Reductase. Acta Crystallogr D. 2005;61:1413–1417. doi: 10.1107/S0907444905024285. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW, Sweet RM, editors. Vol 276. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 17.Sheldrick GM, Schneider TR. In: Methods in Enzymology. Carter CW, Sweet RM, editors. Vol 277. New York: Academic; 1997. pp. 319–343. [Google Scholar]
- 18.Kohanoff JJ, Cachau RE. Multiple proton translocation in biomolecular systems: Concerted to stepwise transition in a simple model. Mol Phys. 2004;102:1007–1014. [Google Scholar]
- 19.Steuber H. Marburg, Germany: Marburg University; 2007. PhD thesis. [Google Scholar]
- 20.Cipriani F, Castagna JC, Wilkinson C, Lehmann MS, Buldt G. A neutron image plate quasi-Laue diffractometer for protein crystallography. Basic Life Sci. 1996;64:423. doi: 10.1007/978-1-4615-5847-7_36. [DOI] [PubMed] [Google Scholar]
- 21.Myles DAA, et al. Neutron Laue diffraction in macromolecular crystallography. Phys B. 1998;1122:241–243. [Google Scholar]
- 22.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 23.Niimura NC, Kurihara T, Maeda KM. Hydrogen and hydration in proteins. Cell Biochem Biophys. 2004;40:351–369. doi: 10.1385/CBB:40:3:351. [DOI] [PubMed] [Google Scholar]
- 24.Parker CL, Ventura ON, Burt SK, Cachau RE. DYNGA: A general purpose QM-MM-MD program. I. Application to water. Mol Phys. 2003;101:2659–2668. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.