Abstract
ssDNA-binding proteins are key components of the machinery that mediates replication, recombination, and repair. Prokaryotic ssDNA-binding proteins share a conserved DNA-binding fold and an acidic C-terminal tail. It has been proposed that in the absence of ssDNA, the C-terminal tail contacts the ssDNA-binding cleft, therefore predicting that the binding of ssDNA and the C-terminal tail is mutually exclusive. Using chemical cross-linking, competition studies, and NMR chemical-shift mapping, we demonstrate that: (i) the C-terminal peptide of the gene 2.5 protein cross-links to the core of the protein only in the absence of ssDNA, (ii) the cross-linked species fails to bind to ssDNA, and (iii) a C-terminal peptide and ssDNA bind to the same overall surface of the protein. We propose that the protection of the DNA-binding cleft by the electrostatic shield of the C-terminal tail observed in prokaryotic ssDNA-binding proteins, ribosomal proteins, and high-mobility group proteins is an evolutionarily conserved mechanism. This mechanism prevents random binding of charged molecules to the nucleic acid-binding pocket and coordinates nucleic acid–protein and protein–protein interactions.
Keywords: gene 2.5 protein, replication
Single-stranded DNA (ssDNA)-binding proteins are a key component of the machinery for replication, recombination, and DNA repair (1). Once assigned to such mundane roles as eliminating secondary structure in ssDNA and protecting DNA from cleavage by nucleases, they are now emerging as key components in coordinating reactions at replication forks (2, 3).
The structures of several prokaryotic ssDNA-binding proteins have been solved (4–9). Although these proteins do not have sequence homology, their structures share a common oligosaccharide/oligonucleotide-binding fold (OB-fold), suggesting that they have a similar mode of action. The OB-fold is comprised of antiparallel β-sheets forming a barrel with a well defined cleft. Structural and mutagenesis data have shown that ssDNA binds within their cleft via stacking and electrostatic interactions. Several invariant aromatic residues surrounded by positively charged amino acids are found in all ssDNA-binding proteins. Although the aromatic residues stack with the DNA bases, the positively charged amino acids contact the phosphate backbone (4–8, 10, 11).
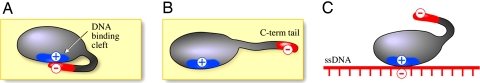
In addition to a common structure, all prokaryotic ssDNA-binding proteins have an acidic C-terminal tail. This C-terminal tail is essential for DNA replication, and phage and bacteria lacking the C-terminal tail fail to grow (11–15). In vitro, ssDNA-binding proteins lacking the C-terminal tail have a higher affinity for ssDNA (12, 15–21). The characterization of the nucleic acid-binding properties of gp32, the ssDNA-binding protein of phage T4 (17–20), led to the proposal of a model for the binding of gp32 to polynucleotides (Fig. 1). The model postulates that the negatively charged C-terminal tail binds electrostatically to the positively charged DNA-binding cleft and that it is displaced upon binding of DNA. Thus, proteins lacking the C-terminal tail have a vacant DNA-binding cleft lacking the electrostatic shield provided by the C-terminal tail, and ssDNA can bind directly to the cleft without having to compete with the C-terminal tail for occupancy of this site.
Fig. 1.
Model for the binding of ssDNA-binding protein to polynucleotides. Based on the studies of the binding of gp32 to various oligo- and polynucleotide substrates, Kowalczykowski et al. (17) proposed that the binding to polynucleotides is accompanied by a conformational change that results in “the pulling apart of a cluster of positively charged residues on the protein that have previously comprised tight anion binding site.” Thus, basic amino acid residues that have been shielded become available for nucleic acid binding. (A) Free protein. Negatively charged residues from C-terminal tail (in red) are contacting positively charged residues (in blue) via electrostatic interactions. (B) Displacement of the C-terminal tail results in the highly accessible DNA-binding cleft. (C) Bound state. The C-terminal tail is displaced, and previously shielded basic residues contact electrostatically the phosphate backbone (in red).
The finding that the C-terminal tail of prokaryotic ssDNA-binding proteins mediates multiple protein–protein interactions at the replication fork in Escherichia coli and in bacteriophage T7- and T4-infected cells further extended the model presented in Fig. 1 (2, 11, 22–24). It was proposed that the C-terminal tail functions as a two-way switch that coordinates and modulates the ssDNA binding and the protein–protein interactions. When the ssDNA-binding protein is bound to ssDNA, then the C-terminal tail is free to electrostatically interact with other proteins and recruit them at the site of the ssDNA and/or modulate their activity (5).
The gene 2.5 protein (gp2.5) is one of the four proteins that comprise the T7 replisome. The three other proteins are the T7 gene 5 DNA polymerase; its processivity factor, E. coli thioredoxin (trx); and the multifunctional gene 4 helicase-primase. The acidic C-terminal tail of gp2.5 is critical for the interactions of the protein with T7 DNA polymerase and helicase-primase as evidenced by their abolishment when the tail is deleted (11, 23). Recently, we have shown that the C-terminal tail of gp2.5 binds to a highly positively charged segment located in the thumb subdomain of the T7 gene 5 DNA polymerase (25). This fragment also is the site of binding of the processivity factor, E. coli trx, and the C terminus of gene 4 helicase-primase. Thus, the C-terminal tail of gp2.5 could potentially interfere with all interaction pairs at the T7 replication fork: directly by being part of the contact interface itself or indirectly by competing with another partner for the same contact surface (11, 23, 26, 27). The multiple interactions of the C terminus of gp2.5 could thus function as one mechanism to coordinate the multiple reactions occurring at the replication fork. Accordingly, gp2.5 is critical for establishing coordinated leading and lagging strand DNA synthesis (28–30).
Recently, we showed that, in addition to its acidic charge, a C-terminal aromatic residue on the tail also is essential for the function of gp2.5. gp2.5 lacking its C-terminal phenylalanine is defective in DNA replication and is unable to grow in E. coli (31). In vitro, gp2.5 is defective in its interaction with T7 DNA polymerase in vitro. Interestingly, the T7 helicase-primase also has an acidic C-terminal tail with a functionally important C-terminal aromatic residue (32). The common features of the C-terminal tails of both proteins provide additional evidence that they are part of a regulatory mechanism involving competition for binding to the same site.
gp2.5 is a dimer in solution. It has been proposed that the C-terminal tail of each monomer could bind in the ssDNA-binding cleft of the other subunit in trans, thus stabilizing the dimer interface, seen in the crystal structure of gp2.5 lacking the C-terminal tail (5). The displacement of the C-terminal tail upon ssDNA binding was in turn proposed to result in the destabilization of the dimer interface, which could lead to protein monomerization (33). Despite the ample evidence that the C terminus of gp2.5 is critical for its function, there is no information about the structure of the tail or its position with respect to the rest of the gp2.5 molecule. Unfortunately, no such information is available for any other acidic C-terminal tail of prokaryotic ssDNA-binding protein.
In the present study, we use gp2.5 as a model system for a prokaryotic ssDNA-binding protein to test the prediction that the acidic C-terminal tail and ssDNA compete for binding to the ssDNA-binding cleft of the molecule. By employing a combination of chemical cross-linking, competition studies, and NMR chemical shift mapping, we demonstrate that the C-terminal peptide of gp2.5 and ssDNA bind to the same overall surface of gp2.5 in a mutually exclusive manner.
Results
The C-Terminal Tail of gp2.5 Cross-Links to the Core of the Protein.
The C-terminal tail of gp2.5 contains multiple amino acids with carboxylic side chains, whereas its predicted site of binding, the DNA-binding cleft, contains multiple residues with amino groups. The 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC)/N-hydroxysulfosuccinimide (sulfo-NHS) cross-linking system (Pierce) activates free carboxyl groups for subsequent reaction with amino group side chains in amino acids (34). Therefore, we chose this cross-linking system because it utilizes the natural chemical composition of the regions of interest of gp2.5 and does not lead to the incorporation of additional components among the cross-linked products.
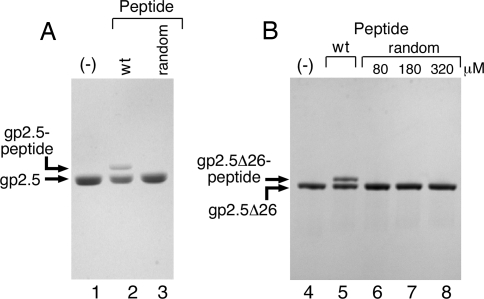
A peptide having a sequence identical to the C-terminal 26 amino acids of gp2.5 (ASKPRDEESWDEDDEESEEADEDGDF) readily cross-links to residues in wild-type gp2.5 (Fig. 2A, lane 2). In these experiments, the cross-linked species are identified by their decreased mobility during electrophoresis through SDS gels. A control peptide containing the same amino acids, but in random arrangement, does not cross-link to residues in gp2.5 (Fig. 2A, lane 3). The same cross-linking reaction was performed with gp2.5Δ26 lacking the C-terminal tail (5). Gp2.5Δ26 behaves similarly to wild type in the cross-linking reaction, producing a cross-linked product with decreased electrophoretic mobility (Fig. 2B). The products of the cross-linking reaction between gp2.5Δ26 and the C-terminal peptide migrate on SDS/PAGE very similarly, if not identically, to full-length gp2.5, demonstrating that only one peptide cross-links to each molecule of gp2.5Δ26. Taken together with the finding that the random peptide does not cross-link to gp2.5Δ26 even when it is in large molar excess (Fig. 2B, lanes 6–8), this observation strongly suggests that the cross-linked product most likely reflects the natural binding position of the acidic C-terminal tail.
Fig. 2.
EDC/sulfo-NHS cross-linking of the acidic C-terminal tail of gp2.5. (A) A peptide (80 μM) corresponding to the C-terminal 26 amino acids (residues 206–232) of gp2.5 was activated with EDC/sulfo-NHS. A control peptide (160 μM) containing a random sequence of the same residues also was activated with the same reagent. Each activated peptide was then incubated with 4 μM wild-type gp2.5 for 20 min at room temperature. The cross-linking reaction was stopped with SDS-loading buffer, and the products were resolved on 4–20% SDS/PAGE (Bio-Rad). (B) EDC/sulfo-NHS-activated wild-type (80 μM) or random (80, 160, and 320 μM) peptide was cross-linked to gp2.5Δ26 protein (4 μM) for 20 min at room temperature. The cross-linking reaction was stopped with SDS/PAGE-loading buffer, and the products were resolved on 4–20% SDS/PAGE (Bio-Rad).
The ability of the activated peptide to compete with the native C-terminal tail and cross-link argues that the C-terminal tail is only transiently and weakly bound to the core of the protein. We chose to perform all subsequent experiments by using gp2.5Δ26 to avoid any competition between the natural C-terminal tail and the C-terminal peptide supplied in trans.
ssDNA Abolishes the Cross-Linking of the C-Terminal Peptide to gp2.5.
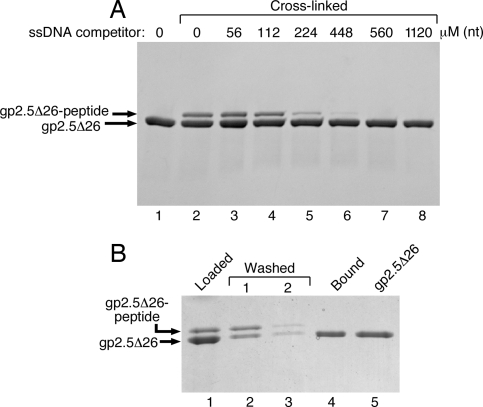
If indeed the C-terminal tail of gp2.5 binds in the ssDNA-binding cleft of the protein, ssDNA should compete with the C-terminal peptide for cross-linking to the protein. gp2.5Δ26 was preincubated with ssDNA (5.4-kb circular ssDNA from bacteriophage ΦX174) for 10 min before activated peptide was added to the reaction. The addition of increasing amounts of ssDNA resulted in abolishment of cross-linking (Fig. 3A). Cross-linking was abolished at 560 μM (nt) ssDNA (Fig. 3A, lane 7).
Fig. 3.
ssDNA-binding and peptide cross-linking are mutually exclusive. (A) Gp2.5Δ26 (4 μM) was preincubated with increasing amounts (56–1,120 μM in nucleotides) of ΦX174 circular ssDNA for 10 min on ice. EDC/sulfo-NHS-activated peptide (80 μM) was added, and the reaction was incubated at room temperature for 20 min. The cross-linking reactions were stopped with SDS/PAGE-loading buffer, and the products were resolved on 4–20% SDS/PAGE (Bio-Rad). (B) Wild-type C-terminal peptide (80 μM) was activated with EDC/sulfo-NHS and cross-linked to 4 μM gp2.5Δ26. One half of the cross-linking reaction was loaded on an ssDNA spin column, subjected to two low-salt buffer washes, and eluted with high-salt buffer as described in Materials and Methods. Half of the cross-linking reaction was loaded on gel as a reference for the species subjected to ssDNA cellulose binding (lane 1).
Cross-Linked Species Do Not Bind ssDNA.
The cross-linking experiments shown in Fig. 3A suggest that the C-terminal tail of gp2.5 binds in the ssDNA-binding cleft of the protein. Therefore, the cross-linked species in which the C-terminal peptide is presumably covalently bound in the DNA-binding cleft are expected to not be able to bind ssDNA. To test this hypothesis, we have examined the ability of gp2.5Δ26 cross-linked to peptide to bind to ssDNA by using an ssDNA cellulose column. The cross-linking reaction loaded on the column (Fig. 3B, lane 1) contains gp2.5Δ26 and gp2.5Δ26-peptide cross-linked complex. After two washes with 200 μl of low-salt buffer each, the bound proteins were eluted with 200 μl of high-salt buffer containing 1 M sodium chloride. The binding properties of the gp2.5Δ26 and gp2.5Δ26-peptide cross-linked complex were determined by SDS/PAGE analysis after concentration of the proteins by precipitation with trichloroacetic acid (TCA) (Fig. 3B). The species of gp2.5Δ26 cross-linked to peptide is present in the protein applied to the column (Fig. 3B, lane 1), but does not bind to the ssDNA, thus appearing predominantly in the first wash of the column (Fig. 3B, lane 2). No cross-linked species is retained by the column as judged by its absence in the high-salt eluate. In contrast, free gp2.5Δ26 protein was retained on the column and eluted with the high-salt buffer (Fig. 3B, lane 4). Control reactions with activated random peptide or no peptide demonstrated that the cross-linking reagents do not affect the ability of gp2.5Δ26 to bind to ssDNA (data not shown). Our results show that only uncross-linked protein is able to bind to ssDNA cellulose, whereas cross-linked species do not bind the column and appear in the low-salt wash. This finding further demonstrates that the binding of ssDNA and the C-terminal tail is mutually exclusive.
NMR Chemical Shift Mapping of the Binding Surfaces of the C-Terminal Peptide and ssDNA.
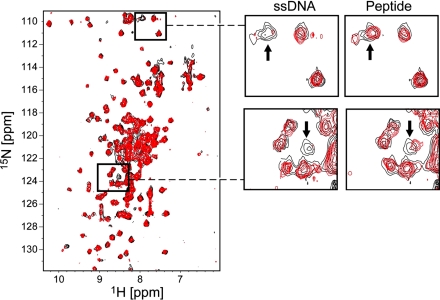
We aimed to test whether the C-terminal peptide of gp2.5 and ssDNA bind to the same surface of the protein by performing a series of NMR chemical shift perturbation experiments to monitor the interaction of gp2.5Δ26 with wild-type C-terminal peptide and ssDNA, respectively. In the 1H-15N heteronuclear single-quantum coherence (HSQC) NMR spectra (Fig. 4), each NH group in the 15N-labeled protein gives rise to a peak in the spectrum at the intersection between the chemical shifts of the respective 15N and 1H nuclei. The exact position of each peak (the chemical shifts of the 15N and 1H nuclei) is influenced by the environment around the NH group in the protein. Any change in this environment (e.g., binding of a ligand in the vicinity of the NH group) affects the peak position. The NMR chemical shift perturbation experiment exploits this phenomenon to determine whether a ligand binds to the labeled protein. If more than one ligand binds to the same protein, NMR chemical shift perturbation experiments can be used to determine whether the same, different, or partially overlapping sets of residues of the 15N-labeled protein are involved in the interactions with different ligands (for a recent review, see ref. 35).
Fig. 4.
ssDNA and C-terminal peptide bind to the same area of gp2.5 core module. (Left) 1H-15N TROSY-HSQC spectra of 15N-labeled gp2.5Δ26 in the presence (red) and the absence (black) of unlabeled 23-mer ssDNA oligonucleotide. (Right) Selected regions of the spectra from Left are shown side by side, with the corresponding regions of the 1H-15N TROSY-HSQC spectra of 15N-labeled gp2.5Δ26 in the presence (red) and the absence (black) of unlabeled 26-mer gp2.5 C-terminal peptide. The arrows mark peaks that are significantly affected by both ssDNA and peptide binding to 15N-labeled gp2.5Δ26.
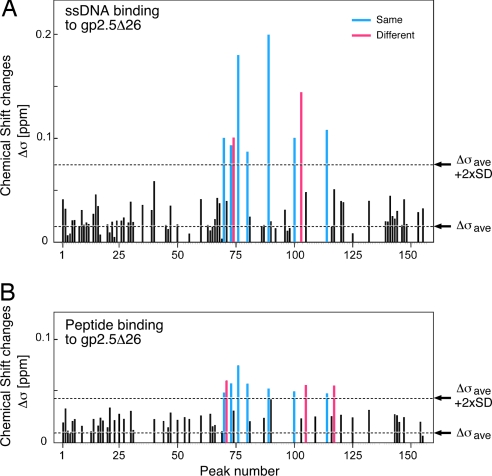
Binding of the C-terminal peptide was in fast exchange on the NMR time scale (peaks gradually moving upon addition of increasing amounts of peptide), indicative of a fast dissociation rate. The binding of the 23-mer ssDNA to gp2.5Δ26 appeared stronger than that of the peptide: in fast-to-intermediate exchange regime on the NMR time scale, with some peaks becoming broadened or completely disappeared (intermediate exchange), indicative of a slower dissociation rate. Upon adding increasing amounts of ssDNA, most of the spectrum of gp2.5Δ26 disappeared (data not shown) presumably due to the formation of high-molecular-weight complexes composed of multiple gp2.5Δ26 molecules bound to the same ssDNA molecule. Therefore, an early titration point with a substiochiometric amount of ssDNA, where the spectral quality was still good, was used for analysis. Examples of peaks with significant chemical shift changes are shown in Fig. 4. Statistical analysis of the observed chemical shifts showed that only a small subset of peaks (Fig. 5, depicted in blue and red) exhibit changes beyond the average background chemical shift change plus two standard deviations. Nine such peaks were observed in the titration with ssDNA (Fig. 5A), whereas 10 such peaks were observed in the titration with gp2.5 C-terminal peptide (Fig. 5B). Seven of those peaks are identical between the two titrations (depicted in blue), showing that the titration of ssDNA and gp2.5 C-terminal peptide affected the same residues from the core module of gp2.5 protein. Because backbone resonance assignments are not available for gp2.5Δ26, NMR could not be used to identify the specific residues of gp2.5Δ26 affected by the interactions. This finding directly demonstrates that ssDNA and the C-terminal tail bind in the same overall area of the surface of the protein. Furthermore, ssDNA binds to gp2.5Δ26 tighter than the C-terminal peptide does, as evidenced by the different magnitude of the chemical shift changes observed upon binding of both ligands (Fig. 5).
Fig. 5.
Chemical shift changes of gp2.5Δ26 spectra upon ligand titration. (A) Plot of chemical shift changes in the spectra of 15N-labeled gp2.5Δ26 upon titration with unlabeled 23-mer ssDNA oligonucleotide. (B) Plot of chemical shift changes in the spectra of 15N-labeled gp2.5Δ26 upon titration with unlabeled gp2.5 C-terminal peptide. Note that the numbers in the graphs are arbitrary peak numbers and do not correspond to the residue numbers in the protein because no backbone NMR resonance assignments are available for gp2.5. The combined chemical shift changes (Δσ) were calculated by using the equation: Δσ = ΔσH + ΔσN/5, where ΔσH and ΔσN are the chemical shift changes in the 1H and 15N dimensions, respectively. Residues in gp2.5Δ26 significantly affected by both ssDNA and peptide are in blue, and those residues significantly affected by only ssDNA or peptide are shown in red.
Discussion
Organisms from all domains of life and many prokaryotic and eukaryotic viruses encode ssDNA-binding proteins as part of the molecular machinery that maintains the integrity of the genome and ensures its replication. Despite the absence of significant sequence homology, all ssDNA-binding proteins share a common structural core, the OB-fold. In addition, prokaryotic ssDNA-binding proteins, mitochondrial ssDNA-binding proteins, and crenaceous archaeal ssDNA-binding proteins have a flexible acidic C-terminal tail (10).
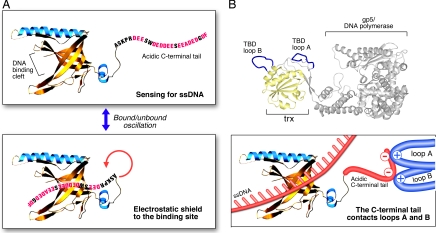
Earlier studies of the nucleic acid-binding properties of full-length and proteolytic fragments of gp32, the ssDNA-binding protein encoded by bacteriophage T4, led to the proposal that, in the absence of DNA, the acidic C-terminal tail binds in the DNA-binding cleft (17). Studies of E. coli and T7 and T4 replisomes revealed that the C-terminal tails of the cognate ssDNA-binding proteins mediate multiple protein–protein interactions (reviewed in refs. 2 and 3). Thus, the current working model (Fig. 6A) for the role of the acidic C-terminal tail is that, in the absence of DNA, it binds to the DNA-binding cleft. Upon DNA binding, the tail is displaced and available for protein–protein interactions. Although the model depicts the C-terminal tail of a gene 2.5 monomer binding within its own DNA-binding cleft, it also is possible for the C-terminal tail of one monomer to bind within the DNA-binding cleft of another monomer to form a dimer, as suggested previously (5, 33).
Fig. 6.
Model for the functional role of the acidic C-terminal tail. (A) The C-terminal tail functions as an electrostatic shield. In the absence of ssDNA, the C-terminal tail oscillates between the bound and unbound states, thus effectively protecting the positively charged DNA-binding cleft from binding to random negatively charged surfaces and sensing for the presence of ssDNA. For clarity, the figure represents only one monomer of a dimer of gp2.5 with C-terminal tail binding within its own DNA-binding cleft and does not take into account the possibility for the C-terminal tail of one monomer to bind in the ssDNA-binding cleft of another monomer. (B) Mechanism of the interaction of the C-terminal tail with the gp5/DNA polymerase. When the DNA-binding cleft is occupied (ssDNA is depicted in red), the negatively charged gp2.5 C-terminal tail (in red) interacts electrostatically with loop A (residues 275–285 of gp5, depicted in blue) and loop B (residues 299–314 of gp5, depicted in blue) of the TBD of the gp5/DNA polymerase. Loops A and B are easily accessible for protein–protein interactions as seen from the crystal structure of the complex of gp5/trx/primer template (omitted for clarity). The C-terminal phenylalanine of gp2.5, which is essential for the interaction with the polymerase, is not depicted because of lack of experimental data for its position.
One prediction from the described model is that the binding of ssDNA and the C-terminal tail to the DNA-binding cleft is mutually exclusive. Using cross-linking and affinity binding, we have demonstrated that the cross-linking (i.e., binding of the acidic C-terminal tail to the core of the gp2.5) prevents the protein from binding to ssDNA. Vice versa, the binding of ssDNA competes successfully (i.e., prevents the binding/cross-linking of a peptide), corresponding to the acidic C-terminal tail. Furthermore, NMR chemical shift mapping demonstrates that both ssDNA and C-terminal peptide contact gp2.5Δ26 in the same area, providing additional evidence that indeed the C-terminal tail binds into the DNA-binding cleft of the protein. The range of the changes in the NMR spectra observed upon titration of ssDNA and wild-type peptide demonstrates that ssDNA binds to gp2.5Δ26 with higher affinity than the C-terminal peptide, a result that is consistent with the C-terminal tail being displaced upon ssDNA binding. In summary, using gp2.5 of bacteriophage T7, we demonstrate that the binding of ssDNA and the acidic C-terminal tail to the core of gp2.5 is mutually exclusive as predicted.
The competition of the DNA and the C-terminal tail for the DNA-binding cleft represents an elegant mechanism that ensures efficient coverage of any ssDNA arising in the cell without posing a road block to the DNA processing machinery. All ssDNA-binding proteins studied up to date are abundant in the cell, thus ensuring the timely coverage of any ssDNA arising in the cell. However, the moderate affinity for ssDNA allows prompt removal of the protein upon the recruitment of the DNA processing machinery that would convert the ssDNA to dsDNA via replication, recombination, or repair. The relatively high cellular concentration of ssDNA-binding protein increases the probability that it will bind randomly to any negatively charged molecule or surface in the cell. Prokaryotic ssDNA-binding proteins avoid that possibility by using their acidic C-terminal tail as a protective electrostatic shield. We propose that the C-terminal tail oscillates between the bound and the unbound position, thus sensing the presence of ssDNA in the cell (Fig. 6A). The oscillation mechanism also can explain why the C-terminal tail can mediate the interaction of gp2.5 with T7 DNA polymerase and helicase-primase in the absence of DNA (11, 23). In the presence of ssDNA, the C-terminal tail is displaced from the DNA-binding cleft because of the higher affinity of the protein for ssDNA. It is possible that the binding of the tail is purely electrostatic, whereas the DNA binding employs electrostatic interactions with the backbone and stacking interactions with the bases, collectively resulting in a stronger interaction.
Further support for the electrostatic shield hypothesis comes from the finding that T4 gp32 can bind dsDNA and RNA, although with much lower affinity (i.e., when negatively charged molecules are available in sufficient concentration, they bind to the positively charged cleft) (16–18, 20, 36, 37). Similar to ssDNA, the removal of the acidic C-terminal tail results in higher affinities for these substrates. Deletion analysis of the C-terminal tail of gp2.5 demonstrated that the gradual removal of charged residues results in the gradual increase of the affinity for ssDNA (31). This observation provides further evidence that the C-terminal tail shields the ssDNA-binding cleft.
Interestingly, T4 gp32 binds specifically to a translational operator in its own mRNA and self-regulates its expression, thus allowing effective control of the abundance of the protein (38). No data are available about such control mechanism for any other ssDNA-binding protein. However, it is logical to expect that the abundance of these proteins should be regulated to ensure the availability of sufficient amounts of protein to cover any transiently arising ssDNA without posing a road block to the DNA processing machinery. However, the electrostatic shield of the acidic C-terminal tail provides a means of protection of the DNA-binding cleft from binding to random charged surfaces.
Similar to prokaryotes, eukaryotes also employ OB-folds as modules of their ssDNA-binding proteins, but their molecules lack an acidic tail. The best studied eukaryotic ssDNA-binding protein, human replication protein A, is composed of several subunits harboring one or more OB-fold modules (39). Most likely, the heteromeric complex of RPA has a sophisticated 3D structure that prevents binding of random charged molecules to the DNA-binding cleft(s) of the protein complexes. Also, the presence of multiple OB-folds within the same complex creates multiple possibilities for modulation of the DNA-binding activity.
The presence of phage and bacterial ssDNA-binding proteins together in the cell during infection is an interesting biological phenomenon. Both proteins bind DNA nonspecifically and are able to remove secondary structure in front of the DNA processing machinery, suggesting that they should be able to substitute for each other. However, at least in the case of the T7/E. coli system, the phage ssDNA-binding protein is essential for infection (40), demonstrating that the host ssDNA-binding protein cannot substitute for that encoded by the phage. Most likely the ability of the phage ssDNA-binding protein to participate in phage-specific protein–protein interactions confers its indispensable function. Essential ssDNA-binding proteins also are found in eukaryotic viruses (e.g., HSV) (41). A model for the mechanism of the interaction of the gp2.5 C-terminal tail with the T7 DNA polymerase is presented in Fig. 6B. The negatively charged C-terminal tail contacts positively charged loops A and B from the trx-binding domain (TBD) of gene 5/T7 DNA polymerase (42). As seen from the structure of the T7 DNA polymerase depicted, these loops are readily accessible for protein–protein interactions.
The combination of the OB-fold and the negatively charged tail also is found in some ribosomal proteins (e.g., S28E, a protein from the small ribosomal subunit) (43, 44) and translation factors (e.g., eIF1A) (45), which bind to RNA in a structure-dependent manner and are relatively abundant in the cell. Although studies on the effect of the acidic tails on the RNA-binding properties of these proteins are not available, it is likely that they use the tails as an electrostatic shield when the proteins are not bound to RNA.
High-mobility group (HMG) proteins are found in chromatin complexes and bind nonspecifically to dsDNA (46). These proteins use the strategy of an acidic electrostatic shield to protect their nucleic acid-binding site and to modulate the dsDNA-binding activity of the molecule. HMG proteins contain a conserved HMG-box domain that binds and bends dsDNA. DNA binding is facilitated by a positively charged N-terminal tail. It was shown that, in the absence of DNA, the negatively charged C-terminal tail of maize HMGB1 protein interacts with the positively charged N-terminal tail, thus protecting the DNA-binding site. When the N-terminal positively charged tail is removed, the acidic C-terminal tail contacts the HMG-box domain (47). Similarly to prokaryotic ssDNA-binding proteins, the removal of the acidic C-terminal tail increases the affinity for DNA. In contrast, phosphorylation of the C-terminal tail decreases the affinity for DNA and enhances the interaction with the positively charged N-terminal tail (47).
In conclusion, it appears that the protection of the positively charged DNA-binding cleft by the electrostatic shield of the negatively charged C-terminal tail is an evolutionary conserved mechanism. This mechanism is used by abundant cellular proteins to prevent the random binding of charged molecules to their nucleic acid-binding pocket, as well as a tool for the coordination of protein–nucleic acids and protein–protein interactions within the macromolecular machinery that ensures the integrity of the genome and its replication.
Materials and Methods
Peptides and DNA.
A peptide corresponding to the 26 C-terminal amino acids of T7 gp2.5 (ASKPRDEESWDEDDEESEEADEDGDF) and a scrambled peptide with the same amino acid composition (RDDEDEDDDSEDFKGAPSEASWEEE), were synthesized by Tufts University Core Facility (Boston, MA). Single-stranded ΦX174 DNA was purchased from New England Biolabs, and ssDNA 23-mer oligonucleotide was purchased from Integrated DNA Technologies.
EDC/Sulfo-NHS Cross-Linking.
EDC and sulfo-NHS were purchased from Pierce. EDC/sulfo-NHS is a zero-length cross-linking system that activates free carboxyl groups and cross-links them to free amino groups. A two-step cross-linking protocol was used as recommended by the manufacturer (34). The activation reaction contained 2 mM EDC, 5 mM sulfo-NHS, 50 mM potassium phosphate (pH 7), 150 mM NaCl, 0.1 mM EDTA, 10% glycerol, and 0.1 mM DTT and was performed at room temperature for 15 min. The excess cross-linking agents were quenched with 20 mM β-mercapto-ethanol (final concentration), and the acti-vated peptide was incubated with gp2.5 at room temperature for 20 min. The cross-linking reaction was terminated by the addition of SDS/PAGE-loading buffer and was immediately analyzed on 4–20% Tris polyacrylamide gel (Bio-Rad).
ssDNA Cellulose-Binding Assay.
ssDNA cellulose (Sigma–Aldrich) was equilibrated in 50 mM potassium phosphate (pH 7), 1 M NaCl, 0.1 mM EDTA, 0.1 mM DTT, and 10% glycerol according to the manufacturer's recommendation. Then 500 μl of 50% slurry was loaded on a spin column and spun for 30 s at 1,000 × g in a microcentrifuge. The column was washed three times with 750 μl of low-salt buffer [50 mM potassium phosphate (pH 7), 0.1 mM EDTA, 0.1 mM DTT, and 10% glycerol]. The cross-linking reaction was loaded on the DNA cellulose and incubated on ice for 30 min. The column was washed two times with 0.2 ml of low-salt buffer and eluted with 0.2 ml of the same buffer containing 1 M NaCl. The proteins in the washes and the eluate were precipitated with ice-cold TCA 10% (wt/vol) final concentration. The precipitates were washed with 70% ethanol, air dried, and dissolved in SDS sample-loading buffer. The samples were analyzed on 4–20% SDS gels (Bio-Rad).
Expression of Isotope-Labeled Proteins for NMR.
The bacterial cultures were grown on minimal medium as described previously (48). 15NH4Cl (Cambridge Isotope Laboratories) was used as the only nitrogen source to achieve uniform 15N-isotope labeling.
NMR Titrations.
15N-labeled gp2.5Δ26 [100–200 μM in a buffer containing 20 mM Tris·HCl (pH 7.0) and 300 mM NaCl] was titrated with increasing concentrations of unlabeled species: 23-mer ssDNA or 26-mer gp2.5 C-terminal peptide. 1H-15N Transverse Relaxation Optimized Spectroscopy-HSQC (TROSY-HSQC) spectra were collected at each point. Experiments were performed at 37°C on a Varian INOVA 500 spectrometer. Higher protein concentration, lower salt concentration, or lower temperature than the ones used caused severe line broadening, which was indicative of the formation of species with high molecular weight. Even at the experimental conditions selected, gp2.5Δ26 appeared to form species larger than 25-kDa monomer. Therefore, 1H-15N TROSY-HSQC and long experiment times had to be used. To reduce losses from relaxation, tNH (the time delay for transfer of magnetization between the 15N and 1H nuclei) was shortened to 2.1 ms. Spectra were processed in nmrPipe (49) and analyzed in CARA. The combined chemical shift changes (Δσ) were calculated by adding the chemical shift changes in the 1H (ΔσH) and 15N (ΔσN) dimensions, after dividing ΔσN by 5, to compensate for its greater magnitude compared with ΔσH. Analysis of titration data were done as described in ref. 35.
ACKNOWLEDGMENTS.
We thank all members of C.C.R.'s laboratory for helpful discussions and Steven Moskowitz for help with figure preparation. This work was supported by U.S. Public Health Services Grants GM54397 (to C.C.R.), GM47467, and CA68268 (to G.W.); National Cancer Institute Howard Temin K01 CA119107 Award (to A.M.); and National Institutes of Health Postdoctoral Fellowship F32GM72305 (to B.M.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 2.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 3.O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 4.Savvides SN, et al. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–1947. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc Natl Acad Sci USA. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 7.Saikrishnan K, et al. Structure of mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: Biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr D. 2005;61:1140–1148. doi: 10.1107/S0907444905016896. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein DA, et al. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc Natl Acad Sci USA. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr ID, et al. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. EMBO J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YT, Richardson CC. Acidic carboxyl-terminal domain of gene 2.5 protein of bacteriophage T7 is essential for protein-protein interactions. J Biol Chem. 1994;269:5270–5278. [PubMed] [Google Scholar]
- 12.Burke RL, Alberts BM, Hosoda J. Proteolytic removal of the COOH terminus of the T4 gene 32 helix-destabilizing protein alters the T4 in vitro replication complex. J Biol Chem. 1980;255:11484–11493. [PubMed] [Google Scholar]
- 13.Hosoda J, Moise H. Purification and physicochemical properties of limited proteolysis products of T4 helix destabilizing protein (gene 32 protein). J Biol Chem. 1978;253:7547–7558. [PubMed] [Google Scholar]
- 14.Williams KR, Konigsberg W. Structural changes in the T4 gene 32 protein induced by DNA polynucleotides. J Biol Chem. 1978;253:2463–2470. [PubMed] [Google Scholar]
- 15.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 16.Pant K, Karpel RL, Rouzina I, Williams MC. Mechanical measurement of single-molecule binding rates: Kinetics of DNA helix-destabilization by T4 gene 32 protein. J Mol Biol. 2004;336:851–870. doi: 10.1016/j.jmb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids: I. Characterization of the binding interactions. J Mol Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 18.Newport JW, Lonberg N, Kowalczykowski SC, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids: II. Specificity of binding to DNA, RNA. J Mol Biol. 1981;145:105–121. doi: 10.1016/0022-2836(81)90336-3. [DOI] [PubMed] [Google Scholar]
- 19.Lonberg N, Kowalczykowski SC, Paul LS, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids: III. Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III). J Mol Biol. 1981;145:123–138. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 20.Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, von Hippel PH. On the thermodynamics and kinetics of the cooperative binding of bacteriophage T4-coded gene 32 (helix destabilizing) protein to nucleic acid lattices. Biophys J. 1980;32:403–418. doi: 10.1016/S0006-3495(80)84964-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyland EM, Rezende LF, Richardson CC. The DNA binding domain of the gene 2.5 single-stranded DNA-binding protein of bacteriophage T7. J Biol Chem. 2003;278:7247–7256. doi: 10.1074/jbc.M210605200. [DOI] [PubMed] [Google Scholar]
- 22.Krassa KB, Green LS, Gold L. Protein-protein interactions with the acidic COOH terminus of the single-stranded DNA-binding protein of the bacteriophage T4. Proc Natl Acad Sci USA. 1991;88:4010–4014. doi: 10.1073/pnas.88.9.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YT, Tabor S, Churchich JE, Richardson CC. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J Biol Chem. 1992;267:15032–15040. [PubMed] [Google Scholar]
- 24.Kong D, Richardson CC. Role of the acidic carboxyl-terminal domain of the single-stranded DNA-binding protein of bacteriophage T7 in specific protein-protein interactions. J Biol Chem. 1998;273:6556–6564. doi: 10.1074/jbc.273.11.6556. [DOI] [PubMed] [Google Scholar]
- 25.Hamdan SM, et al. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci USA. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notarnicola SM, Mulcahy HL, Lee J, Richardson CC. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J Biol Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 27.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J Biol Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Chastain PD, II, Griffith JD, Richardson CC. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage t7 replication proteins. J Mol Biol. 2002;316:19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Chastain PD, II, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 30.Nakai H, Richardson CC. Leading and lagging strand synthesis at the replication fork of bacteriophage T7. Distinct properties of T7 gene 4 protein as a helicase and primase. J Biol Chem. 1988;263:9818–9830. [PubMed] [Google Scholar]
- 31.Marintcheva B, Hamdan SM, Lee SJ, Richardson CC. Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single-stranded DNA-binding protein. J Biol Chem. 2006;281:25831–25840. doi: 10.1074/jbc.M604601200. [DOI] [PubMed] [Google Scholar]
- 32.Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J Biol Chem. 2006;281:25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- 33.Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J Biol Chem. 2006;281:38689–38696. doi: 10.1074/jbc.M608460200. [DOI] [PubMed] [Google Scholar]
- 34.Grabarek Z, Gergely J. Zero-length crosslinking procedure with the use of active esters. Anal Biochem. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 35.Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- 36.Pant K, Karpel RL, Rouzina I, Williams MC. Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: Single molecule force spectroscopy measurements. J Mol Biol. 2005;349:317–330. doi: 10.1016/j.jmb.2005.03.065. [DOI] [PubMed] [Google Scholar]
- 37.Pant K, Karpel RL, Williams MC. Kinetic regulation of single DNA molecule denaturation by T4 gene 32 protein structural domains. J Mol Biol. 2003;327:571–578. doi: 10.1016/s0022-2836(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 38.Borjac-Natour JM, Petrov VM, Karam JD. Divergence of the mRNA targets for the Ssb proteins of bacteriophages T4 and RB69. Virol J. 2004;1:4. doi: 10.1186/1743-422X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bochkarev A, Bochkareva E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Kim YT, Richardson CC. Bacteriophage T7 gene 2.5 protein: An essential protein for DNA replication. Proc Natl Acad Sci USA. 1993;90:10173–10177. doi: 10.1073/pnas.90.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Challberg MD. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamdan SM, et al. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 43.Wu B, et al. Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum. Protein Sci. 2003;12:2831–2837. doi: 10.1110/ps.03358203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aramini JM, et al. Solution NMR structure of the 30S ribosomal protein S28E from Pyrococcus horikoshii. Protein Sci. 2003;12:2823–2830. doi: 10.1110/ps.03359003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 46.Thomas JO, Travers AA. HMG1 and 2, and related “architectural” DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 47.Thomsen MS, Franssen L, Launholt D, Fojan P, Grasser KD. Interactions of the basic N-terminal and the acidic C-terminal domains of the maize chromosomal HMGB1 protein. Biochemistry. 2004;43:8029–8037. doi: 10.1021/bi0499009. [DOI] [PubMed] [Google Scholar]
- 48.Weber DJ, et al. NMR docking of a substrate into the X-ray structure of staphylococcal nuclease. Proteins. 1992;13:275–287. doi: 10.1002/prot.340130402. [DOI] [PubMed] [Google Scholar]
- 49.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]