Abstract
Kinetic partitioning is predicted to be a general mechanism for proteins to fold into their well defined native three-dimensional structure from unfolded states following multiple folding pathways. However, experimental evidence supporting this mechanism is still limited. By using single-molecule atomic force microscopy, here we report experimental evidence supporting the kinetic partitioning mechanism for mechanical unfolding of T4 lysozyme, a small protein composed of two subdomains. We observed that on stretching from its N and C termini, T4 lysozyme unfolds by multiple distinct unfolding pathways: the majority of T4 lysozymes unfold in an all-or-none fashion by overcoming a dominant unfolding kinetic barrier; and a small fraction of T4 lysozymes unfold in three-state fashion involving unfolding intermediate states. The three-state unfolding pathways do not follow well defined routes, instead they display variability and diversity in individual unfolding pathways. The unfolding intermediate states are local energy minima along the mechanical unfolding pathways and are likely to result from the residual structures present in the two subdomains after crossing the main unfolding barrier. These results provide direct evidence for the kinetic partitioning of the mechanical unfolding pathways of T4 lysozyme, and the complex unfolding behaviors reflect the stochastic nature of kinetic barrier rupture in mechanical unfolding processes. Our results demonstrate that single-molecule atomic force microscopy is an ideal tool to investigate the folding/unfolding dynamics of complex multimodule proteins that are otherwise difficult to study using traditional methods.
Keywords: energy landscape, force spectroscopy, multiple pathways, single-molecule studies, unfolding intermediate
Protein folding and unfolding are fundamental processes inside the cell. From an unfolded and presumably random coil-like state, most proteins must fold into the well defined three-dimensional structures, which are unique to each protein, to be biologically functional. The folding and unfolding processes are complex and may involve multiple pathways (1, 2), which are believed to be governed by the general kinetic partitioning mechanism (3, 4), although experimental proof supporting this mechanism is still limited (5–8). Small, single-domain proteins are often used as model systems to investigate folding/unfolding dynamics. However, certain proteins are complex such that they are composed of smaller modules that can behave quite independently, thus presenting more complex folding/unfolding dynamics. The coupling between modules plays important roles in defining the overall conformational dynamics of these proteins (9–11). T4 lysozyme is an excellent model system in this aspect. T4 lysozyme is 164 residues long and composed of 10 α-helices and four short β-strands (12) (Fig. 1A). It has been widely studied for >30 years and the availability of high-resolution structures of hundreds of T4 lysozyme mutants makes it especially appealing (13). Although traditional ensemble studies showed that T4 lysozyme unfolds in an apparent two-state fashion (14, 15), it has been well recognized that two subdomains exist in T4 lysozyme: an α/β N-terminal lobe and an all α C-terminal lobe. The unique feature of T4 lysozyme is that the N-terminal helix A forms part of the C-terminal lobe, resulting in coupling between the two subdomains. The two subdomains have distinct thermodynamic stability giving rise to the possibility for T4 lysozyme to unfold from at least two regions (16). Recent fragment studies confirmed the subdomain architecture of T4 lysozyme (17–19). Here, we use single-molecule atomic force microscopy (AFM) techniques to investigate the unfolding dynamics and pathways of T4 lysozyme.
Fig. 1.
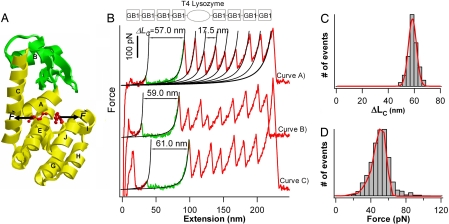
Majority of T4 lysozyme molecules unfold in an apparent all-or-none fashion. (A) Tertiary structure of T4L* (PDB ID code 1L63). The N-terminal lobe is green, and the C-terminal lobe and the helix A are yellow. Residues 1 and 164, from which T4 lysozyme is being pulled to unfold, are shown as ball and sticks. Arrows indicate the force acting on T4L*. (B Upper) Schematic illustration of the polyprotein chimera (GB1)4-T4L*-(GB1)4 used in single-molecule AFM experiments. (B Lower) Typical force-extension curves of polyprotein (GB1)4-T4L*-(GB1)4. The mechanical unfolding events of the well characterized GB1 domains (red) occurred at ≈180 pN with ΔLC of ≈18 nm and serve as fingerprints to discern signatures of the mechanical unfolding of T4L*. The unfolding of T4L* always precedes the unfolding of GB1 domains and is characterized by unfolding forces of ≈50 pN and ΔLC of ≈60 nm, which corresponds to the complete unfolding of T4L* (green). The mechanical unfolding of the majority of T4L* molecules occurs in an apparent two-state fashion. Black lines correspond to WLC fits to the experimental data. (C) Histogram of ΔLC for T4L* molecules that unfold in two-state fashion. Red line is the Gaussian fit to the experimental data with average ΔLC of 59.0 ± 4.0 nm (n = 1,269). (D) Unfolding force histogram for T4L* molecules that unfold in two-state fashion (pulling speed: 400 nm/s, n = 1,269). Red line is the Monte Carlo fit, using an α0 of 0.055 s−1 and a Δxu of 0.75 nm.
Single-molecule AFM exploits the stretching force as a “denaturant” to destabilize the protein and force proteins to undergo force-induced unfolding reaction along the reaction coordinate defined by the stretching force, providing a glimpse of the particular cross-section of the energy landscape underlying protein unfolding (20–22). Because of its superb ability to trigger and monitor the mechanical unfolding of individual protein molecules, single-molecule AFM has evolved into a powerful tool to investigate the folding and unfolding dynamics of proteins at the single-molecule level (21, 23–25). Most of the single-molecule AFM studies carried out to date focused on small, single-domain proteins (23, 24, 26, 27) and many of them were considered as two-state folders in traditional ensemble studies. Despite the relative simplicity of these model systems, single-molecule AFM revealed information about the conformational dynamics of proteins, such as stable unfolding intermediate states and distinct alternative unfolding pathways, which are invisible in ensemble studies (28–33). The unique topology makes T4 lysozyme an ideal model system to use single-molecule AFM to investigate its mechanical unfolding behaviors. By using a solid-state polymerized T4 lysozyme polyprotein, the mechanical unfolding of T4 lysozyme was investigated in detail by stretching from its two residues 21 and 124 (34). Here, we use single-molecule AFM to stretch a cysteine-free pseudo wild-type T4 lysozyme (T4L*) and its circular permutant PERM1 (35) from their N and C termini to investigate their mechanical unfolding dynamics and the role of subdomain coupling in their mechanical unfolding. Our results showed that, on pulling from its N and C termini, the mechanical unfolding of T4L* displays kinetic partitioning and proceeds by multiple parallel unfolding pathways: the majority of T4L* unfold in an apparent two-state fashion, whereas ≈13% of T4L* unfold in three-state fashion involving partially unfolded intermediate states. In addition, the three-state unfolding pathways show great diversity in the individual unfolding trajectory. We demonstrate that the interaction of the N-terminal helix A, which forms part of the C-terminal lobe, with the rest of C-terminal lobe is critical for the mechanical stability of T4L*. The unfolding intermediate states are local energy minima in the free-energy landscape and are likely to associate with the residual structures present in the two subdomains. Such complex kinetic behaviors reflect the stochastic nature of kinetic barrier rupture in mechanical unfolding processes, and provide direct evidence for the kinetic partitioning of the mechanical unfolding pathways of T4 lysozyme.
Results
T4 Lysozyme Exhibits Distinct Multiple Unfolding Pathways.
To characterize the mechanical unfolding of T4L* by using single-molecule AFM, we engineered (GB1)4-T4L*-(GB1)4 polyprotein chimera, in which T4L* was flanked by (GB1)4 at both ends (Fig. 1B Upper). In the polyprotein chimera, the well characterized GB1 domains serve as fingerprints for identifying single-molecule stretching events and discerning the signatures of the mechanical unfolding of T4L*. The mechanical unfolding of GB1 is characterized by contour-length increments ΔLC of ≈18 nm and unfolding force of ≈180 pN (26). Stretching polyprotein chimera (GB1)4-T4L*-(GB1)4 results in force-extension curves with a characteristic saw-tooth pattern appearance corresponding to the mechanical unfolding of (GB1)4-T4L*-(GB1)4 (Fig. 1B). Fig. 1B, curves A and B, presents two examples that show the stretching and unfolding of a full-length polyprotein chimera. In these two curves, we observed eight unfolding events occurring at ≈180 pN with ΔLC of ≈18 nm as measured by fitting the worm-like-chain (WLC) model of polymer elasticity (36) to consecutive unfolding force peaks. These eight unfolding events can be easily identified as the mechanical unfolding of the eight GB1 domains that flank T4L* in the chimera. Hence, the unfolding event that precedes the unfolding of GB1 domains and occurs at ≈50 pN must correspond to mechanical unfolding of T4L*. Because the polyprotein chimera were picked up randomly along the contour length of the molecule, the majority of force-extension curves correspond to the stretching and unfolding of part of the polyprotein. In these curves, if we observed five or more unfolding events of GB1 in a force-extension curve, we are certain that the unfolding event before the unfolding events of GB1 domains must correspond to the stretching and unraveling of T4L* (Fig. 1B, curve C). It is evident that, to obtain these force-extension curves with at least five GB1 unfolding events, the polyprotein must have been picked up by attaching the AFM tip to one of the GB1 domains, and there should be no direct interaction between the AFM tip and T4L*, avoiding any potential modification of the native state of T4L* by the AFM tip [supporting information (SI) Fig. 5]. Fitting the WLC model to the unfolding events of T4L* measures an average ΔLC of 59.0 ± 4.0 nm (avg ± SD, n = 1,269) for the mechanical unfolding of T4L* from its N and C termini (Fig. 1C).
T4L* comprises 164 aa residues and is 59 nm long on being unfolded and fully extended (164 aa × 0.36 nm/aa). The distance between the N and C termini in the folded state is ≈0.8 nm (PDB ID code 1L63). Hence, a complete mechanical unfolding of T4L* will result in a ΔLC of 58.2 nm (59.0–0.8 nm), which is in close agreement with the experimentally determined ΔLC, corroborating our conclusion that the unfolding events of ΔLC of ≈59 nm correspond to the mechanical unfolding of T4L*. The close match between the observed and predicted ΔLC indicates that the unfolding events of T4L* correspond to the complete unfolding of T4 lysozyme in an apparent all-or-none fashion within the force resolution of our AFM experiments (≈20 pN) (see SI Text).
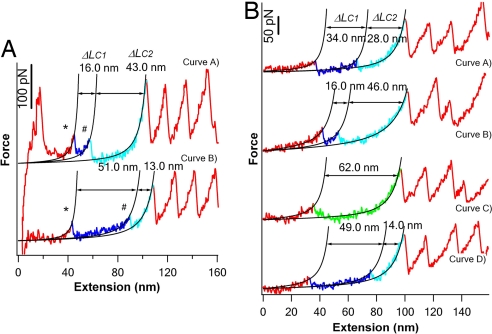
In ≈13% of unfolding events of T4L*, we observed that T4 lysozyme unfolded in a complex three-state fashion involving unfolding intermediate state: instead of the all-or-none unfolding process (as those shown in Fig. 1B), the mechanical unfolding of some T4L* occurred in two steps characterized by ΔLC1 and ΔLC2, respectively (Fig. 2A). Although different unfolding trajectories may show different patterns of ΔLC1 and ΔLC2, the sum of ΔLC1 and ΔLC2 gave a total ΔLC of ≈60 nm, which is consistent with the complete unfolding of one folded T4L*. It is evident that, after crossing the first kinetic barrier (corresponding to the first unfolding peaks, indicated by *), T4L* are kinetically “trapped” into unfolding intermediate states that are thermodynamically and mechanically stable, at least on the time scale of subseconds, and should correspond to local energy minima along the unfolding pathways of T4L*. The subsequent unfolding of such unfolding intermediate states results in the second unfolding force peaks as indicated by #. These results indicate that T4L* can unfold by multiple parallel pathways. For clarity, we will discuss the features of the two-state unfolding and the three-state unfolding pathways separately.
Fig. 2.
T4 lysozyme can unfold by multiple parallel three-state unfolding pathways. (A) Typical force-extension curves of T4L* showing three-state unfolding behaviors. The initial partial unfolding events, which correspond to the unfolding event from the native state to the unfolding intermediate state, are shown in blue, and the subsequent unfolding events, which correspond to the unfolding of the intermediate state to the fully unfolded state, are cyan. WLC fits (black lines) to the experimental data reveal distinct patterns of ΔLC1 and ΔLC2. (B) A series of force-extension curves of the same T4L* measured during repeated stretching–relaxation experiments. The same T4L* molecule exhibited distinct mechanical unfolding pathways, including the two-state pathway and multiple distinct three-state unfolding pathways. For all of the events showing three-state unfolding behaviors, the sum of ΔLC1 and ΔLC2 is close to ≈60 nm, which is in agreement with that expected from the complete unfolding of T4L*.
Two-State Unfolding of T4 Lysozyme Is Characterized by a Long Unfolding Distance to the Transition State.
T4L* that unfold in a two-state fashion show a narrow distribution in their unfolding forces (Fig. 1D) with an average of 50 ± 13 pN (n = 1,269) at a pulling speed of 400 nm/s. The unfolding forces of T4L* show a weak dependency on pulling speeds (SI Fig. 6): the average unfolding force of T4L* increases to 59 pN at a pulling speed of 1,000 nm/s, which is similar to that when T4L* is unraveled from its residues 21 and 124 (34) and in good agreement with theoretical prediction (37). By using a standard Monte Carlo procedure (38) and assuming that the location of the transition state does not move with different pulling speeds, we fit the unfolding-force histogram and the pulling-speed dependence of the unfolding forces to estimate the unfolding rate constant at zero force α0 and the unfolding distance Δxu between the folded state and the transition state along the reaction coordinate. We found that the experimental data can be reproduced well by using a α0 of 0.055 s−1 and a Δxu of 0.75 nm. This result suggests that the mechanical resistance to unfolding is distributed over a distance of 0.75 nm, in contrast to the smaller Δxu and highly localized mechanical resistance observed for elastomeric proteins, such as I27 (38) and ubiquitin (31). The measured Δxu is similar to that reported by Yang et al. (34) for T4 lysozyme being unraveled from residues 21 and 124.
The observed ΔLC and Δxu for the two-state unfolding of T4L* suggest that the mechanical unfolding barrier lies close to the N- and C-termini. To extend T4L*, helix A, which is at the N terminus of the entire sequence but part of C-terminal lobe, will have to be detached first from the remainder of C-terminal lobe. Hence, it is likely that the mechanical unfolding energy barrier for the two-state unfolding corresponds to breaking the interactions connecting helix A to the reminder of the C-terminal lobe. After crossing the main unfolding barrier, the rest of T4L* (N-terminal lobe and the rest C-terminal lobe) unfolds readily. However, to obtain detailed information about the molecular events during the unfolding process and pinpoint the exact location of the energy barrier, molecular dynamics simulations will be needed.
Three-State Unfolding of T4 Lysozyme Shows Diversity in Unfolding Pathways.
In contrast to the majority of T4L* that unfold in a well defined all-or-none fashion, ≈13% of T4L* unfold in three-state fashion involving an unfolding intermediate state. Such a three-state unfolding scheme does not follow a well defined pathway, instead it showed diversity and variability in their unfolding pathways. Fig. 2A shows two force-extension curves in which T4L* unfolds in two steps but with significantly different ΔLC1. For example, T4L* in the top curve unfolds in two steps: the first step is a partial unfolding event resulting in ΔLC1 of 16 nm, and the second step corresponds to the subsequent unraveling of the remainder of T4 lysozyme resulting in ΔLC2 of 43 nm. The sum of ΔLC1 and ΔLC2 gave a total ΔLC of ≈60 nm, which corresponds to the ΔLC resulting from the complete unfolding of T4L*. In contrast, the three-state unfolding of T4L* in the bottom curve results in ΔLC1 of 51 nm and ΔLC2 of 13 nm. ΔLC is an intrinsic structural parameter that provides information about the location of kinetic barriers. The distinct patterns of ΔLC1 and ΔLC2 suggest the existence of distinct unfolding pathways and the different location of the kinetic barriers for unfolding.
The diversity in unfolding pathways for T4L* was also observed in the same T4L* molecule during repeated stretching–relaxation cycles. A few representative force-extension curves resulted from such an experiment are shown in Fig. 2B (relaxation curves are not shown), where the T4L* molecule exhibits both three-state and two-state unfolding behaviors. The choice of a given unfolding pathway is not deterministic, reflecting the stochastic nature of kinetic barrier crossing (39). For the events showing three-state unfolding behavior, all of the T4L* molecules show a combined ΔLC of ≈60 nm, whereas ΔLC1 and ΔLC2 show different patterns. This observation indicates that the different unfolding pathways, both two-state unfolding and the diverse three-state unfolding schemes, are intrinsic properties of T4 lysozyme defined by its underlying energy landscape along the reaction coordinate defined by the stretching force, and therefore the same T4L* molecule can sample distinct unfolding pathways.
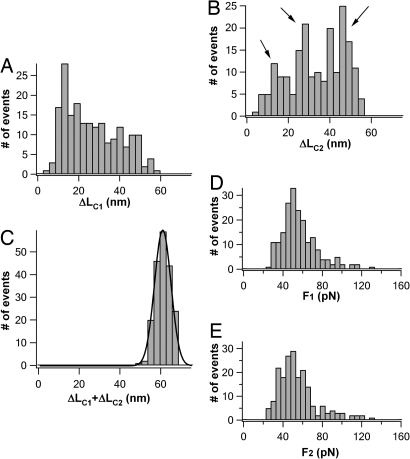
The diversity of the three-state unfolding pathways is further illustrated by the histogram of ΔLC1 and ΔLC2 compiled from 196 T4L* molecules that unfold in three-state fashion (Fig. 3 A and B). For comparison, the corresponding histogram for ΔLC1+ΔLC2 is also shown (Fig. 3C). It is clear that ΔLC1 and ΔLC2 show broad distributions that are clearly beyond the experimental error of the ΔLC measurements in our experiments. Therefore, the broadness in the distribution of ΔLC1 and ΔLC2 reflects the intrinsic diversity of the three-state unfolding schemes manifested by T4L* molecules. From limited three-state unfolding events, it seems that there exist three preferred pathways with ΔLC2 being ≈14 nm, ≈29 nm, and ≈46 nm (as indicated by arrows), respectively, suggesting that the second kinetic energy barrier for the three-state unfolding is located ≈45 nm, 30 nm, and 13 nm away from its resting length between N and C termini. However, because of the uncertainties of using the WLC model to fit the low unfolding force events of T4L*, our resolution in ΔLC measurements is rather limited (as indicated by the relatively large bin size of 3 nm in Fig. 3 A–C). Such a limited resolution made it difficult to accurately determine the distribution of unfolding pathways.
Fig. 3.
Histogram of ΔLC1 (A) and ΔLC2 (B) in three-state unfolding trajectories. For comparison, the histogram of ΔLC1+ΔLC2, which was measured from the same unfolding trajectories of T4L*, is shown in C. It is evident that ΔLC1 and ΔLC2 show broad distribution, indicating broad distribution of unfolding pathways for T4L*. Solid line in C is the Gaussian fit to the experimental data with an average ΔLC1+ΔLC2 of 61.0 ± 4.0 nm (n = 196). (D and E) Unfolding-force histograms of the first (D) and second (E) unfolding events of T4L* observed in three-state unfolding pathways.
Despite the diversity in the three-state unfolding pathways, the unfolding forces of the two unfolding force peaks are surprisingly similar to each other, and to that for two-state unfolding. The unfolding forces for the two unfolding force peaks show a narrow distribution with an average force of ≈50 pN (Fig. 3 D and E).
Possible Pathways for Three-State Unfolding.
T4L* is made of two subdomains and has been shown to exhibit various intermediate states along both folding and unfolding pathways in chemical denaturation studies (40–45). However, in the mechanical unfolding experiments, the unfolding of T4L* proceeds along a well defined reaction coordinate set by the stretching force, which is quite different from that in chemical folding/unfolding studies. Hence, it is unlikely that the mechanical unfolding intermediates observed here directly correspond to those observed in the chemical folding/unfolding studies. Indeed, our single-molecule AFM studies show that mechanical unfolding pathways exhibit features that are quite different from that for chemical unfolding pathway. In T4L*, the N-terminal helix A is part of the C-terminal lobe. Therefore, on stretching T4L* from its N and C termini, helix A will have to be detached first from the remainder of the C-terminal lobe to extend T4L*. Hence, regardless of the two-state or three-state unfolding pathways, disruption of the interaction between helix A and the remainder of the C-terminal lobe constitutes the first barrier for mechanical unfolding of T4L* and the necessary condition for the observation of the unfolding of the N-terminal lobe. In contrast, the unfolding of the N-terminal lobe occurs before the unfolding of the A helix in the chemical unfolding pathways of T4L* (43–45).
In the mechanical unfolding of T4L*, the observation of multiple three-state unfolding pathways indicates that, after crossing the first barrier, there are many potential contacts, being native or newly formed in the two subdomains, that can lead to local energy minima and potentially provide mechanical resistance to unfolding in a stochastic fashion. By using ΔLC as a probe, it is possible to estimate the location of the kinetic barrier during the three-state unfolding (32). For example, the unfolding pathway of ΔLC1 and ΔLC2 of 30/29 nm is consistent with the unfolding of helix A plus the N-terminal lobe followed by subsequent unraveling of the remaining C-terminal lobe. However, because of broad distribution of ΔLC1 and ΔLC2 and the degeneracy in ΔLC1/ΔLC2 due to the fact that different unfolding pathways may result in similar ΔLC1/ΔLC2, it remains difficult to accurately determine the location of kinetic barrier in three-state unfolding processes. Extended AFM studies with much improved resolution in ΔLC measurements, in combination with complementary structural characterization experiments, will be required to characterize the unfolding intermediate states in detail.
Interactions Between Helix A and Reminders of the C-Terminal Lobe Play a Critical Role in Determining the Mechanical Stability of T4 Lysozyme.
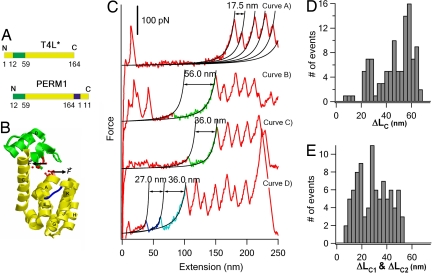
Our results indicated that the interaction between helix A with the C-terminal lobe may hold the key to the mechanical stability of T4L*. To further investigate the role of helix A on the mechanical unfolding of T4L*, we use single-molecule AFM to study the mechanical unfolding of a well characterized T4L* circular permutant PERM1. In PERM1, helix A is shuffled to the C terminus of the overall sequence (Fig. 4A), leading to the decoupling of the two subdomains (35). PERM1 was shown to have an almost identical structure (35) to T4L* (Fig. 4B).
Fig. 4.
Mechanical unfolding behaviors of circular permutant PERM1. (A and B) The sequence (A) and three-dimensional structure (B) of PERM1 are shown. In the circular permutant PERM1, the sequence of helix A is relocated to the C terminus of the whole sequence, and the two subdomains are decoupled. For clarity, the N-terminal lobe is in green and the C-terminal lobe is in yellow. The new N and C termini, from which PERMI is pulled to unfold, are shown in red and ball-and-stick representation. (C) Representative force-extension curves of polyprotein chimera (GB1)4-PERM1-(GB1)4. The mechanical unfolding of PERM1 shows diverse unfolding behaviors. PERM1 in curve A did not show clear unfolding force peaks, indicating that it unfolded at low forces below our detection limit. PERM1 in curves B and C corresponds to the two-state unfolding of a fully folded or partially folded PERM1. Curve D shows a three-state unfolding event of PERM1. (D) Histogram of ΔLC for the PERM1 molecules that unfold in two-state fashion (n = 107). (E) Histogram of ΔLC1 and ΔLC2 for PERM1 molecules that unfold in three-state fashion (n = 45).
Using a similar strategy, we constructed (GB1)4-PERM1-(GB1)4 to characterize the mechanical unfolding of PERM1. If we observed five or more unfolding events of GB1, we are certain that the given force-extension curve contains the signature of mechanical unfolding of PERM1. In contrast to T4L*, the unfolding of PERM1 displays even greater diversity (Fig. 4C): ≈20% of the force-extension curves (n = 190) did not show any unfolding peak corresponding to the unfolding of PERM1, indicating that these PERM1 molecules unfold at forces below our detection limit (≈20 pN); ≈56% of force-extension curves showed a single unfolding force peak with a broad distribution of ΔLC from 20 nm to 60 nm (Fig. 4D); and ≈24% of unfolding events showed a three-state unfolding (Fig. 4E).
In those unfolding events that only display one unfolding force peak, approximately half of the unfolding events have ΔLC of ≈60 nm, corresponding to the two-state complete unfolding of PERM1. However, 54 of 107 molecules show ΔLC that is much smaller than 60 nm, suggesting that part of PERM1 unfold at low forces before the observed unfolding event (Fig. 4C).
Compared with T4L*, there is a significant increase in the number of PERM1 that unfold at forces <20 pN, and the number of PERM1 that unfold from an already partially unfolded intermediate state. These results strongly indicate that the shuffling of helix A from the N terminus to C terminus significantly weakens T4L*. Although the two subdomains in circular permutant PERM1 remain relatively intact, the coupling of the two subdomains by helix A is no longer present. And the mechanical unfolding barrier, which corresponds to the disruption of helix A from the rest of C-terminal lobe, is no longer the dominant unfolding barrier. Hence, a shift of the mechanical unfolding barrier made it easier to unfold PERM1. Similar to the mechanical unfolding of T4L*, a significant percentage of PERM1 unfold in three-state fashion with broad distribution of ΔLC. Together with the observation that the unfolding of many PERM1 only show partial unfolding, we conclude that many local interactions in T4 lysozyme may result in local energy minima along the mechanical unfolding pathway. These results highlight the critical importance of the coupling of helix A to the C-terminal lobe on the mechanical stability of T4L*. This observation is consistent with recent observations that coupling helix A to the C-terminal lobe is key to the overall thermodynamic stability of T4L* (18, 45).
Discussion
The statistical mechanics description of protein folding has provided tremendous insights into the protein-folding mechanism (1, 46). Although there is only limited experimental data (5–8), it is now generally accepted that proteins can fold into their well defined native structure following multiple folding pathways by kinetic partitioning mechanisms (4). Recent developments in single-molecule techniques have made it possible to directly probe the folding and unfolding dynamics of proteins at the single-molecule level, providing the possibility to directly probe the kinetic partitioning mechanism. Here, our single-molecule mechanical unfolding trajectories revealed that T4 lysozyme unfolds by multiple distinct unfolding pathways: 87% of T4L* unfold in an all-or-none fashion involving overcoming a dominant unfolding kinetic barrier, and ≈13% of T4L* unfold in a three-state fashion and exhibit variability and diversity in their individual unfolding pathways. These mechanical unfolding trajectories provide direct evidence supporting the kinetic partitioning mechanism for the mechanical unfolding of T4L*.
Two possible scenarios are feasible to explain the kinetic partitioning observed for the mechanical unfolding of T4L*. In the first scenario, there is one well defined native conformation for T4L*. The frustration on the free-energy landscape results in direct (two-state unfolding) and indirect pathways to the unfolded state, leading to the kinetic partitioning observed here for the mechanical unfolding pathways. Along the indirect pathways, T4L* can enter into various local energy minima, which are stable enough to be captured in our single-molecule unfolding trajectories.
The second scenario is associated with the heterogeneity of the native conformations of T4L*. Although there is no direct experimental evidence, it was suggested that the N-terminal lobe of T4L* probably undergoes hinge-bending motion in solution (47), which could potentially lead to more than one native conformation. If this scenario holds, the observed heterogeneity of the mechanical unfolding pathways of T4L* could be explained by the unfolding from distinct native conformations of T4L*. In addition, this model could also provide direct evidence for a kinetic partitioning mechanism for the folding pathways of T4L*: if we assume the existence of multiple native conformations of T4L*, the repetitive unfolding–refolding experiments shown in Fig. 2B would indicate that T4L*, from the same unfolded and extended conformation, folded into different native conformations following distinct folding pathways. Considering that there is no direct experimental evidence for the heterogeneity of native conformations for T4L*, we think that the kinetic partitioning observed here is more likely to originate from the first mechanism. Nonetheless, the common theme of both scenarios is the frustration in the free-energy landscape, which is the key to kinetic partitioning mechanism.
The broad distribution of unfolding pathways suggests that the three-state unfolding behavior is not originating from well defined unfolding intermediates. Instead, there are many local interactions/contacts in T4L* that can “trap” the protein into an unfolding intermediate state along its unfolding pathways. Such complex multiple unfolding pathways reported here have much resemblance to that of the mechanical unfolding and folding of Tetrahymena thermophila ribozyme, where a wealth of unfolding and folding pathways observed in optical tweezers experiments are predominantly determined by local interactions (39, 48). This similarity between T4L* and ribozymes provides good supporting evidence for the proposal that the kinetic partitioning mechanism is a common theme in the folding of proteins and RNAs (4, 49).
Moreover, similar to the complexity in the mechanical unfolding pathways of T4L*, complex folding/unfolding intermediate states were also observed in chemical folding/unfolding of T4L*(40, 43–45, 50). However, because of the different reaction coordinates in the mechanical and chemical unfolding pathways, significant differences in the structure of mechanical and chemical unfolding intermediate states do exist. Although mechanical and chemical unfolding pathways of T4L* are different from each other, information from the mechanical and chemical unfolding experiments do reflect the intrinsic properties of the different regions of the underlying energy landscape. It is likely that the unique two subdomain structure of T4L* and the associated domain–domain interactions play important roles in defining such a complex energy landscape that gives rise to the complex unfolding kinetics of T4L*. Although T4L* displays two-state unfolding behaviors probed by traditional methods, it has been well recognized that there exist two subdomains in T4L* that have distinct thermodynamic stability and show possibilities of unfolding from at least two regions (16). Recent fragment studies confirmed the subdomain architecture of T4L* (18, 45). In addition, a continuum of stability was observed by native-state hydrogen exchange to occur throughout each subdomain that may give rise to a variety of folding and unfolding intermediate states (40, 43, 44, 50). In the mechanical unfolding of T4L*, we observed that the interaction between the N-terminal helix A and the reminder of the C-terminal lobe and the coupling between the two subdomains by helix A provide the dominant resistance to the mechanical unfolding. And various unfolding intermediate states observed in three-state unfolding correspond to various partially unfolded structures of the two subdomains after the main barrier-crossing event. These results highlight the critical importance of the domain–domain coupling and their interactions on the folding/unfolding kinetics of a multidomain protein. We anticipate that single-molecule mechanical manipulation exemplified here will provide a general tool and strategy to thoroughly investigate the mechanical unfolding kinetics of complex multidomain proteins.
Materials and Methods
Protein Engineering.
The plasmids that encode the T4L* protein and its circular permutant PERM1 were generous gifts from Brian W. Matthews (University of Oregon, Eugene, OR) and Martin Sagermann (University of California, Santa Barbara, CA). Polyprotein chimera (GB1)4-T4L*-(GB1)4 and (GB1)4-PERM1-(GB1)4 were constructed by using a method described in ref. 38. Polyproteins were overexpressed in DH5α strain and purified from supernatant by using Ni2+-affinity chromatography. The polyproteins were kept at 4°C in PBS buffer at a concentration of ≈200 μg/ml.
Single-Molecule Atomic Force Microscopy.
Single-molecule AFM experiments were carried out on a custom-built atomic force microscope as described in ref. 26. The details for analyzing force-extension curves and classifying unfolding events of T4L* are provided in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Steve Plotkin for stimulating discussion and Dr. Deepak Sharma, Yi Cao, and Canaan Lam for technical assistance. This work was supported by the Natural Sciences and Engineering Research Council of Canada, Canada Research Chairs program, and Canada Foundation for Innovation. H.L. is a Michael Smith Foundation for Health Research Career Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706775105/DC1.
References
- 1.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Sali A, Shakhnovich E, Karplus M. How does a protein fold? Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 3.Guo ZY, Thirumalai D. Kinetics of protein-folding: Nucleation mechanism, time scales, and pathways. Biopolymers. 1995;36:83–102. [Google Scholar]
- 4.Thirumalai D, Klimov DK, Woodson SA. Kinetic partitioning mechanism as a unifying theme in the folding of biomolecules. Theor Chem Acc. 1997;96:14–22. [Google Scholar]
- 5.Kiefhaber T. Kinetic traps in lysozyme folding. Proc Natl Acad Sci USA. 1995;92:9029–9033. doi: 10.1073/pnas.92.20.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright CF, Lindorff-Larsen K, Randles LG, Clarke J. Parallel protein-unfolding pathways revealed and mapped. Nat Struct Biol. 2003;10:658–662. doi: 10.1038/nsb947. [DOI] [PubMed] [Google Scholar]
- 7.Zaidi FN, Nath U, Udgaonkar JB. Multiple intermediates and transition states during protein unfolding. Nat Struct Biol. 1997;4:1016–1024. doi: 10.1038/nsb1297-1016. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez IE, Kiefhaber T. Hammond behavior versus ground state effects in protein folding: Evidence for narrow free energy barriers and residual structure in unfolded states. J Mol Biol. 2003;327:867–884. doi: 10.1016/s0022-2836(03)00171-2. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain AK, Handel TM, Marqusee S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat Struct Biol. 1996;3:782–787. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- 10.Main ER, Stott K, Jackson SE, Regan L. Local and long-range stability in tandemly arrayed tetratricopeptide repeats. Proc Natl Acad Sci USA. 2005;102:5721–5726. doi: 10.1073/pnas.0404530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huyghues-Despointes BM, Scholtz JM, Pace CN. Protein conformational stabilities can be determined from hydrogen exchange rates. Nat Struct Biol. 1999;6:910–912. doi: 10.1038/13273. [DOI] [PubMed] [Google Scholar]
- 12.Weaver LH, Matthews BW. Structure of bacteriophage T4 lysozyme refined at 1.7 A resolution. J Mol Biol. 1987;193:189–199. doi: 10.1016/0022-2836(87)90636-x. [DOI] [PubMed] [Google Scholar]
- 13.Matthews BW. Structural and genetic analysis of the folding and function of T4 lysozyme. FASEB J. 1996;10:35–41. doi: 10.1096/fasebj.10.1.8566545. [DOI] [PubMed] [Google Scholar]
- 14.Chen BL, Baase WA, Schellman JA. Low-temperature unfolding of a mutant of phage T4 lysozyme. 2. Kinetic investigations. Biochemistry. 1989;28:691–699. doi: 10.1021/bi00428a042. [DOI] [PubMed] [Google Scholar]
- 15.Wetzel R, Perry LJ, Baase WA, Becktel WJ. Disulfide bonds and thermal stability in T4 lysozyme. Proc Natl Acad Sci USA. 1988;85:401–405. doi: 10.1073/pnas.85.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llinas M, Gillespie B, Dahlquist FW, Marqusee S. The energetics of T4 lysozyme reveal a hierarchy of conformations. Nat Struct Biol. 1999;6:1072–1078. doi: 10.1038/14956. [DOI] [PubMed] [Google Scholar]
- 17.Llinas M, Marqusee S. Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci. 1998;7:96–104. doi: 10.1002/pro.5560070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cellitti J, et al. Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment. Protein Sci. 2007;16:842–851. doi: 10.1110/ps.062628607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Bertelsen E, Benvegnu D, Alber T. Circular permutation of T4 lysozyme. Biochemistry. 1993;32:12311–12318. doi: 10.1021/bi00097a006. [DOI] [PubMed] [Google Scholar]
- 20.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 21.Fisher TE, Oberhauser AF, Carrion-Vazquez M, Marszalek PE, Fernandez JM. The study of protein mechanics with the atomic force microscope. Trends Biochem Sci. 1999;24:379–384. doi: 10.1016/s0968-0004(99)01453-x. [DOI] [PubMed] [Google Scholar]
- 22.Dietz H, Berkemeier F, Bertz M, Rief M. Anisotropic deformation response of single protein molecules. Proc Natl Acad Sci USA. 2006;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brockwell DJ. Force denaturation of proteins: An unfolding story. Curr Nanosci. 2007;3:3–15. [Google Scholar]
- 24.Carrion-Vazquez M, et al. In: Advanced Techniques in Biophysics. Arrondo J, Alonso A, editors. New York: Springer; 2006. pp. 163–236. [Google Scholar]
- 25.Fernandez JM, Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y, Lam C, Wang M, Li H. Nonmechanical protein can have significant mechanical stability. Angew Chem Int Ed Engl. 2006;45:642–645. doi: 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- 27.Sharma D, et al. Single-molecule force spectroscopy reveals a mechanically stable protein fold and the rational tuning of its mechanical stability. Proc Natl Acad Sci USA. 2007;104:9278–9283. doi: 10.1073/pnas.0700351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marszalek PE, et al. Mechanical unfolding intermediates in titin modules. Nature. 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Huang HH, Badilla CL, Fernandez JM. Mechanical unfolding intermediates observed by single-molecule force spectroscopy in a fibronectin type III module. J Mol Biol. 2005;345:817–826. doi: 10.1016/j.jmb.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Schwaiger I, Kardinal A, Schleicher M, Noegel AA, Rief M. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat Struct Mol Biol. 2004;11:81–85. doi: 10.1038/nsmb705. [DOI] [PubMed] [Google Scholar]
- 31.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci USA. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc Natl Acad Sci USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J Biol Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- 34.Yang G, et al. Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme. Proc Natl Acad Sci USA. 2000;97:139–144. doi: 10.1073/pnas.97.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagermann M, Baase WA, Mooers BH, Gay L, Matthews BW. Relocation or duplication of the helix A sequence of T4 lysozyme causes only modest changes in structure but can increase or decrease the rate of folding. Biochemistry. 2004;43:1296–1301. doi: 10.1021/bi035702q. [DOI] [PubMed] [Google Scholar]
- 36.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 37.Klimov DK, Thirumalai D. Native topology determines force-induced unfolding pathways in globular proteins. Proc Natl Acad Sci USA. 2000;97:7254–7259. doi: 10.1073/pnas.97.13.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrion-Vazquez M, et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onoa B, et al. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gassner NC, et al. Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding. Biochemistry. 1999;38:14451–14460. doi: 10.1021/bi9915519. [DOI] [PubMed] [Google Scholar]
- 41.Desmadril M, Yon JM. Evidence for intermediates during unfolding and refolding of a two-domain protein, phage T4 lysozyme: Equilibrium and kinetic studies. Biochemistry. 1984;23:11–19. doi: 10.1021/bi00296a003. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Dahlquist FW. Detection and characterization of an early folding intermediate of T4 lysozyme using pulsed hydrogen exchange and two-dimensional NMR. Biochemistry. 1992;31:4749–4756. doi: 10.1021/bi00135a002. [DOI] [PubMed] [Google Scholar]
- 43.Kato H, Feng H, Bai Y. The Folding pathway of T4 lysozyme: The high-resolution structure and folding of a hidden intermediate. J Mol Biol. 2007;365:870–880. doi: 10.1016/j.jmb.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato H, Vu ND, Feng H, Zhou Z, Bai Y. The folding pathway of T4 lysozyme: An on-pathway hidden folding intermediate. J Mol Biol. 2007;365:881–891. doi: 10.1016/j.jmb.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cellitti J, Bernstein R, Marqusee S. Exploring subdomain cooperativity in T4 lysozyme II: Uncovering the C-terminal subdomain as a hidden intermediate in the kinetic folding pathway. Protein Sci. 2007;16:852–862. doi: 10.1110/ps.062632807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinner AR, Sali A, Smith LJ, Dobson CM, Karplus M. Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem Sci. 2000;25:331–339. doi: 10.1016/s0968-0004(00)01610-8. [DOI] [PubMed] [Google Scholar]
- 47.Faber HR, Matthews BW. A mutant T4 lysozyme displays five different crystal conformations. Nature. 1990;348:263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- 48.Li PT, Bustamante C, Tinoco I., Jr Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 50.Jacobsen K, Hubbell WL, Ernst OP, Risse T. Details of the partial unfolding of T4 lysozyme on quartz using site-directed spin labeling. Angew Chem Int Ed Engl. 2006;45:3874–3877. doi: 10.1002/anie.200600008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.