Abstract
A common integration site, cloned from MoMuLV-induced rat T cell lymphomas, was mapped immediately upstream of Not dead yet-1 (Ndy1)/KDM2B, a gene expressed primarily in testis, spleen, and thymus, that is also known as FBXL10 or JHDM1B. Ndy1 encodes a nuclear, chromatin-associated protein that harbors Jumonji C (JmjC), CXXC, PHD, proline-rich, F-box, and leucine-rich repeat domains. Ndy1 and its homolog Ndy2/KDM2A (FBXL11 or JHDM1A), which is also a target of provirus integration in retrovirus-induced lymphomas, encode proteins that were recently shown to possess Jumonji C-dependent histone H3 K36 dimethyl-demethylase or histone H3 K4 trimethyl-demethylase activities. Here, we show that mouse embryo fibroblasts engineered to express Ndy1 or Ndy2 undergo immortalization in the absence of replicative senescence via a JmjC domain-dependent process that targets the Rb and p53 pathways. Knockdown of endogenous Ndy1 or expression of JmjC domain mutants of Ndy1 promote senescence, suggesting that Ndy1 is a physiological inhibitor of senescence in dividing cells and that inhibition of senescence depends on histone H3 demethylation.
Keywords: cancer, histone demethylase, immortalization, senescence, insertional mutagenesis
Mutations caused by provirus integration into the genome play a critical role in the induction and progression of retrovirus-induced neoplasms (1–3). Given that provirus integration is practically random, the detection of an integrated provirus within a given DNA region in multiple tumors has been interpreted to suggest that the mutation caused by this provirus endows the affected cell with a selective advantage over its neighbors. Based on this understanding, the identification of common integration sites in retrovirus-induced tumors has been used as an effective tool to identify novel oncogenes (1–3).
Core histones contain a “histone fold” globular domain, which is responsible for histone–DNA and histone–histone interactions and N- and C-terminal tails. Histone tails contain sites that are targets of various posttranslational modifications, including phosphorylation, acetylation, methylation, and ubiquitination (4). Posttranslational modifications of histone tails regulate the interaction of nucleosomes with other nucleosomes and with linker DNA and direct the folding of chromatin into a higher-order structure (5). The same modifications regulate the binding of various nonhistone chromatin-associated proteins (“the histone code hypothesis”) (4, 6). As a result, enzymes involved in the posttranslational modification of histone tails, in combination with chromatin remodeling enzymes (7), regulate transcription and other chromatin-dependent activities. Modification of histone tails is a dynamic process with all modifications being transient (4). The most recently discovered enzyme group responsible for the reversal of a histone modification is that of histone demethylases (8–10).
Histone methylation on lysine and arginine residues contributes to the regulation of gene expression, dosage compensation, and epigenetic memory (4, 11). The functional consequences of histone methylation depend on the methylation site. Thus, whereas methylation at H3K9, H3K27, and H4K20 is usually associated with transcriptional repression, methylation at H3K4, H3K36, and H3K79 is associated with transcriptional activation (11). The stoichiometry of histone methylation at a given site is also functionally important, because the binding of transcriptional regulators to methylated histones is stoichiometry-dependent (12). There are three distinct classes of histone demethylases (10). The largest of these classes consists of enzymes containing a JmjC domain, a homolog of the cupin metalloenzymes (10, 13). The JmjC domain-containing enzymes catalyze demethylation through an oxidative reaction that depends on two cofactors, iron Fe(II) and α-ketoglutarate. Specifically, these enzymes catalyze the hydroxylation of mono-, di- or trimethylated lysine in histone tails, giving rise to an unstable hydroxyl–methyl group, which is released spontaneously as formaldehyde (9, 10).
Some JmjC domain-containing histone demethylases have been linked to cancer (14–16). However, the mechanisms of their oncogenic activities are not well understood. Here, we present evidence that the two members of the JHDM1 protein family, Ndy1 (FBXL10 or JHDM1B) and Ndy2 (FBXL11 or JHDM1A), contribute to the induction and/or progression of MoMuLV-induced T cell lymphomas in rodents. Furthermore, upon overexpression, both proteins immortalize mouse embryonic fibroblasts (MEFs) in culture. Moreover, knockdown of Ndy1 and expression of Ndy1 dominant-negative mutants promote senescence, suggesting that Ndy1 is a physiological inhibitor of senescence in dividing cells. Immortalization depends on the JmjC domain and perhaps on JmjC domain-mediated histone demethylation. Finally, the immortalization activity of Ndy1 depends on targeting the Rb and p53 pathways.
Results
Ndy1 Is a Common Target of Provirus Integration in MoMuLV-Induced Rat T Cell Lymphomas.
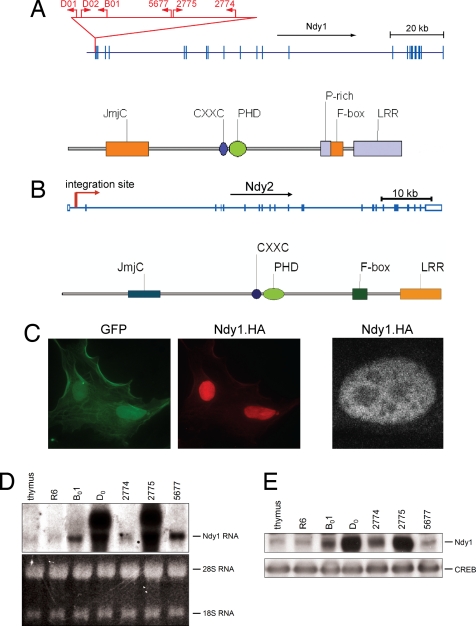
A genome-wide screen of 44 MoMuLV-induced T cell lymphomas for novel targets of proviral DNA integration yielded clones of 149 independent provirus integrations. Six of these integrations, cloned from five independent tumors, had targeted a gene we named Ndy1, also known as FBXL10 or JHDM1B (9, 10, 16, 17). Two of them were cloned from a single tumor, D0, suggesting that this tumor consists of at least two populations of cells, both of which carry an integrated provirus in this locus. All of the integrations in the vicinity of Ndy1 detected to date were located within a region 140 bp upstream of one of several transcription initiation sites used by this gene. The transcriptional orientation of two of the five integrated proviruses was the same as that of the Ndy1 gene (Fig. 1A). A single provirus insertion was also detected immediately upstream of the Ndy1 homolog Ndy2, also known as FBXL11 or JHDM1A (Fig. 1B). Interestingly, provirus insertions were also detected upstream of two more genes encoding JmjC domain-containing proteins, Phf2 and Phf8 [supporting information (SI) Fig. 6]. This article focuses on the characterization of Ndy1 and Ndy2.
Fig. 1.
Provirus insertion activates the Ndy1 gene. (A Upper) Sites and orientation of provirus integration at the 5′ end of the Ndy1 gene. The numbers above the arrows showing the sites of provirus integration identify the tumors in which the integrations were detected. (Lower) Domain structure of the Ndy1 protein. (B Upper) Site and orientation of provirus integration at the 5′ end of the Ndy2 gene in tumor #4. (Lower) Domain structure of the Ndy2 protein. (C) NIH 3T3 cells were infected with a MigR1 construct of Ndy1.HA. (Left) Two GFP-positive, infected cells. (Center) The same cells stained with an anti-HA antibody. (Right) Ndy1.HA-expressing cell stained with an anti-HA antibody and visualized by confocal microscopy. The darker spots correspond to nucleoli. (D Upper) Northern blot of total cell RNA derived from normal rat thymus and the indicated tumors (R6 to 5677) probed with a full-length rat Ndy1 cDNA probe. (Lower) Ethidium bromide staining of the gel (loading control). (E) Western blot of nuclear cell lysates from normal thymus and the indicated tumors probed with anti-Ndy1 and anti-CREB antibodies as indicated.
Ndy1encodes a protein that contains a JmjC domain (9, 10, 16, 17), a CXXC zinc finger, a PHD zinc finger, a proline-rich region (PRR), an F-box, and a leucine-rich repeat (LRR) (Fig. 1A and ref. 10). Ndy1 is localized in the nucleus of transiently transfected HEK293 cells (Fig. 1C Left and Center) and stably infected NIH 3T3 cells (Fig. 1C Right), and it is tightly associated with an insoluble fraction that is highly enriched in histones (SI Fig. 7B). The chromatin association of this protein is compatible with the results of recent studies showing that both Ndy1 and its homolog Ndy2 function as demethylases of histone H3 dimethylated at K36 or histone H3 trimethylated at K4 (9, 17).
Northern and Western blotting of various rat tissues revealed that Ndy1 is expressed primarily in testis, spleen, and thymus (SI Fig. 7A). Moreover, Northern and Western blotting carried out on normal rat thymus and several MoMuLV-induced rat T cell lymphomas showed that Ndy1 expression is increased in all tumors that harbor a provirus in the Ndy1locus. Highest levels of Ndy1 expression were detected in tumors in which the orientation of the integrated provirus was the same as the orientation of the Ndy1gene (tumors D0 and 2775) (Fig. 1 D and E). However, these tumors also express high levels of aberrant mRNA transcripts encoding an Env-Ndy1 nonnuclear chimeric protein (SI Fig. 8).
Sequence comparison of Ndy1 cDNA clones in the GenBank database revealed that there are multiple forms of the Ndy1 mRNA that are generated through differential transcriptional initiation or alternative splicing. The protein predicted to be encoded by one of these transcripts lacks the JmjC domain and contains a unique N-terminal region (NM_013910, SI Fig. 9). This protein will be referred to as the short form of Ndy1. Experiments presented in this article used the v1 variant of Ndy1 (NM_001003953).
MEFs Engineered to Express Exogenous Ndy1 Bypass Replicative Senescence and Undergo Immortalization.
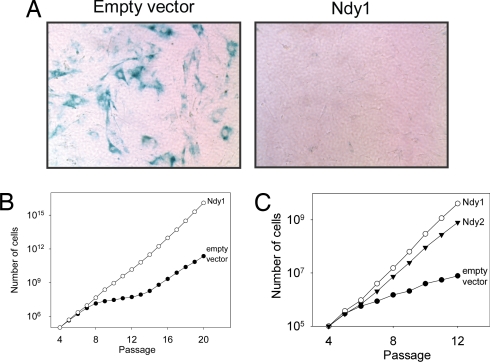
To determine the phenotypic effects of Ndy1 expression, MEFs were infected with an Ndy1-MigR1 retrovirus construct or with the empty MigR1 retrovirus vector (SI Fig. 10). After several passages, the cells infected with the empty vector were senescing, whereas the MigR1-Ndy1-infected cells continued to divide without signs of senescence. Staining the cells at passage 11 for β-galactosidase (Fig. 2A) confirmed this observation. Plotting the cumulative cell number at each passage (Fig. 2B) revealed that, whereas the proliferation of the vector-infected cells practically stops between passages 8 and 13, the proliferation of Ndy1-expressing cells continues uninterrupted. At the end, both the MigR1 and the MigR1-Ndy1-infected cells undergo immortalization. However, only the MigR1-Ndy1-infected cells bypass replicative senescence. Further studies revealed that not only Ndy1 but also Ndy2 promotes immortalization of MEFs in culture (SI Fig. 10 and Fig. 2C). Moreover, passaging of cells infected with a MigR1-Env-Ndy1 retrovirus revealed that the cytoplasmic Env-Ndy1 protein does not immortalize MEFs, suggesting that the nuclear localization of the protein is required for the immortalization phenotype (SI Figs. 8 and 11).
Fig. 2.
MEFs expressing Ndy1 or Ndy2 bypass replicative senescence and undergo immortalization. (A) β-Galactosidase staining of 11th-passage MEFs infected with the MigR1 retrovirus vector (Left) or with a MigR1-Ndy1construct (Right). (B and C) MEFs infected with the MigR1 vector or MigR1-Ndy1 and MigR1-Ndy2 constructs. The graph shows the cumulative number of cells in each culture at sequential passages.
Histone Demethylase Activities of Ndy1 and Ndy2 Are Required for Immortalization.
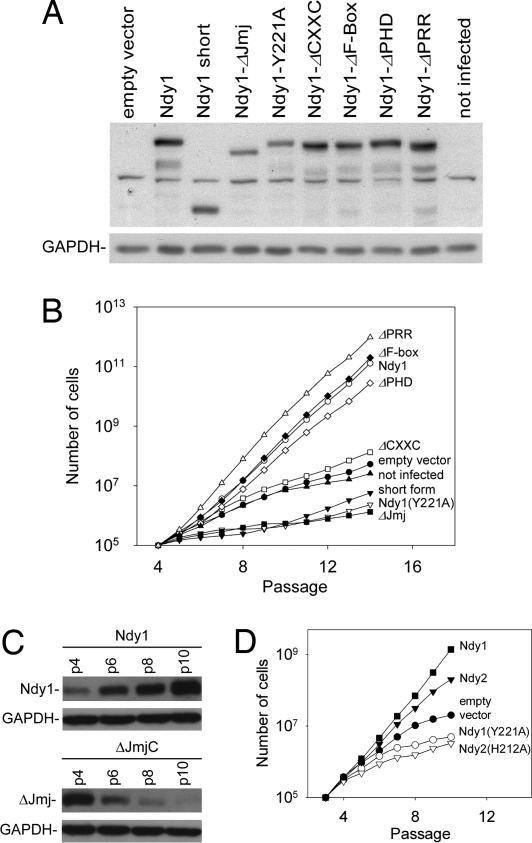
Ndy1 is a multidomain protein that may exhibit multiple functional activities that singly or in combination may contribute to the observed immortalization phenotype. To map the domain responsible for immortalization, we addressed the immortalizing activity of all single-domain deletion mutants of Ndy1 (Fig. 3A). Regarding the JmjC domain, in addition to the deletion mutant, we used the physiologically expressed short form of Ndy1 that lacks the JmjC domain and the point mutants Ndy1 Y221A and Ndy1 H283Y. The Y221A mutation was based on a similar mutation that inhibits the biological activity of the Saccharomyces pombe protein Epe1 (18–21), whereas the H283Y mutation modifies the Fe(II)-binding pocket of the JmjC domain, which is required for the demethylase activity. Passaging the stably infected MEFs separated them into three distinct groups that differed regarding the emergence of senescence. One group included noninfected and empty vector-infected cells as well as cells infected with the CXXC mutant, which began to show evidence of senescence by passage 7–8. The second group included the ΔJmjC deletion mutant, the short form of Ndy1, and the two JmjC domain point mutants, which began to show evidence of senescence at a very early passage, suggesting that endogenous Ndy1 inhibits senescence and that these mutants interfere with the physiological function of the endogenous protein. The third group included wild-type Ndy1 and all of the remaining mutants, which continued to immortalize MEFs (Fig. 3B). The preceding data were further supported by two additional observations: (i) Western blots of passaged MEFs infected with MigR1-Ndy1.hemagglutinin (HA) or MigR1-ΔJmjC Ndy1.HA constructs, revealed that Ndy1.HA expression increases, whereas ΔJmjC Ndy1.HA expression decreases with each passage, indicating that Ndy1.HA-expressing cells are positively selected, and ΔJmjC Ndy1.HA-expressing cells are counterselected (Fig. 3C); and (ii) β-galactosidase staining of 11th passage MEFs infected with wild-type or mutant Ndy1 stained strongly only the cells infected with constructs of the JmjC domain mutants (SI Fig. 12).
Fig. 3.
Immortalization depends on the histone demethylase activities of Ndy1 and Ndy2. (A) Western blots of total cell lysates derived from MEFs infected with MigR1 or with wild-type or mutant MigR1-Ndy1.Myc. The blots were probed with anti-Ndy1 or with anti-GAPDH (loading control) antibodies as indicated. Ndy1-ΔPRR carries a deletion of the prolene-rich region, upstream of the F-box (see Fig. 1A). The LRR deletion mutant was also tested in separate experiments, and it was shown to immortalize MEFs as efficiently as the wild-type protein (data not shown). (B) MEFs shown in A were passaged in culture. Graphs show the cumulative number of cells at each passage. The Ndy1.H283Y mutant was also tested in separate experiments, and it was shown to have the same dominant-negative phenotype as the Ndy1.Y221A mutant (data not shown). (C) Western blots of cell lysates from cells infected with MigR1-Ndy1 or MigR1-ΔJmjC Ndy1 were probed with the anti-Ndy1 antibody. (D) MEFs infected with the indicated constructs were passaged in culture. Cumulative numbers of cells at each passage are indicated.
To determine whether MEF immortalization by Ndy2 is also JmjC domain-dependent, we repeated the preceding experiment with Ndy2, and the Ndy2 JmjC domain mutant H212A, which does not bind Fe(II) and has no demethylase activity (9). Wild-type Ndy1, the Ndy1 mutant Y221A, and the empty vector were used as controls. The results of this experiment confirmed that MEF immortalization by Ndy2 also depended on a functional JmjC domain (Fig. 3D).
Endogenous Ndy1 Is a Physiological Inhibitor of Senescence.
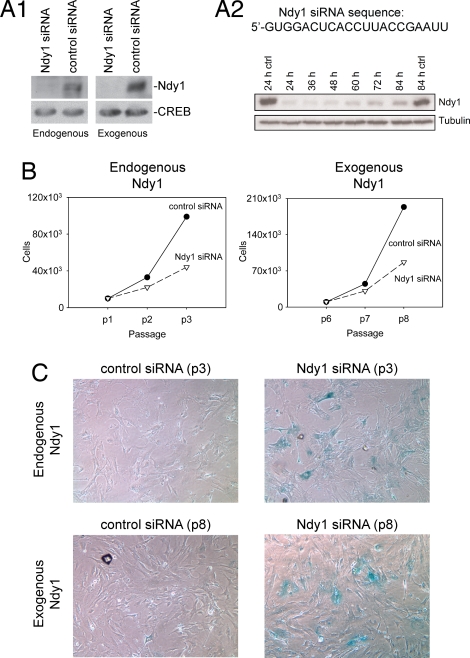
To determine whether endogenous Ndy1 is a physiological regulator of senescence, passage-3 wild-type MEFs and passage-8 MEFs overexpressing Ndy1 were transfected with Ndy1 or control siRNA. The down-regulation of both the endogenous and the exogenous Ndy1 in the transfected cells was confirmed by Western blotting (Fig. 4A1 and A2). The transfected cells were passaged twice, and cell proliferation was monitored by counting the cells at each passage. Cell morphology and β-galactosidase staining were recorded after the second passage (Fig. 4 B and C). The data confirmed that the endogenous Ndy1 indeed protects dividing cells from replicative senescence.
Fig. 4.
Endogenous Ndy1 physiologically inhibits replicative senescence in MEFs. (A) The Ndy1 and scrambled siRNAs were transfected into wild-type or MigR1-Ndy1.HA-infected MEFs. Western blots of transfected cell lysates were probed with the anti-Ndy1 antibody. (A1) Western blot of cell lysates harvested 48 h after the transfection. (A2) Time course of exogenous Ndy1 starting with cell lysates harvested 24 h after transfection. (B) Early-passage wild-type and MigR1-Ndy1.HA-infected MEFs were transfected with Ndy1 or control siRNAs, and they were passaged twice every 72 h. siRNA transfection was repeated after the first passage at the 72-h time point. Cells were counted, and the numbers were plotted as indicated. (C) Early-passage wild-type and MigR1-Ndy1.HA-infected MEFs were transfected with Ndy1 or control siRNA, and they were stained for β-galactosidase.
Ndy1 Does Not Prevent Senescence in IMR90 Cells.
To determine whether Ndy1 also promotes immortalization of primary human fibroblasts, IMR90 cells were infected with MigR1 or MigR1-Ndy1. After passage 20, the proliferation of vector-infected IMR90 cells slowed down, whereas the proliferation of MigR1-Ndy1-infected IMR90 cells continued, suggesting that Ndy1 inhibits early senescence in these cells. However, the proliferation of MigR1-Ndy1-infected cells also slowed down by passage 26, and the cells failed to undergo immortalization (SI Fig. 13). Given that human cells go into senescence because of telomere shortening (22, 23), these data indicate that Ndy1 does not protect cells from telomere erosion, although it inhibits the DNA damage response elicited by the erosion (24).
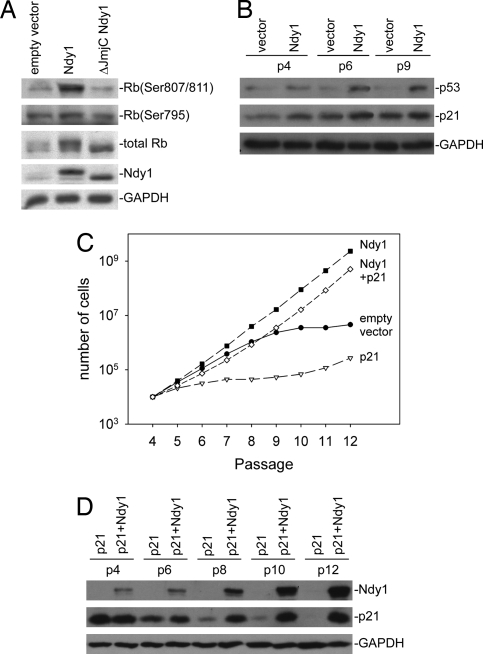
Ndy1 Promotes MEF Immortalization by Targeting the Rb and p53 Pathways.
The activation of the Ink4a/ARF locus and the response to DNA damage, which are the main factors promoting senescence, target both the p53 and Rb pathways (25–27). To determine which pathway may be targeted by Ndy1, we examined the phosphorylation of Rb at Ser-807/811 and Ser-780 and the expression of p53 and its target p21CIP1 in early- and late-passage MEFs infected with MigR1 and MigR1-based constructs of Ndy1 and ΔJmjC-Ndy1. The results (Fig. 5A) showed that Ndy1 selectively promotes the phosphorylation of Rb at Ser-807/811. However, it also up-regulates p53 and its target p21CIP1 (28) (Fig. 5B). Overexpression of Ndy1 in p53+/+ and p53−/− HCT116 cells revealed that the induction of p21CIP1 by Ndy1 is p53-dependent (SI Fig. 14).
Fig. 5.
Ndy1 promotes immortalization by targeting the Rb and p53 pathways. (A) Western blots of MEFs infected with the indicated constructs were probed with the indicated antibodies. (B) Passaged MEFs infected with the indicated constructs express high levels of p53 and its target p21CIP1. Western blots of cell lysates harvested from passaged cells, probed with the indicated antibodies are shown. (C) MEFs infected with the indicated constructs were passaged in culture. Cumulative numbers of cells at each passage are indicated. (D) Western blots of passaged cells infected with the indicated constructs were probed with the indicated antibodies.
The preceding data suggest that Ndy1 immortalizes cells by targeting the Rb pathway and that, in the presence of Ndy1, the activation of the p53/p21CIP1 pathway does not inhibit cell proliferation. To address this hypothesis, we infected MEFs with a MigR1-GFP construct of Ndy1, a MigR1-RFP construct of p21CIP1, a combination of the two, or with the MigR1-GFP empty-vector control. Passaging revealed that cells overexpressing p21CIP1 senesce rapidly, whereas cells overexpressing both p21CIP1 and Ndy1 immortalize nearly as efficiently as cell overexpressing only Ndy1 (Fig. 5C). Probing Western blots of cell lysates harvested at the indicated passages with antibodies against p21CIP1 or Ndy1 revealed that, in the absence of Ndy1, p21CIP1-overexpressing cells were strongly counterselected. However, in the presence of Ndy1, they were positively selected (Fig. 5D), suggesting that Ndy1 expression indeed abrogates the cell cycle inhibitory activity of p21CIP1.
Discussion
In this report, we showed that Ndy-overexpressing cells bypass replicative senescence and undergo immortalization via a JmjC domain-dependent process. In addition, we showed that knocking down Ndy1 and expression of dominant-negative mutants of Ndy1 promotes senescence, suggesting that Ndy1 is a physiological inhibitor of senescence in dividing cells. Ndy2, a homolog of Ndy1, also promotes immortalization of MEFs. Immortalization was linked to the selective phosphorylation of Rb (25) and the selective abrogation of the prosenescence activity of p21CIP1 (26, 29).
Ndy1 and Ndy2 are multidomain chromatin-associated proteins (9, 10). Systematic deletion of all of the known domains of Ndy1 revealed that the JmjC and the CXXC domains are the only ones required for immortalization. However, whereas the JmjC domain mutants exhibit a dominant-negative, prosenescence phenotype, the CXXC motif mutants do not. These findings were interpreted to suggest that the JmjC domain provides the functional activity for immortalization and that the CXXC motif may control immortalization by regulating the DNA targeting of the protein. In the absence of the correct DNA binding, the function of the protein is impaired. However, the mutant does not have a dominant-negative phenotype because it does not interfere with function of the endogenous protein (17). The preceding hypothesis is supported by our finding that the Env-Ndy1 hybrid protein, which is localized primarily in the cytoplasm, also lacks both immortalizing and dominant-negative prosenescence activities.
Ndy1 and Ndy2 possess JmjC domain-dependent histone H3 demethylase activities. Ndy2, and perhaps Ndy1, demethylate histone H3 dimethylated at K36 (9). Moreover, Ndy1 demethylates histone H3 trimethylated at K4 (17). JmjC domain-dependent demethylation is an oxidative reaction that requires Fe(II) and α-ketoglutarate as cofactors (9). The JmjC domain residues that coordinate the binding of these cofactors have been mapped (10). Mutation of some of these sites in Ndy1 and Ndy2 gave rise to proteins that exhibited dominant-negative prosenescence, rather than immortalizing phenotypes, indicating that the histone demethylase activities of these proteins are required for immortalization.
Cellular senescence is due to a number of factors, including the progressive shortening of telomeres, the activation of the Ink4a/ARF locus, and telomere shortening-independent DNA damage (28). MEFs and human fibroblasts differ with regard to the relative importance of telomere shortening in the induction of senescence in culture. Thus, the primary cause of senescence in human, but not in mouse fibroblasts, is telomere erosion (22, 23), which is recognized as DNA damage (24). Given that Ndy1 prevented early senescence but failed to immortalize IMR90 cells, we suggest that it interferes with the withdrawal from the cell cycle induced by telomere shortening, but it does not prevent telomere shortening per se.
The shortening of telomeres and the activation of the Ink4a/ARF locus may be developmentally programmed in dividing cells (28). In addition they can be induced in response to DNA damage, which plays a central role in the progression into senescence (24, 30). In dividing cells, the DNA damage response is activated by telomere shortening, oxidative stress, the aberrant firing of replication origins, or by activated oncogenes (28). Signaling pathways activated by DNA damage target the Rb and p53 pathways and induce reversible or irreversible cell cycle arrest or apoptosis (25–27). To determine the mechanism by which Ndy1 prevents senescence, we examined the effects of its overexpression on the Rb and p53 pathways. The results revealed that Ndy1 promotes the phosphorylation of Rb at Ser-807/811. Given that phosphorylation at this and other sites relieves the transcriptional repression activity of Rb and promotes progression through the G1 phase of the cell cycle, these data provide an explanation for the immortalizing activity of Ndy1. The precise mechanism by which Ndy1 regulates the phosphorylation of Rb remains to be determined. It is possible that it may regulate the activity of cyclin-dependent kinases, which phosphorylate Rb by altering the expression or posttranslational modification of the components of cyclin/cdk complexes or, more likely, by repressing the expression of cdk inhibitors. Alternatively, it may directly modify Rb, thus altering its ability to be phosphorylated by cdks.
Examination of the p53 pathway in Ndy1-overexpressing MEFs revealed that p53 and its target p21CIP1 were significantly up-regulated. Given that p53 and p21CIP1 promote cell cycle arrest, senescence, and apoptosis (29, 31), this finding was surprising. Further studies, however, revealed that, although Ndy1 does not inhibit globally the DNA damage response, it selectively abrogates the prosenescence phenotype of p21CIP1. The mechanism by which Ndy1 inhibits p21CIP1 remains to be determined. It is possible that it may alter directly or indirectly the cyclin/cdk inhibitory activity of p21CIP1. Alternatively, it may alter, again directly or indirectly, the cdk-independent transcriptional activities of p21CIP1 (32).
The data presented in the report strongly suggest that Ndy1 is a molecule that promotes oncogenesis. Thus, Ndy1 is overexpressed as a result of provirus integration in retrovirus-induced lymphomas. Moreover, it inhibits senescence, which is a potent tumor-protective process, as suggested by genetic animal models and by clinical studies on tumors and precancerous lesions (28, 30, 33, 34). Finally, it is overexpressed in human lymphomas and mammary adenocarcinomas (http://source.stanford.edu). However, recent studies from other laboratories have suggested that Ndy1 may function as a tumor suppressor (16, 17). First, it appears to protect the genome against mutations (16, 35). In addition, it inhibits cell growth and proliferation when overexpressed in some tumor cell lines, such as HELA cells (17). Finally, it is expressed at very low levels in aggressive glioblastomas (17). To reconcile these facts, we propose that Ndy1 and Ndy2 may function both as oncogenes and as tumor suppressor genes and that the final balance of their prooncogenic and antioncogenic activities may be context-dependent. Thus, in lymphomas and mammary adenocarcinomas that express high levels of Ndy1, the Ndy1 protein may have a tumor-promoting role, whereas in glioblastomas, in which aggressiveness correlates with low levels of expression, Ndy1 may function as a tumor suppressor.
In summary, data presented in this report, identify a previously unrecognized function of JmjC domain-containing proteins and provide another link between epigenetic regulation and cancer.
Materials and Methods
Cloning of Ndy1 and Ndy2 from MoMuLV-Induced Rat T Cell Lymphomas.
Newborn Fisher-344 rats were injected with 105 PFUs of MoMuLV intraperitoneally, monitored for tumor development, and killed before death. Provirus integration sites were cloned from tumor cell DNA by inverse PCR as described (2) (SI Text).
Plasmid Constructs.
All retroviral constructs were based on the retrovirus vector MigR1, a variant of MigR1 in which the green fluorescent protein gene was replaced by the red fluorescent protein gene and pBabe-puro. Ndy1, Ndy2, and p21CIP1 were cloned within the multiple cloning sites of these vectors and tagged at the carboxyl-terminus with either myc or HA tags. Retroviral constructs of Ndy1 deletion mutants were generated from the wild-type construct by overlap extension PCR (SI Text).
The point mutants Ndy1 Y221A, Ndy1 H283Y, and Ndy2 H212A (mouse) were generated from the wild-type mouse cDNA by using the Quikchange XL mutagenesis kit (cat no 200517; Stratagene) (SI Text).
Cell Culture, siRNA, and Senescence Assays.
MEFs were isolated from 13.5-day C57BL/6 mouse embryos. MEFs, IMR90, and HEK293T cells were cultured in Dulbecco's modified Eagle's MEM supplemented with 10% FBS, penicillin and streptomycin, and nonessential amino acids. The human colon cancer cell lines HCT-116(p53−/−) and HCT-116(p53+/+) (from B. Vogelstein, Johns Hopkins Oncology Center, Baltimore) (36) were grown in McCoy's 5a medium supplemented with 10% FBS. To generate HCT-116 cells stably overexpressing Ndy1, we infected the parental HCT-116 cells with pBabe-Ndy1.Myc or pBabe-puro, and we selected the infected cells for puromycin resistance (2 μg/ml) for 2 days.
MEFs overexpressing Ndy1 or Ndy2 and MEFs in which Ndy1 was knocked down with siRNA (1433-GUGGACUCACCUUACCGAAUU-1454) were monitored for senescence by light microscopy and β-galactosidase staining and by cell counting at each passage (SI Text). Transfection of siRNA was carried out by using Lipofectamine 2000 (Invitrogen). Knockdown of Ndy1 was confirmed by Western blotting and real-time RT-PCR.
Gene Expression Analysis: Northern Blotting, Real-Time RT-PCR, Antibodies, and Western Blotting.
Northern blotting and real-time RT-PCR were carried out by using standard procedures (SI Text). To measure the expression of the Ndy1 protein in both tumors and MEFs, Western blots of nuclear cell lysates were probed with anti-Ndy1, anti-Myc, or anti-HA antibodies (SI Text). The polyclonal antibody against Ndy1 was raised by injecting rabbits with the peptide 907-KMRRKRRLVNKELSKC-921, which maps between the PHD2 and F-Box domains. The position of the N-terminal and C-terminal amino acids was based on the sequence of the mouse protein. The antibody recognizes the mouse, human, and rat Ndy-1 (data not shown).
Immunostaining.
Early-passage MEFs were infected with the empty MigR1 vector or a MigR1-Ndy1.HA construct. After fixation with paraformaldehyde (4% for 10 min), cells were washed, permeabilized (0.2% Triton X-100), and stained with a mouse monoclonal anti-HA antibody.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Yujiang Shi, Phil Hinds, Rick Van Etten, Christos Polytarchou, and Antonios Makris for helpful discussions and Bert Vogelstein for kindly providing the HCT116 cells. This work was supported by National Institutes of Health Grant R01CA109747. Some of the core services were provided by the Tufts–New England Medical Center Gastroenterology Research on Absorptive and Secretory Processes (GRASP) Center, supported by National Institutes of Health Grant P30 DK 34928.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711865105/DC1.
References
- 1.Tsichlis PN, Lazo PA. Virus–host interactions and the pathogenesis of murine and human oncogenic retroviruses. Curr Top Microbiol Immunol. 1991;171:95–171. doi: 10.1007/978-3-642-76524-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Gilks CB, Bear SE, Grimes HL, Tsichlis PN. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth after activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, et al. Leukaemia disease genes: Large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 5.Hansen JC. Conformational dynamics of the chromatin fiber in solution: Determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Kwon CS, Wagner D. Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 11.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 12.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 13.Clissold PM, Ponting CP. JmjC: Cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci. 2001;26:7–9. doi: 10.1016/s0968-0004(00)01700-x. [DOI] [PubMed] [Google Scholar]
- 14.Yamane K, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Cloos PA, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 18.Ayoub N, et al. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol. 2003;23:4356–4370. doi: 10.1128/MCB.23.12.4356-4370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Verdel A. Destabilizing heterochromatin: Does Swi6/HP1 make the choice? Mol Cell. 2006;22:709–710. doi: 10.1016/j.molcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007;26:4670–4682. doi: 10.1038/sj.emboj.7601892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 23.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 24.von Zglinicki T, Saretzki G, Ladhoff J, d'Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126:111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 25.Genovese C, Trani D, Caputi M, Claudio PP. Cell cycle control and beyond: Emerging roles for the retinoblastoma gene family. Oncogene. 2006;25:5201–5209. doi: 10.1038/sj.onc.1209652. [DOI] [PubMed] [Google Scholar]
- 26.Sharpless NE, DePinho RA. p53: Good cop/bad cop. Cell. 2002;110:9–12. doi: 10.1016/s0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 27.Sharpless NE. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Choudhury AR, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- 30.Blasco MA. Telomeres and human disease: Ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 31.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 32.Perkins ND. Not just a CDK inhibitor: Regulation of transcription by p21(WAF1/CIP1/SDI1). Cell Cycle. 2002;1:39–41. [PubMed] [Google Scholar]
- 33.Dimri GP. What has senescence got to do with cancer? Cancer Cell. 2005;7:505–512. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pothof J, et al. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.