Abstract
Trypanosoma brucei is a kinetoplastid flagellate, the agent of human sleeping sickness and ruminant nagana in Africa. Kinetoplastid flagellates contain their eponym kinetoplast DNA (kDNA), consisting of two types of interlocked circular DNA molecules: scores of maxicircles and thousands of minicircles. Maxicircles have typical mitochondrial genes, most of which are translatable only after RNA editing. Minicircles encode guide RNAs, required for decrypting the maxicircle transcripts. The life cycle of T. brucei involves a bloodstream stage (BS) in vertebrates and a procyclic stage (PS) in the tsetse fly vector. Partial [dyskinetoplastidy (Dk)] or total [akinetoplastidy (Ak)] loss of kDNA locks the trypanosome in the BS form. Transmission between vertebrates becomes mechanical without PS and tsetse mediation, allowing the parasite to spread outside the African tsetse belt. Trypanosoma equiperdum and Trypanosoma evansi are agents of dourine and surra, diseases of horses, camels, and water buffaloes. We have characterized representative strains of T. equiperdum and T. evansi by numerous molecular and classical parasitological approaches. We show that both species are actually strains of T. brucei, which lost part (Dk) or all (Ak) of their kDNA. These trypanosomes are not monophyletic clades and do not qualify for species status. They should be considered two subspecies, respectively T. brucei equiperdum and T. brucei evansi, which spontaneously arose recently. Dk/Ak trypanosomes may potentially emerge repeatedly from T. brucei.
Keywords: dourine, mitochondria, protozoa, RNA editing, surra
Trypanosoma brucei is a kinetoplastid flagellate responsible for human sleeping sickness and the ruminant disease nagana in Africa. The mitochondrial (mt) or kinetoplast DNA (kDNA) of trypanosomatids consists of a huge network of interlocked circular DNA molecules of two types: maxicircles and minicircles. The former encode classical mt genes, the transcripts of most of which are rendered translatable only upon the uridine insertion/deletion type of RNA editing. Minicircles encode small RNA molecules termed guide (g) RNAs, necessary for decoding encrypted maxicircle transcripts (1–3).
The kDNA is one of the largest mt genomes known. A sophisticated machinery ensures faithful replication of the kDNA of T. brucei, which is represented by thousands of minicircles (each ≈1.0 kb in size) and dozens of maxicircles (≈23 kb) (4, 5). Because it supplies the substrate gRNAs required for RNA editing, the possibility that T. brucei would lose its kDNA would seem unlikely. Yet a partial or even full loss of kDNA, termed dyskinetoplastidy (Dk) and akinetoplastidy (Ak), occurs in nature and can be induced in the laboratory (6).
T. brucei has a complex life cycle. It proliferates in vertebrates as the bloodstream stage (BS), which relies on glycolysis and has down-regulated mitochondria; in the tsetse fly vector, it exists as the procyclic stage (PS) with a fully active organelle (7). The partial or total loss of kDNA locks the trypanosome in the BS form, because the information in the kDNA network is essential for the PS (2). The reduction of a heteroxenous life cycle to a monoxenous one has had dramatic consequences, such as the elimination of the tsetse vector (or any other similar insect vector) from the life cycle, which paradoxically allowed trypanosomes to leave the African tsetse belt and spread to other continents (8). Recombination or genetic exchange does not occur in Dk/Ak trypanosomes (6, 9).
Dk/Ak trypanosomes are categorized into three species: Trypanosoma equiperdum, Trypanosoma evansi, and Trypanosoma equinum (10). T. equinum has now been synonymized with T. equiperdum (11, 12) and will not be further discussed herein. A widely accepted paradigm holds that T. evansi evolved, via T. equiperdum, when camels infected with T. brucei moved to tsetse-free areas (10). T. equiperdum and T. evansi are responsible for dourine and surra, respectively, economically significant diseases infecting horses, camels, and water buffaloes, transmitted in the BS by bloodsucking insects or coitus (9). Surra is the most widespread and serious disease of camels. Apart from causing different clinical diseases, T. equiperdum (dourine) and T. evansi (surra) differ only in that the former contains at least fragments of kDNA maxicircles (Dk), missing from T. evansi (Ak). Hundreds of studies on the epidemiology, pathology, and prevalence of both diseases have been published (6, 9, 13). Yet the existence of T. equiperdum has been questioned, because no new strains have been isolated for a long time (ref. 13, but see refs. 14 and 15).
We have performed a multifarious characterization of representative strains of T. equiperdum and T. evansi. Our results reveal that both species are strains of T. brucei, which independently lost parts of their kDNA. Neither the Dk nor Ak trypanosomes are monophyletic clades, and thus they would not qualify for species status. We suggest they be considered as subspecies, respectively, T. brucei equiperdum and T. brucei evansi. Our findings provide important insights into the mt functions and reveal the enormous plasticity of T. brucei.
Results
It has been shown that some T. equiperdum strains contain a deletion in their kDNA maxicircle (16–18), whereas ambiguity remained whether other strains have an intact or disrupted maxicircle (19). We first sequenced the maxicircle coding region in two strains of T. equiperdum. The analysis of strain STIB818 (Table 1) confirmed the occurrence of a 9-kb deletion, flanked by truncated 12S rRNA and cytochrome oxidase subunit 1 (cox1) genes; all other retained genes are intact [supporting information (SI) Fig. 8C]. The second strain, STIB842, is a representative of strains with the whole assortment of maxicircle genes, as shown by a PCR-based screen of nine T. equiperdum strains (Table 2; SI Table 3). Sequencing confirmed the presence of all apparently functional maxicircle genes in STIB842 (SI Fig. 8B), with 97% nucleotide identity with T. brucei (SI Fig. 8A). No maxicircle genes could be amplified from three other strains labeled as T. equiperdum (American Type Culture Collection nos. ATCC30023, STIB784, and AnTat 4.1; Table 2), yet the presence of maxicircle genes is the only molecular feature that would distinguish T. equiperdum from T. evansi (6, 9, 12). The absence of the maxicircle genes in all tested T. evansi strains was anticipated (Table 2).
Table 1.
Strains of Trypanosoma spp. investigated in this study
| Strains | Species | Host | Origin | Date |
|---|---|---|---|---|
| 29–13* | T. brucei | Cattle | Tanzania | 1956 |
| EATRO HN | T. brucei | n.a. | n.a. | n.a. |
| STIB 920 | T. brucei | Hartebeest | Tanzania | 1971 |
| BoTat1.1 | T. equiperdum | Horse | Morocco | 1924 |
| STIB 842 | T. equiperdum | n.a. | n.a. | n.a. |
| OVI | T. equiperdum | Horse | South Africa | 1975 |
| STIB 841 | T. equiperdum | n.a. | South Africa | n.a. |
| STIB 818 | T. equiperdum | Horse | China | 1979 |
| ATCC30019 | T. equiperdum | n.a. | France | 1903 |
| ATCC30023 | T. equiperdum | n.a. | France | 1903 |
| AnTat4.1 | T. equiperdum | n.a. | n.a. | n.a. |
| STIB 784 | T. equiperdum | n.a. | n.a. | n.a. |
| STIB 810 | T. evansi | Water buffalo | China | 1985 |
| STIB 805 | T. evansi | Water buffalo | China | 1985 |
| STIB 807 | T. evansi | Water buffalo | China | 1979 |
| CPOgz1 | T. evansi | Water buffalo | China | 2005 |
| GDH | T. evansi | Horse | China | 1964 |
| GDB2 | T. evansi | Water buffalo | China | 1981 |
| GXM | T. evansi | Mule | China | 1979 |
| JSB2 | T. evansi | Water buffalo | China | 1986 |
| AS131M | T. evansi | Horse | Philippines | 2006 |
| SS73M | T. evansi | Water buffalo | Philippines | 2006 |
| SS143M | T. evansi | Water buffalo | Philippines | 2006 |
| Ted3 | T. evansi | Dog | Brazil | 1992 |
| Teh2 | T. evansi | Horse | Brazil | 1996 |
| Teh3 | T. evansi | Horse | Brazil | 1996 |
| RoTat1.2 | T. evansi | Water buffalo | Indonesia | 1982 |
| Stock Kazakh | T. evansi | Camel | Kazakhstan | 1995 |
| Stock Viet | T. evansi | Water buffalo | Vietnam | 1998 |
| KETRI 2479 | T. evansi | Camel | Kenya | 1980 |
n.a., information not available.
*Derived from strain Lister 427.
Table 2.
Presence/absence of maxicircle-encoded genes in selected Trypanosoma spp. strains
| Species | Strains | ND7 | A6 | CO2 | ND5 | 12S rRNA | ND7-Cyb | MURF1-ND1 | ND1-MURF2 | MURF2-CO1 | ND4-ND5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T. brucei | STIB 920 | + | + | + | + | + | + | + | + | + | + |
| T. equiperdum | BoTat1.1 | + | + | + | + | + | + | + | + | + | + |
| T. equiperdum | STIB 842 | + | + | + | + | + | + | + | + | + | + |
| T. equiperdum | OVI | + | + | + | + | + | ND | ND | + | + | ND |
| T. equiperdum | STIB 841 | + | + | + | + | + | + | + | + | + | + |
| T. equiperdum | STIB 818 | − | − | − | + | + | − | − | − | − | + |
| T. equiperdum | ATCC30019 | − | − | − | + | +* | −* | −* | −* | −* | + |
| T. equiperdum | ATCC30023† | − | − | − | − | − | − | − | − | − | − |
| T. equiperdum | STIB 784† | − | − | − | − | − | − | − | − | − | − |
| T. equiperdum | AnTat4.1 | − | − | − | − | − | − | − | − | − | − |
| T. evansi | STIB 810 | − | − | − | − | − | − | − | − | − | − |
| T. evansi | STIB 805† | − | − | − | − | − | − | − | − | − | − |
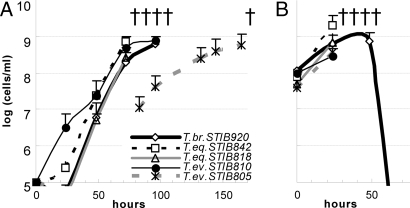
To rule out the possibility that some T. equiperdum strains are mislabeled T. brucei strains, mice were experimentally infected with the T. brucei strain STIB920, T. equiperdum strains STIB818 and STIB842, and T. evansi strains STIB805 and STIB810. With the exception of Ak STIB805, all strains showed very similar dynamics of parasitaemia and invariably killed the mice within 3–4 days (Fig. 1A). To test the ability of the studied trypanosomes to lose kDNA, mice were injected with ethidium bromide (EtdBr) in the early phase of infection. Indeed, all tested Dk strains were prone to the loss: 90% of cells had no detectable kDNA within 48 h (SI Fig. 9B). The absence of kDNA had no effect on pathogenicity, because the parasitaemia progressed equally with and without EtdBr treatment (Fig. 1B). However, the T. brucei STIB920 infection performed with EtdBr treatment was not lethal for the mice, because the parasites were eliminated from the blood (Fig. 1B).
Fig. 1.
Parasitaemia growth curves. (A) Growth curve of parasitaemia caused by representative strains in mice. The cell density in blood is indicated. (B) Growth curve of parasitaemia of the same strains as in A in mice with concurrent i.p. injection of 10 μg of EtdBr per gram of weight.
To corroborate that the studied T. equiperdum are morphologically indistinguishable T. brucei, transformation from the BS to PS was performed in vitro in the SDM-79 medium supplemented with citrate and cis-aconitate (20). Although the transformation of T. brucei STIB920 started within 12 h, and at 60 h virtually all cells had transformed into the PS, no transformation was observed in the Dk strains, which all perished after 4 days (SI Fig. 10).
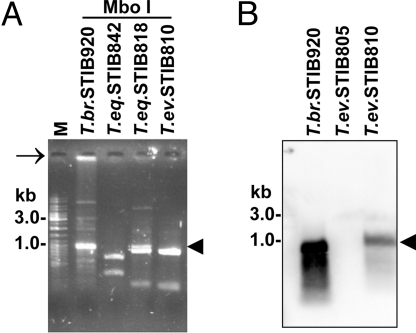
An extensive restriction analysis to assess minicircle complexity revealed sequence heterogeneity of the T. brucei minicircles and their apparent homogeneity in T. equiperdum and T. evansi (Fig. 2A and SI Fig. 11). Note that the T. equiperdum strains with full-size maxicircles (STIB841 and STIB842) (Table 2) exhibit the same extent of minicircle homogenization as other Dk strains (Fig. 2A). The lack of minicircle heterogeneity was confirmed by sequence analysis, because all 14 and 7 AsuII-linearized minicircles of strains STIB818 (class A T. equiperdum) and STIB842 (class B T. equiperdum), respectively, were virtually identical within each strain (SI Fig. 12). The class A minicircles contain three putative gRNA genes, whereas a nonoverlapping set of five gRNAs is encoded by class B minicircles (SI Fig. 11). With the whole T. brucei minicircle population as a template for random hexamer labeling, Southern hybridization confirmed the total absence of minicircles in the Ak strains of T. evansi (Fig. 2B).
Fig. 2.
Minicircle homogenization (A) and/or absence (B). (A) EtdBr-stained agarose gel showing restriction analysis of kDNA minicircles from representative strains using MboI (see also SI Fig. 11). Linearized minicircles are indicated by an arrowhead. Note the presence of kDNA network in the slot of the T. brucei lane (arrow). (B) Southern blotting of total DNA isolated from representative strains and digested with restriction enzyme TaqI. The membrane was hybridized with a radiolabeled probe prepared from total T. brucei minicircles.
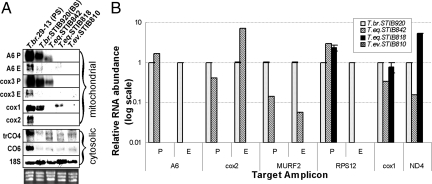
To assess the consequences of the alterations of the mini- and maxicircle components of kDNA on the RNA level, we analyzed the transcription and editing of selected mt transcripts. Using probes specific for preedited, edited, and never-edited mRNAs, we performed Northern blot analysis of ATPase subunit 6 (A6) and cytochrome oxidase subunits 1–3 (cox1, -2, and -3). All probes gave the strongest signal in the PS T. brucei 29–13 cells. The preedited A6 and cox3 and never-edited cox1 mRNAs were also abundant in the BS T. brucei strain STIB920 (Fig. 3A). However, the only mt transcripts detected in the Dk trypanosomes were the preedited A6 and cox3 mRNA in the T. equiperdum strain STIB842 and never-edited cox1 in STIB818, both known to contain the respective genes in their maxicircles (Table 2).
Fig. 3.
Transcript levels (A) and RNA editing (B). (A) Levels of mt and cytosolic transcripts detected by Northern blotting in representative strains of T. brucei, T. equiperdum, and T. evansi. P, preedited; E, edited; A6, ATP synthase subunit 6; cox1–3, cytochrome oxidase subunits 1–3; trCO4, trypanosome cytochrome oxidase subunit 4; CO6, cytochrome oxidase subunit 6; 18S, 18S ribosomal RNA. As a control, the gel was stained with EtdBr to visualize rRNA bands. (B) RNA editing of some mRNAs is affected in the Dk/Ak cells. Real-time PCR analysis of preedited, edited, and never-edited mRNAs, performed in triplicate on cDNAs. For each target amplicon, the relative change in RNA abundance was determined by using cytosolic transcripts of β-tubulin and 18S rRNA (data not shown) as internal references, because their transcription was not affected. The relative abundance of each examined transcript was plotted on a logarithmic scale: 1.0 represents the level in BS of T. brucei STIB920; A6, ATPase subunit 6; cox2, cytochrome oxidase subunit 2; MURF2, maxicircle unknown reading frame 2; RPS12, ribosomal protein S12; never-edited cox1 mRNA; and ND4, NADH-dehydrogenase subunit 4.
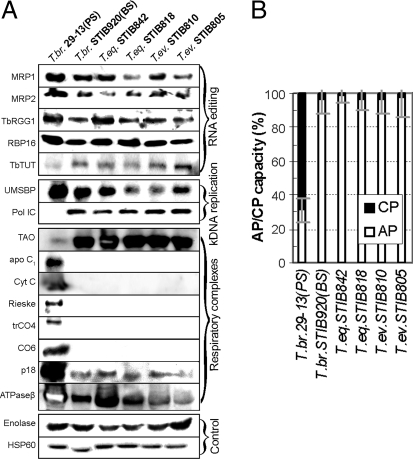
To determine whether the mt mRNAs in the full-size maxicircle strain STIB842 undergo editing, we performed quantitative real-time (q)PCR on cDNA from these cells with primers directed against selected preedited [A6; ribosomal protein S12 (RPS12); cox2; maxicircle unknown reading frame 2 (MURF2)], edited (A6; RPS12; cox2; MURF2), and never-edited transcripts [cox1; NADH-dehydrogenase subunit 4 (ND4) (SI Table 3)]. All reactions were performed in triplicate, including those for determination of the expression of nuclear-encoded β-tubulin and 18S rRNA transcripts, used as references for calculations of the relative abundances of the mt mRNAs. The levels of all examined transcripts in the PS and BS of T. brucei strain 29–13 were as described (2), and the A6, MURF2, RPS12, and cox2 mRNAs were edited (Fig. 3B). However, although the T. equiperdum STIB842 has a full set of maxicircle genes, editing was confined to transcripts that require only maxicircle-encoded gRNAs, namely cox2 and MURF2 (Fig. 3B). The T. equiperdum strain STIB818 has a 9-kb deletion in maxicircle but still retains genes for RPS12, cox1, and ND4; the mRNAs testify to the ongoing transcription of the maxicircle (Fig. 3A). Only preedited RPS12 mRNA was detected in this strain (Fig. 3B). Because missing minicircle-encoded gRNAs have restricted editing in the STIB842 strain to the usage of maxicircle-encoded gRNA genes, we wondered which components of the editing machinery are present in these Dk cells. We confirmed the presence of all tested proteins involved in editing (MRP1, MRP2, TbRGG1, RBP16, and TbTUT1) in the essayed strains, including Ak T. evansi STIB805 (Fig. 4A). Moreover, even in the total absence of minicircles in this strain (Fig. 2B), the minicircle-binding protein UMSBP and the mt DNA polymerase IC are still produced and imported into the organelle (Fig. 4A).
Fig. 4.
Protein levels (A) and respiration (B). (A) Levels of mt and cytosolic proteins detected by Western blotting in strains of T. brucei, T. equiperdum, and T. evansi. MRP1/2, mtRNA-binding proteins 1/2; TbRGG1, editing-associated RGG1 protein; RBP16, RNA-binding protein 16; TbTUT, terminal uridyl transferase; UMSBP, universal minicircle sequence binding protein; POL IC, mt DNA polymerase IC; apo C1, apocytochrome Cyt C1; Cyt C, cytochrome c; Rieske, Rieske Fe-S protein; trCO4, trypanosome cytochrome oxidase subunit 4; CO6, cytochrome oxidase subunit 6; p18, ATP synthase subunit b; ATPase β, ATP synthase subunit β; hsp60, heat shock protein 60. (B) Relative contribution of TAO-mediated pathway (AP) and cytochrome-mediated pathway (CP) in T. brucei PS and BS, and the Dk/Ak strains. The amount of O2 consumption inhibited by KCN was measured as the CP capacity. The mean and the SD values of three experiments are shown.
We tested the presumption that the metabolism of Dk/Ak trypanosomes is similar to that of the BS of T. brucei. Trypanosome alternative oxidase (TAO), which is strongly up-regulated in the BS, was revealed to be similarly abundant in the Dk/Ak cells (Fig. 4A). Antibodies against subunits of ATPase (F1 subunit β and F0 subunit b) recognized their target in all lysates, but this essential complex was down-regulated in T. evansi (Fig. 4A). Proteins confined to the PS T. brucei (see also Northern blot in Fig. 3A), such as the nuclear-encoded cox6, Rieske Fe-S protein, trypanosome-specific subunit of cytochrome oxidase (trCO4), cytochrome c (CytC), and apocytochrome c1 (apoC1), were absent from the BS, regardless of their species affiliation (Fig. 4A). Both insect (PS) and mammalian stages (BS) of T. brucei differ strikingly in their sensitivity to KCN and salicylhydroxamic acid (SHAM), the inhibitors of respiratory complexes and TAO, respectively. The BS of T. brucei STIB920 and the T. equiperdum and T. evansi strains use TAO as their sole electron acceptor (Fig. 4B; SI Fig. 13).
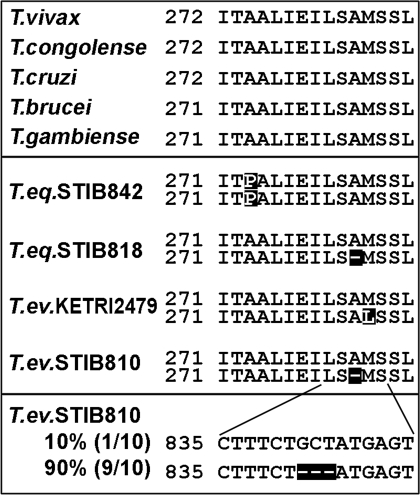
Because the membrane potential was shown to be upheld by F1-ATP synthase in the DK strain 164, which contains a mutated γ subunit to compensate for the loss of the putative A6 subunit (21), we sequenced the gene for the γ subunit in 19 Dk/Ak strains. In the relevant region, which is highly conserved among all known members of the genus Trypanosoma (Fig. 5Top), the Dk/Ak strains show either a deletion or a mutation. With one exception (STIB842), the deletion or mutation was present only in one allele of what is a single-copy gene in T. brucei (Fig. 5 Middle). Sequence analysis of the γ subunit cDNAs in STIB810 confirmed that both alleles are transcribed, but 90% mRNAs are derived from the mutated allele, indicating it is more highly expressed (Fig. 5 Bottom). Membrane potential was virtually identical in all tested Dk/Ak strains and T. brucei, showing that a noncanonically composed ATP synthase (Fig. 4A) is sufficient for its maintenance (SI Fig. 14).
Fig. 5.
Amino acid (Top and Middle) and nucleotide (Bottom) alignment of a highly conserved part of the γ subunit of ATP synthase.
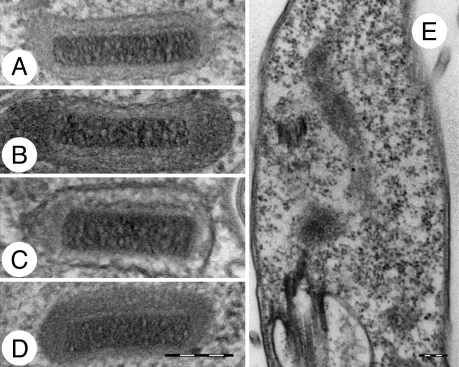
Neither the deletion nor the absence of the maxicircle kDNA has an impact on kinetoplast morphology, which retains its characteristic disk-like structure in the Dk strains (Fig. 6 A–D). The only cells in which electron microscopy failed to detect any kDNA disk were the Ak strains, which completely lack minicircles (Fig. 6E).
Fig. 6.
Electron microscopy of kinetoplasts in fixed cells. (A) T. brucei STIB920, (B) T. equiperdum STIB842, (C) T. equiperdum STIB818, (D) T. evansi STIB810, and (E) T. evansi STIB805. (Scale bar, 200 nm.)
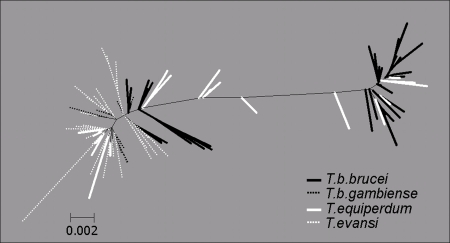
We addressed the controversial issue of taxonomy and species assignment of the Dk/Ak trypanosomes using the spliced leader (SL) RNA gene. We sequenced 59 nonreiterative SL RNA genes with their intergenic regions from 21 Dk/Ak strains. Because pronounced intragenomic variability was confined to the most variable part of the intergenic region, the amplicons could always be unambiguously aligned. Sequences were aligned with a complete set of 26 SL RNA genes from T. brucei 927, incomplete sets of newly sequenced (14 SL RNA sequences), and available SL RNA genes for T. brucei strains 29–13, EATRO-HN and STIB920, and T. brucei gambiense, analyzed by neighbor joining and shown as an unrooted tree (Fig. 7). Because of a limited amount of phylogenetic information in the ≈1.4-kb region, bootstrap support is generally low. However, two distant clusters of the T. brucei SL RNA sequences obtained from the EATRO-HN and 29–13 strains are intermingled with T. equiperdum sequences. The T. evansi sequences are confined to half of the tree but are interspersed with T. equiperdum and T.b. gambiense SL RNA sequences (Fig. 7).
Fig. 7.
Neighbor-joining clustering of the SL RNA repeats from Trypanosoma species. Alignment was performed by using CLUSTALW with gap opening weight = 15 and gap extension weight = 6.6.
Discussion
For >100 years T. brucei, T. equiperdum, and T. evansi have been considered separate species, based on differences in the mode of transmission, host range and pathogenicity, and longstanding understanding that T. equiperdum retains at least a part of maxicircle kDNA, whereas T. evansi completely lost it (6, 9, 12). Our data show that the Dk/Ak strains of T. equiperdum and T. evansi have evolved from T. brucei after losing the capacity to faithfully replicate their kDNA.
RNA editing is essential in T. brucei for both PS and BS (22). However, the selective pressure to retain the full array of minicircle gRNA genes, a prerequisite for fully functional editing, will no longer exist in the absence of the tsetse fly infective stage (PS). This possibility exists because respiratory complexes III and IV are down-regulated in the BS, and many of the mt-encoded genes are subunits of these complexes. A notable exception is the putative A6 subunit of ATP synthase, which is essential for maintaining membrane potential in both PS and BS (21, 23). Its mRNA is edited by ≈20 gRNAs located on numerous kDNA minicircles, whereas hundreds of gRNAs needed for decoding of at least 11 mRNAs in the PS are unnecessary for the BS (2, 3). Therefore, in the mammalian stage, the minicircles may no longer be essential. Although unfaithful kDNA replication in the PS will result in the loss of essential gRNA(s) (24), leading to death, the BS may lose many gRNAs, because most encode information not essential in this stage, with the exception of the A6-specific gRNAs.
The loss of gRNA genes entails the inability of the parasite to transform into the insect PS. Therefore, transmission can only be by haematophagous insects or blood exchange during coitus. The subsequent homogenization of minicircles will lead to the loss of the A6-specific gRNAs, disrupting editing of the still essential A6 subunit. This pressure will likely select cells with a mutation in the γ subunit of ATP synthase, enabling the parasite to compensate for the lack of the A6 subunit, thus allowing survival. Computer simulations of minicircle-sequence class plasticity show that many unnecessary classes may be lost within a few hundred generations, whereas it may take tens of thousands of generations before the last few classes are lost (24), showing how vulnerable trypanosomes are in their vertebrate host to the loss of gRNA genes unnecessary for the BS (see below).
Our data show that the first step toward Dk is the homogenization of minicircles, followed by some deletion in the maxicircles. Because we found no strains with maxicircles that progressively accumulated mutations, we assume that fragility of the mt genome makes it prone to deletions. The maxicircles are thereafter lost altogether, and the demise of kDNA is completed by a total elimination of minicircles in the Ak cells. A consequence of minicircle homogenization is that the mt RNAs cannot be properly edited. Even with the number of minicircle-encoded gRNA genes dropping from several hundreds in T. brucei to only approximately eight in the Dk strains, the protein machinery underlying editing remains intact, using maxicircle-encoded gRNAs. This is the case in the maturation of cox2 and MURF2 mRNAs, which are edited by cis- and trans-acting maxicircle gRNAs, respectively. In the latter case, editing is drastically reduced, presumably because the MURF2 gRNAs are also supplied by minicircle genes. Unexpectedly, components of all of the characterized multiprotein complexes involved in editing are still present in the Ak strains, which lack any nucleic acids in their organelle, a situation mimicked in laboratory-induced Dk T. brucei (25).
RNA editing and kDNA replication are baroque processes, requiring dozens to hundreds of proteins (2, 3, 5). However, it is the unfaithful replication and/or segregation of minicircles that is the trigger of kDNA loss. As with the editosome proteins, replication proteins such as mtDNA polymerase IC and UMSBP remain abundantly present even in the absence of their substrates. (Note that the absence of maxicircles does not impact the structure of the kDNA disk.) Without selective pressure, the now-useless organellar nucleic acids are doomed, with the weird twist that proteins involved in their replication, transcription, and editing remain despite an apparent redundancy. Because not even the Ak strains are devoid of these proteins, dozens of such proteins continue to be synthesized and imported into the DNA-lacking mitochondrion, testifying to a stunning lack of “communication” between the organelle and the nucleus in these primitive flagellates.
This situation may be an indication that the Dk/Ak trypanosomes diverged from T. brucei fairly recently, because they still retain genes rendered unnecessary by the loss of the mt genome. However, another factor may account for the persistence of these unneeded genes. In contrast to other eukaryotes, trypanosome protein-coding genes are organized as polycistronic transcription units (26). Comparative analysis of the genomes of T. brucei, T. cruzi, and Leishmania major has revealed a tendency for maintaining gene order (27). It may be difficult to lose specific editing and kDNA replication/maintenance genes without major rearrangements of the chromosomes.
We have discovered intermediate steps in the process of kDNA loss, which was unexpected because T. brucei is an ancient protist, split from T. cruzi ≈100 Mya (28), whereas the loss of nonessential parts of the kDNA may have happened within decades (24). One would expect T. brucei either with intact kDNA or totally devoid of it. This conundrum can be resolved by postulating a continuous appearance of new strains outside Africa with nonfunctional kDNA that at some point reach the Ak state. The loss of kDNA, combined with the lack of the genetic recombination in the insect vector, amounts to a selective disadvantage of these strains, eventually leading to their elimination and replacement by newly evolved strains following the same path. An alternative explanation would be that humans have caused this process recently, by transporting infected animals out of the African tsetse belt. The transformation into the Dk and eventually the Ak cells enabled these flagellates to spread out of Africa and become the most widespread pathogenic trypanosome (8).
No substantial differences have been found between T. brucei and the Dk/Ak strains other than in the kDNA (9, 12, 29). The SL RNA intergenic region is a multicopy marker suitable for discrimination between closely related trypanosomatid isolates (30, 31). The SL RNA sequences derived from T. equiperdum and T. evansi strains are intermingled with the T. brucei and T.b. gambiense sequences, none of which form a species-specific cluster. The paraphyly of T. equiperdum and T. evansi is further supported by the strain-specific mutations in the highly conserved γ subunit and by the fact that the T. equiperdum strains differ in the last abundant minicircle class. These independent genetic traits suggest that any strain of T. brucei can serve as a source of a Dk/Ak lineage.
The absence of monophyly and the short genetic distance from T. brucei strains imply that the species category is inappropriate for T. equiperdum and T. evansi. The available evidence suggests they are the equivalents in T. brucei of the petite mutants of Saccharomyces cerevisiae and other yeasts. Similarly as the petite mutants, the Dk/Ak cells are respiratory-deficient, have an accelerated mutation rate in the γ subunit of ATP synthase, and a disrupted mt genome, which can be easily and completely lost upon EtdBr treatment (32). In the petite mutants of yeast, it has been postulated that allele-specific mutations in the α and γ subunits result in the formation of an aberrant ATP synthase that can prevent proton leakage, which allows their survival (33). A similar effect of one such mutation has been experimentally confirmed (21).
We propose that the Dk/Ak mutants occur spontaneously and frequently in T. brucei, which is the case for the petite mutants in S. cerevisiae. In both systems, the mutants do not represent a monophyletic assembly but rather strains arising independently and by (slightly) different mechanisms. It would not be practical for the veterinary community to label T. equiperdum and T. evansi as petite mutants of T. brucei. It has been proposed that single or a few gene(s) conferring the ability to occupy a new niche is sufficient to justify the rank of subspecies in a trypanosome (12). We propose that the rank of subspecies may be assigned in the case of the absence, rather than presence, of some gene(s). In any case, T. equiperdum and T. evansi do not constitute monophyletic groups. We propose to rename them T.b. equiperdum and T.b. evansi. Our data strongly suggest that T.b. equiperdum and T.b. evansi represent intermediate stages of the continuous adjustments of T. brucei cells to life without kDNA.
Materials and Methods
Strains, DNA Analysis, and Light and Electron Microscopy.
Trypanosomes were purified from mice by DE52 DEAE cellulose (Whatman) (ref. 34, with slight modifications). The kDNA network was isolated by sucrose gradient ultracentrifugation (35). MspI, MboI, HindIII, EcoRI, BamHI, HhaI, AsuII, TaqI, HaeIII, and RsaI were used for restriction analysis. Linearized mini- and maxicircles were ligated into pBlueScript SK− vector and sequenced (EU155057-EU155060 and EU185799-EU185800). Light and electron microscopy was as in ref. 36.
Southern, Northern, and Western Blot Analyses.
Total DNA digested with TaqI was resolved in agarose gel, blotted, and hybridized with a radiolabeled probe. Total RNA was isolated by using Trireagent (Sigma); 10 μg was loaded on a 1% formaldehyde agarose gel, blotted, and cross-linked. Hybridization with a radiolabeled probe was as in ref. 37. Cell lysates were analyzed on a 12% SDS/PAGE gradient gel, blotted, and probed with a panel of antibodies following conditions described elsewhere (37, 38).
Sequence Analysis of ATP Synthase Subunit γ and SL RNA.
Subunit γ of ATP synthase (EU185790–EU185798) and SL RNA (GeneBank accession nos. EU180634–EU180706) were amplified and sequenced as in refs. 21 and 31.
qPCR.
Total RNA was isolated from cells and treated with the Turbo DNA-free kit (Ambion). Then cDNA was synthesized by using the SuperScript III reverse transcriptase (Invitrogen) with random hexamers. qPCR analysis was as in ref. 39. The relative abundance of RNAs was determined as in ref. 40.
Growth Curve of Strains and Post-Dyskinetoplastic Treatment.
Mice were randomly grouped, and each group of four mice was infected with different strains through i.p. inoculation. Selected mice with parasitemia of ≈108 cells per milliliter were inoculated with 10 μg/g of EtdBr (41). Parasitemia was assayed by counting trypanosomes from tail blood. Blood smears were stained with DAPI to determine the percentage of Dk/Ak cells.
Measurement of Respiration Rate and Membrane Potential.
BS cells were collected via the DE52 column, washed, and resuspended in PSG at 1 × 107 cells per ml. Oxygen consumption and membrane potential of PS cells were as in ref. 37. Conditions for BS cells were adapted: incubation temperature increased to 37°C, inhibitors were added at 1.5-min intervals, and final concentration of SHAM was raised to 0.09 mM.
Transformation of Bloodstream Trypanosomes to PS.
BS cells from infected blood were cultivated as in ref. 20. For the next 4 days, living cells were observed by fluorescence microscopy after 15-min staining with 250 nM Mitotracker red at 27°C.
Supplementary Material
ACKNOWLEDGMENTS.
R. Brun (Swiss Tropical Institute, Basel), F. Claes (Institute of Tropical Medicine, Antwerp, Belgium), A. Dargantes (University of Gent, Merelbeke, Belgium), W. Gibson (University of Bristol, Bristol, U.K.), L. McInnes (Murdoch University, Murdoch, Perth, Western Australia), M. M. G. Teixeira (University of Sao Paulo, Sao Paulo, Brazil), and J.-L. Zhou (Shanghai Veterinary Research Institute, Shanghai, China) kindly provided strains or DNA; and P. T. Englund (Johns Hopkins University, Baltimore), M. D. Ginger (Lancaster University, Lancaster, U.K.), S. L. Hajduk (University of Georgia, Athens), G. C. Hill (Vanderbilt University, Nashville, TN), M. M. Klingbeil (University of Massachusetts, Amherst), P. A. M. Michels (University Catholique Louvain, Brussels), L. K. Read (State University of New York, Buffalo), J. Shlomai (Hebrew University, Jerusalem), L. Simpson (University of California, Los Angeles), and L. Vanhamme (Université Libre Brussels, Gosselies, Belgium) kindly provided antibodies. This work was supported by the Czech Republic (Grant 204/06/1558), the Czech Academy of Sciences (Grant A500960705), the Czech Ministry of Education (Grants LC07032 and 2B06129, to J.L.), and the National Science Foundation of China (Grants 30570245 and 30670275, to Z.-R.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711799105/DC1.
References
- 1.Simpson AGB, Stevens JR, Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Lukeš J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 3.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Shlomai J. The structure and replication of kinetoplast DNA. Curr Mol Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- 5.Liu BY, Liu YN, Motyka SA, Agbo EEC, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Schnaufer A, Domingo GJ, Stuart K. Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol. 2002;32:1071–1084. doi: 10.1016/s0020-7519(02)00020-6. [DOI] [PubMed] [Google Scholar]
- 7.Besteiro S, Barrett MP, Riviere L, Bringaud F. Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol. 2005;21:185–191. doi: 10.1016/j.pt.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Lun Z-R, Desser SS. Is the broad range of hosts and geographical distribution of Trypanosoma evansi attributable to the loss of maxicircle kinetoplast DNA? Parasitol Today. 1995;11:131–133. doi: 10.1016/0169-4758(95)80129-4. [DOI] [PubMed] [Google Scholar]
- 9.Brun R, Hecker H, Lun Z-R. Trypanosoma evansi and T. equiperdum: distribution, biology, treatment and phylogenetic relationship. Vet Parasitol. 1998;79:95–107. doi: 10.1016/s0304-4017(98)00146-0. [DOI] [PubMed] [Google Scholar]
- 10.Hoare CA. The Trypanosomes of Mammals. Oxford: Blackwell Scientific; 1972. [Google Scholar]
- 11.Ventura RM, et al. Molecular and morphological studies of Brazilian Trypanosoma evansi stocks: The total absence of kDNA in trypanosomes from both laboratory stocks and naturally infected domestic and wild mammals. J Parasitol. 2000;86:1289–1298. doi: 10.1645/0022-3395(2000)086[1289:MAMSOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Gibson W. Resolution of the species problem in African trypanosomes. Int J Parasitol. 2007;37:829–838. doi: 10.1016/j.ijpara.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Claes F, Buscher P, Touratier L, Goddeeris BM. Trypanosoma equiperdum: master of disguise or historical mistake? Trends Parasitol. 2005;21:316–321. doi: 10.1016/j.pt.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Li F-J, Lai D-H, Lukeš J, Chen X-G, Lun Z-R. Doubts about Trypanosoma equiperdum strains classed as Trypanosoma brucei or Trypanosoma evansi. Trends Parasitol. 2006;22:55–56. doi: 10.1016/j.pt.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Vanhollebeke B, et al. Human Trypanosoma evansi infection linked to a lack of apolipoprotein L-1. N Engl J Med. 2006;355:2752–2756. doi: 10.1056/NEJMoa063265. [DOI] [PubMed] [Google Scholar]
- 16.Frasch ACC, et al. The kinetoplast DNA of Trypanosoma equiperdum. Biochim Biophys Acta. 1980;607:397–401. doi: 10.1016/0005-2787(80)90150-1. [DOI] [PubMed] [Google Scholar]
- 17.Lun Z-R, Brun R, Gibson W. Kinetoplast DNA and molecular karyotypes of Trypanosoma evansi and Trypanosoma equiperdum from China. Mol Biochem Parasitol. 1992;50:189–196. doi: 10.1016/0166-6851(92)90215-6. [DOI] [PubMed] [Google Scholar]
- 18.Shu HH, Stuart K. Mitochondrial transcripts are processed but are not edited normally in Trypanosoma equiperdum (ATCC 30019), which has kDNA sequence deletion and duplication. Nucleic Acids Res. 1994;22:1696–1700. doi: 10.1093/nar/22.9.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riou GF, Saucier J-M. Characterization of the molecular components in kinetoplast-mitochondrial DNA of Trypanosoma equiperdum. J Cell Biol. 1979;82:248–263. doi: 10.1083/jcb.82.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brun R, Schönenberger M. Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z Parasitenkd. 1981;66:17–24. doi: 10.1007/BF00941941. [DOI] [PubMed] [Google Scholar]
- 21.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnaufer A, et al. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 23.Brown SV, Hosking P, Li JL, Williams N. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot Cell. 2006;5:45–53. doi: 10.1128/EC.5.1.45-53.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson L, Thiemann OH, Savill NJ, Alfonzo JD, Maslov DA. Evolution of RNA editing in trypanosome mitochondria. Proc Natl Acad Sci USA. 2000;97:6986–6993. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domingo GJ, et al. Dyskinetoplastic Trypanosoma brucei contains functional editing complexes. Eukaryot Cell. 2003;2:569–577. doi: 10.1128/EC.2.3.569-577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tschudi C, Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988;7:455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Sayed NM, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 28.Stevens JR, Rambaut A. Evolutionary rate differences in trypanosomes. Infect Genet Evol. 2001;1:143–150. doi: 10.1016/s1567-1348(01)00018-1. [DOI] [PubMed] [Google Scholar]
- 29.Artama WT, Agey MW, Donelson JE. DNA comparisons of Trypanosoma evansi (Indonesia) and Trypanosoma brucei spp. Parasitology. 1992;104:67–74. doi: 10.1017/s0031182000060819. [DOI] [PubMed] [Google Scholar]
- 30.Thomas S, Westenberger SJ, Campbell DA, Sturm NR. Intragenomic spliced leader RNA array analysis of kinetoplastids reveals unexpected transcribed region diversity in Trypanosoma cruzi. Gene. 2005;352:100–108. doi: 10.1016/j.gene.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Maslov DA, Westenberger SJ, Xu X, Campbell DA, Sturm NR. Discovery and barcoding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J Eukaryot Microbiol. 2007;54:57–65. doi: 10.1111/j.1550-7408.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 32.Chen XJ, Clark-Walker GD. The petite mutations in yeasts: 50 years on. Int Rev Cytol. 2000;194:197–237. doi: 10.1016/s0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- 33.Chen XJ, Clark-Walker GD. Specific mutations in α and γ-subunits of F1-ATPase affect mitochondrial genome integrity in the petite-negative yeast Kluyveromyces lactis. EMBO J. 1995;14:3277–3286. doi: 10.1002/j.1460-2075.1995.tb07331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanham SM, Godfrey DG. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cell. Exp Parasitol. 1970;28:521–534. doi: 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Morga D, Englund PT. The structure of replicating kinetoplast DNA networks. J Cell Biol. 1993;123:1069–1079. doi: 10.1083/jcb.123.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yurchenko V, Lukeš J, Xu X, Maslov DA. An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae). J Euk Microbiol. 2006;53:103–111. doi: 10.1111/j.1550-7408.2005.00078.x. [DOI] [PubMed] [Google Scholar]
- 37.Horváth A, et al. Down-regulation of the nuclear-encoded subunits of the complexes III, IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol Microbiol. 2005;58:116–130. doi: 10.1111/j.1365-2958.2005.04813.x. [DOI] [PubMed] [Google Scholar]
- 38.Vondrušková E, et al. RNA Interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- 39.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential Rnase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci USA. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agbe SA, Yielding KL. Effect of verapamil on antitrypanosomal activity of drugs in mice. Acta Trop. 1993;55:11–19. doi: 10.1016/0001-706x(93)90044-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.