Abstract
Acaryochloris marina is a unique cyanobacterium that is able to produce chlorophyll d as its primary photosynthetic pigment and thus efficiently use far-red light for photosynthesis. Acaryochloris species have been isolated from marine environments in association with other oxygenic phototrophs, which may have driven the niche-filling introduction of chlorophyll d. To investigate these unique adaptations, we have sequenced the complete genome of A. marina. The DNA content of A. marina is composed of 8.3 million base pairs, which is among the largest bacterial genomes sequenced thus far. This large array of genomic data is distributed into nine single-copy plasmids that code for >25% of the putative ORFs. Heavy duplication of genes related to DNA repair and recombination (primarily recA) and transposable elements could account for genetic mobility and genome expansion. We discuss points of interest for the biosynthesis of the unusual pigments chlorophyll d and α-carotene and genes responsible for previously studied phycobilin aggregates. Our analysis also reveals that A. marina carries a unique complement of genes for these phycobiliproteins in relation to those coding for antenna proteins related to those in Prochlorococcus species. The global replacement of major photosynthetic pigments appears to have incurred only minimal specializations in reaction center proteins to accommodate these alternate pigments. These features clearly show that the genus Acaryochloris is a fitting candidate for understanding genome expansion, gene acquisition, ecological adaptation, and photosystem modification in the cyanobacteria.
Keywords: comparative microbial genomics, photosynthesis, oxygenic phototrophs, evolution
The cyanobacteria are oxygenic photosynthetic prokaryotes that span a tremendous variety of light-available environments, ranging from hot springs to ice cores, tropical forests to polar tundra, and desert crusts to the open ocean. They play important roles for carbon and nitrogen cycles in each of these environments where they modify their morphology, metabolism, and light-harvesting systems for survival in their respective niches. To date, genomes of 55 cyanobacterial strains in 21 genera have been completed or are under construction (National Center for Biotechnology Information). These cyanobacterial genomes are diverse in size, ranging from 1.66 to 9.1 Mbp. The differences may arise from the expansion or reduction of genome size as the result of adaptation to the light or nutrient conditions in their particular niches. For example, the 1.66-Mbp genome of Prochlorococcus marinus strain CCMP1986 (MED4) seems to contain a minimal set of genes for survival in the stable oligotrophic open ocean (1). In contrast, the 7.21-Mbp genome of Nostoc sp. PCC7120 (also known as Anabaena) is characteristic of the genetic adaptability of filamentous cyanobacteria that thrive under both free-living and symbiotic conditions where they fix nitrogen; form a number of distinct cell morphologies; and grow photoautotrophically, photoheterotrophically, and chemoheterotrophically (2).
The cyanobacterium Acaryochloris marina was found to be the only oxygenic photoautotroph that uses the pigment chlorophyll (Chl) d as the predominant photosynthetic pigment, with only trace amounts of Chl a (3–5). Although A. marina produces phycobiliproteins (PBPs), they do not form phycobilisomes (PBS), a typical structure for cyanobacterial peripheral antenna, but instead accumulate rod-shaped PBP complexes (6). Previous work has shown that chlorophylls in both reaction centers, photosystem (PS)I and PSII, have been replaced by Chl d, which absorbs light with a wavelength up to 30 nm red-shifted from Chl a (7, 8). Although it was anticipated that the Chl d-PSII in A. marina would harvest insufficient energy to cleave water molecules during oxygen evolution, Shevela et al. (9) showed that the redox potential of the A. marina PSII-special pair was within 0.1 eV of that in other cyanobacteria and would thus not present an energetic barrier to water oxidation.
Acaryochloris ecotypes have been found in marine environments in close association with other oxygenic phototrophs such as Prochloron (associate with colonial ascidians) (3, 5, 10), eukaryotic macroalgae (11, 12), and in a microbial mat in the Salton Sea, a saline and highly eutrophic California lake (13). In each environment, the photosynthetically available radiation is likely completely used by organisms that absorb light using Chl a and/or Chl b. By using Chl d, A. marina thrives in these environments with low visible light intensity but high near-infrared intensity where no other photosynthetic organisms absorb strongly. Recent metagenomic analyses indicate that Acaryochloris is also distributed as a member of epilithic and/or endolithic communities in Antarctic rocks and in limestone from a Mayan archeological site in Mexico (14–16). These results clearly show the global population of Acaryochloris species has a range of lifestyles from free-living to symbiotic and marine to terrestrial.
We report here the complete genome sequence of A. marina str. MBIC11017, the first A. marina strain isolated from the Prochloron-dominated colonial ascidian Lissoclinum patella off the tropical coast of the Palau islands (3, 5). This represents a previously uncharacterized genome sequence for a Chl d-containing organism.
Results and Discussion
Genome Properties.
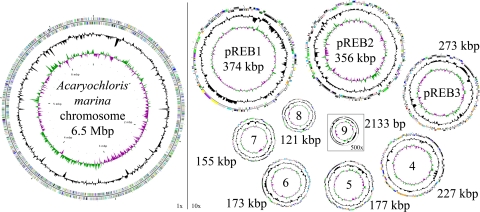
The genome of A. marina was sequenced to completion by a combined pyrosequencing/Sanger approach (see Materials and Methods). It consists of a single circular chromosome and nine distinct plasmids (Table 1 and Fig. 1). The genome has an average G+C content of 47.0% and a coding capacity of 84.1%. The largest category of ORFs (35%) are those that code for hypothetical proteins with little homology to any proteins or motifs. An additional 17% of ORFs code for hypothetical proteins with some conservation among other species. Although this large number of proteins without an assigned function is higher than most sequenced organisms, it is similar to that found in Nostoc sp. PCC7120 (2).
Table 1.
General features of the A. marina genome
| Genome | pREB1 | pREB2 | pREB3 | pREB4 | pREB5 | pREB6 | pREB7 | pREB8 | pREB9 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome size | 6,503,723 | 374,161 | 356,087 | 273,121 | 226,680 | 177,162 | 172,728 | 155,110 | 120,693 | 2,133 | 8,361,598 |
| G + C content | 47% | 47% | 45% | 45% | 46% | 45% | 47% | 46% | 45% | 43% | 47% |
| Open reading frames | 6,342 | 392 | 417 | 384 | 279 | 224 | 192 | 174 | 120 | 4 | 8,528 |
| Pseudogenes | 14 | 5 | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 26 |
| Coding density | 85% | 85% | 84% | 76% | 85% | 82% | 83% | 83% | 78% | 67% | 84% |
| Average gene length | 867 | 814 | 713 | 541 | 693 | 647 | 744 | 737 | 785 | 286 | 824 |
| Ribosomal RNAs | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| Transfer RNAs | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 69 |
| Percent CDS without similarities | 28% | 44% | 50% | 52% | 50% | 66% | 50% | 61% | 67% | 100% | 35% |
| Percent conserved hypothetical | 18% | 15% | 13% | 15% | 14% | 12% | 18% | 17% | 7% | 0% | 17% |
| Insertion elements | 285 | 30 | 28 | 19 | 9 | 27 | 16 | 21 | 51 | 1 | 487 |
| Copy number (approximate) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
Fig. 1.
Circular representation of the A. marina chromosome and plasmids. The different rings represent (from outer to inner) all genes and insertion elements, color-coded by functional category (rings 1 and 2), deviation from average GC content (ring 3), and GC skew (ring 4). All plasmids are represented at ×10 scale for visualization except pREB9 at ×500. Color codes are as follows: turquoise, small-molecule biosynthesis; yellow, central or intermediary metabolism; orange, energy metabolism; red, signal transduction; light blue, DNA metabolism; blue, transcription; purple, protein synthesis/fate; dark green, surface-associated features; gray, miscellaneous features; pink, phage and insertion elements; light green, unknown function; dark gray, conserved hypothetical proteins; black, hypothetical proteins; and brown, pseudogenes.
Although sequence comparisons indicate that none of the plasmids in A. marina arise from very recent duplication events, several of the plasmids do share significant regions of homologous sequence. The largest homology is that between pREB7 and pREB8, with ≈29% of pREB7 (or 38% of pREB8) nucleotides sharing >75% identity. The matching regions range from <100 to >8,500 bp with an average of 1,800 bp. Several plasmids also share a few very large homologous regions (>10 kbp) but do not share a significant global sequence identity. The internal homology between all A. marina plasmids exceeds that seen between its plasmids and those from other bacteria, suggesting that no recent lateral transfer events are responsible for any of the plasmids.
Although all tRNA, rRNA, and ribosomal proteins are encoded by the main chromosome, several of the plasmids code for key metabolic proteins [supporting information (SI) Table 2]. Notable among these are: uridine kinase and aliphatic amidase (pREB2), all phycocyanin (PC)-related genes (pREB3), hoxEFHUWY/hypABCDEF genes for a full complement of bidirectional hydrogenase subunits (pREB4), cobalamin-independent metE methionine synthase and nrdAB ribonucleotide-diphosphate reductase (pREB6) that clearly arose from lateral transfer, and a full second set of genes for an alternate ATP synthase (discussed below). A summary of notable plasmid-encoded proteins can be found in SI Table 2.
The A. marina genome contains a large number of genes for the adaptive regulation of biological activities. Over 170 genes were identified as members of the two-component regulatory system, consisting of sensory kinases and response regulators. Although this number is higher than that in most cyanobacteria, it is still lower than the Nostocales (2). A. marina also contains a number of genes encoding members of the LuxR, LysR, AraC, and TetR transcription factor families. The large number of these regulatory mechanisms indicates that the genome expansion in A. marina is not superfluous but could play a very important role in its adaptation to specialized environmental niches.
Genome Expansion.
The A. marina genome is considerably larger than that in most other single-celled cyanobacteria [see Synechocystis sp. PCC6803 (17)], especially that in other sequenced marine strains [see Prochlorococcus (1, 18) and marine Synechococcus strains (19, 20)]. The high percentage of ORFs with no homologous matches (compared with other sequenced cyanobacteria) suggests that a large part of A. marina codes for either novel proteins or pseudogenes. Large genomes in other bacteria do not tend to accumulate so many hypothetical ORFs (21). A large number of putative transposases (≈350) could account for much of this genetic mobility. Transposase coding regions were in near proximity (1 kb) to predicted transposable regions for <20% of the genes, significantly lower than the 70% found in Nostoc sp. PCC7120 (2). This is partly because of the vast disparity in transposable elements in the two similarly sized genomes, with nearly 1,300 detected in Nostoc and only 500 in A. marina. Surprisingly, a large number of transposases and integrases in A. marina cluster together in regions with no detectable mobile sequences. Further investigation into the mechanisms of genetic mobility in A. marina and other cyanobacteria may clarify this apparent disparity.
Despite the smaller number of transposable elements in A. marina, the genome contains a significant level of duplication. A pairwise comparison of the A. marina chromosome nucleotide sequence (vs. itself) shows 18.7% of the sequence (not including the original, duplicated sequence) has a homology of greater than E = 1×10−10 to another location on the chromosome. This is significantly higher than the 11.2% and 5.8% duplication in Synechocystis PCC6803 and Nostoc sp. PCC7120, respectively. A. marina actually has far fewer duplicated regions than Nostoc, but its contiguous matching regions are much longer, averaging 743 vs. 134 bp. Markov clustering of protein families reveals a similar trend, where 11.6% of the protein families in A. marina contain duplicated copies, accounting for 46.7% of all proteins, considerably higher than Synechocystis and Nostoc at 8.4% (28.1%) and 9.8% (36.1%), respectively.
Another possible influence on the expansion is the presence of duplicate copies of recA, an important multifunctional DNA repair and recombination enzyme found in nearly every organism (reviewed in ref. 22). There are an astounding seven distinct copies of this gene (recA) found in the A. marina genome, far greater than the previously published record of two for Myxococcus xanthus (23). Four of the recA genes are located on four separate plasmids (pREB1, -2, -4, and -5), and the other three are on the main chromosome. Multiple copies of umuD (4x) and umuC (3x), two sets of which are plasmid-encoded, and two putative proteins with a lambda-phage cI motif are also likely correlated with the extra recA (22, 24). The presence of multiples copies of recA and related enzymes could account for gene duplication and/or integration of foreign genes that would lead to genome expansion.
Based on the comparative analysis of many genomes, Konstantinidis et al. (21) hypothesized that species with large genomes may dominate noncompetitive environments where resources are scarce but diverse in nature. This hypothesis appropriately fits the nature of Acaryochloris species. By using far-red light that is not absorbed by other aerobic photoautotrophs Acaryochloris species fill a noncompetitive niche where they are apparently free to specialize their metabolic library.
ATP Synthase.
One interesting case of the idiosyncratic plasmid gene library in A. marina is the inclusion of a second full set of ATP synthase genes on plasmid pREB4 (AM1_D0157-67). These genes are arranged into a unique operon and the individual proteins do not clearly fit into any of the described families (SI Fig. 5) (25). This unusual operon is conserved with full synteny in a remarkable array of organisms, including cyanobacteria, archaea, planctomycetes, chlorobi, and proteobacteria (SI Fig. 5). This alternate ATP synthase was briefly noted in the genome publication for Syntrophus aciditrophicus (26), where it was predicted to be involved in Na+-transport. However, it is clear that this class of ATP synthase does not share any greater degree of similarity to Na+-transport systems in Ilyobacter tartaricus and others (27) than to other ATP synthases. The primary sequences of these proteins are so divergent from other ATP synthases that a thorough biochemical study is needed to confidently propose a functional role.
Chlorophyll Biosynthesis.
The most significant feature of A. marina is its ability to produce Chl d as a major photosynthetic pigment, which accounts for up to 99% of all cellular chlorophyll (28, 29). Molecular and biochemical work has yet to identify any candidate “Chl d synthase” genes. A. marina contains close homologs to all known chlorophyll a biosynthesis genes. The two proteins responsible for the biosynthesis of Chl a from protoporphyrin IX, magnesium-protoporphyrin IX monomethyl ester oxidative cyclase (AcsF) and chlorophyll synthase (ChlG) (30), are highly homologous to those in other cyanobacteria, including a common conserved duplication of acsF. This indicates that Chl d is likely synthesized from chlorophyllide a or Chl a.
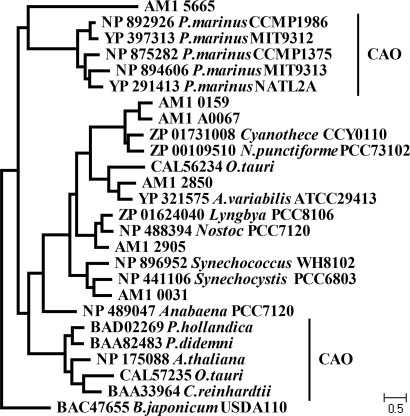
Both Chl b and Chl d contain a formyl side-chain, although at different positions (C-7 and C-3) in the chlorophyll macrocycle. This suggests that a putative Chl d synthase could be a member of the superfamily encompassing a wide range of aromatic ring-degradation proteins including the plant and Prochlorococcus enzymes responsible for the synthesis of Chl b from chlorophyllide a, chlorophyllide a oxygenase (CAO) (Fig. 2) (31). Five putative proteins containing a CAO-type Rieske-FeS motif are encoded by A. marina. Most of the candidate genes in A. marina fall into orthologous clusters with other hypothetical cyanobacterial proteins (Fig. 2). Only one protein, AM1_5665, does not have any significant homologs and diverges early with consistently long branch lengths.
Fig. 2.
Phylogenetic relationship of CAO superfamily proteins constructed using maximum-likelihood. Acaryochloris sequences are listed by their locus tag. Note that the phylogenetic space between Prochlorococcus and plant CAO enzymes includes a number of undescribed proteins implicated in degradation of aromatic ring molecules including chlorophyll.
Anaerobic bacteria use a method of oxygenation using enzymes that proceed via radical chemistry and use S-adenosylmethionine (SAM) to transfer oxygen from water rather than O2 as in aerobic species (32). Such enzymes could provide another means for Chl d biosynthesis. A. marina codes for 12 proteins with putative radical SAM motifs, far more than expected from an oxygen-producing cyanobacterium. Of these 12, two (AM1_5023 and AM1_5798) share very little homology with other sequenced cyanobacteria.
The Chl d biosynthesis pathway may not be as simple as expected. Unlike the formyl side chain of Chl b at C-7 in the chlorin macrocycle, which is derived from a methyl group, the formyl group of Chl d at C-3 is derived from a vinyl group. This loss of a carbon is chemically difficult and could require multiple enzymes for a two-step oxidative cleavage of the double bond. Although it is possible that the means of Chl d synthesis could be completely unrelated to anything familiar in chlorophyll chemistry, it is far more likely that an enzyme has been recruited from related pathways. Major sources of interest are the large pool of proteins orthologous to “Chl degradation” and aromatic ring breakage, which likely provided the origin for CAO, and other poorly understood enzymes, such as the phylogenetically aberrant divinyl chlorophyllide reductase (33), which shows an unusual phylogenetic topology in A. marina (AM1_2394), where it branches near the alphaproteobacteria (SI Fig. 6). This enzyme is of special interest, because A. marina appears to contain a (phylogenetically) normal copy of the newly discovered cyanobacterial-type divinyl reductase (Ayumi Tanaka, personal communication).
Carotenoid Biosynthesis.
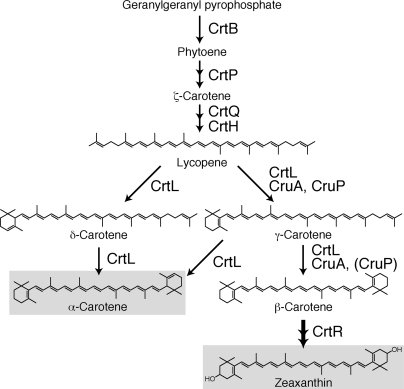
Most cyanobacteria and higher plants contain β-carotene as a primary carotenoid found in both PSI and PSII (Fig. 3). In the case of A. marina, α-carotene was detected instead of β-carotene, and zeaxanthin, an oxidative product of β-carotene, was identified as a major carotenoid (4, 13). A similar carotenoid composition has been reported only in Prochlorococcus species that contain atypical, divinyl-Chls, and α-carotene, β-carotene, and other carotenoids (34). Surprisingly, only α-carotene was detected in the purified PSI and PSII complexes from A. marina (7, 8).
Fig. 3.
A schematic of the putative carotenoid biosynthesis pathway in Acaryochloris. The reaction catalyzed by CrtR passes through the intermediate β-cryptoxanthin.
The absence of β-carotene challenges the established understanding of carotenoid biosynthesis and the role of reaction center carotenoids. Higher plants also synthesize both α- and β-carotene; however, α-carotene is used primarily as a precursor of lutein, and only β-carotene is found in reaction centers. Future biochemical work may indicate whether a preferential interaction between Chl d and α-carotene (rather than β-carotene) could account for its exclusivity in the reaction center.
The chromosome of A. marina codes for 11 proteins predicted to be associated with the biosynthesis of α-carotene and zeaxanthin (SI Table 3). These genes are distributed throughout the chromosome, with no clear operons and no related genes found in plasmid DNA. There are two copies each of crtH, cruA, and crtQ; however, phylogenetic analyses indicate that these genes are more closely related to those in filamentous cyanobacteria than the other α-carotene-containing Prochlorococcus species.
A. marina is the only cyanobacterium found to contain both cruA and cruP, responsible for γ- and β-carotene synthesis, in addition to lycopene cyclase (crtL) (35, 36). One class of cyclase, CrtL-e for ε-cyclase, was identified as the enzyme responsible for α-carotene synthesis in Prochlorococcus (34). However, the A. marina CrtL groups closer to another CrtL found in marine Synechococcus and Prochlorococcus (34, 35) that acts as both a β- and ε-cyclase. In A. marina, this enzyme may catalyze the formation of both α- and β-carotene synthesis (Fig. 3). There is no clear differentiation that would explain the near-complete conversion of β-carotene to zeaxanthin by β-carotene hydroxylase (CrtR), the enzyme responsible for zeaxanthin biosynthesis. We suggest a putative carotene biosynthesis pathway (Fig. 3) that accommodates the distinct complement of the carotenoid biosynthesis enzymes and their end products in A. marina.
Light-Harvesting Systems.
Two major accessory light-harvesting and protection systems exist in cyanobacteria, PBPs (reviewed in ref. 37), and accessory chlorophyll-binding proteins (CBPs) (38–40). Like other “alternative Chl” cyanobacteria, A. marina does not construct the supramolecular PBP assemblies, the PBSs. However, the presence of both PBP aggregates and CBPs has been reported in A. marina (6, 41).
Genes for PBP biosynthesis and assembly in the A. marina genome are summarized in SI Table 4. There are multiple copies of genes encoding PC core and linker proteins (cpcA-cpcG) but no genes for phycoerythrin. Unexpectedly, we found that the only PBP-related genes found on the main chromosome were three copies of apcA and one copy of apcB. All other PBP-related genes were found on plasmid pREB3 in a number of large gene clusters. No apcE gene, which encodes a core-membrane linker peptide, was identified. Phylogenetic analysis suggests that the three copies of cpcG genes in A. marina belong to the CpcG2 group (data not shown), consistent with the fact that A. marina possesses a minor component of allophycocyanin (APC) cores (6).
The detected rod-shaped PBP structures, composed of four ring-shaped subunits, are a likely result of linked PC and/or APC subunits without the core-membrane linker required to construct typical PBS assemblies. The presence of a gene coding for a single rod-core linker (CpcG), which connects a central core to the radiating rods in a PBS, could form the assemblage of PC and APC subunits. It is unlikely that the presence of only α and β subunits of APC is sufficient to form a core structure similar to that in other cyanobacteria. The biochemical identification of all proteins present in PBP assemblies will serve to clarify the role of these peptides in the unique A. marina light-harvesting antenna.
Although most cyanobacteria contain IsiA, a CBP that is produced during iron-deficiency stress, most Prochlorococcus species have a number of other CBPs (Pcb) for additional light harvesting. Although the cloning and characterization of two CBP-like genes was previously reported in A. marina (42), the genome revealed an additional eight CBP genes (for a total of 10) and one isiA that will require further biochemical study.
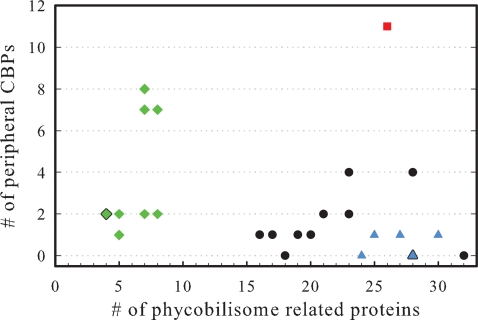
In all cyanobacterial systems studied thus far, the expression of PBP and CBP systems is mutually exclusive. Prochlorococcus, which lack or have only very primitive PBPs, constitutively express their CBP systems (40). Conversely, the expression of the CBPIII (IsiA) is activated only under iron-stress conditions during which PBP expression is strictly repressed (43). The exclusive expression of these systems has evolved into a chasm separating the relative gene content in these species (Fig. 4). Low-light Prochlorococcus strains encode a large number of CBPs (44) and have lost nearly all genes responsible for PBPs. The evolutionarily and environmentally related marine Synechococcus strains encode a large number of PBP-related proteins with very few CBP genes. Only A. marina bucks this trend with its large number of both types of genes that show evidence of concurrent expression (Fig. 4) (6, 45). This indicates that A. marina may be adapted to varying availability of green/orange vs. far-red light. The genetic rift between these two cyanobacterial lifestyles is an intriguing issue that warrants additional investigation.
Fig. 4.
The relationship between the usage of PBPs and accessory CBPs in cyanobacteria. Green diamonds represent Prochlorococcus species, blue triangles represent marine Synechococcus species, a red square represents Acaryochloris, and black circles represent all other cyanobacteria. Points including two or more species are bordered in black.
Photosynthetic Proteins.
A. marina contains a full complement of genes to code for functional cyanobacterial photosystems (SI Table 5). Only genes coding for the small PSI subunits PsaI and PsaX were not detected. Crystallographic analysis suggests that these small single-transmembrane proteins are associated with the stabilization of PSI trimers (46), although a stable trimer was isolated without them in Gloeobacter violaceus (47).
Like several other cyanobacteria, A. marina contains three copies of psbA (the D1 core subunit of PSII) and three copies of psbD (the D2 core subunit of PSII). Two of the psbA sequences (AM1_2166 and AM1_2889) share 97% nucleotide and 100% amino acid identity, whereas the third (AM1_0448) shares only 61% amino acid identity and is more closely related to a divergent PsbA in Anabaena variabilis (YP_324615) and Crocosphaera watsonii (ZP_00515211). Two of the psbD sequences (AM1_1083 and AM1_4084) share 99% nucleotide and 100% amino acid identity, whereas the third (AM1_6045) shares 93% amino acid identity; these likely arose from internal duplications within A. marina. The primary sequence of both D1 and D2 is highly conserved with other cyanobacteria and Chl d likely binds naturally into Chl a-binding sites (48, 49).
Several other genes coding for photosystem-associated proteins have been duplicated in A. marina. Two of four unique copies of psbU (AM1_G0114 and AM1_D0138) are plasmid-encoded, with a protein identity ranging from 36% to 82%. Both psaK and psbE share a conserved duplication found in many other cyanobacteria, with 32% and 68% identity, respectively. Last, all genes responsible for coding cytochrome b6f are present in single copies, with the exception of petH (x3) and petJ (x2).
Conclusions
Although several marine Synechococcus and Prochlorococcus species have been sequenced (1, 18), there is a paucity of genome information for marine cyanobacteria belonging to the traditional “cyanophyte” clade. Our completion of the genome sequence of A. marina, a marine organism with a novel epiphytic/mutualistic lifestyle and a unique light-acclimation method (in Chl d), provides this genetic infrastructure to understand the relationship between marine cyanobacterial species. The number of surprising features, ranging from genome size and gene duplication to recA copy number and light-harvesting protein complement, accentuates the need for complete sequence coverage. Because the incorporation of Chl d in A. marina has eliminated a major source of competitive stress, its expansive genome serves as a model for understanding the relationship between metabolic capacity and niche adaptation.
Materials and Methods
DNA Preparation.
A. marina was grown as described (29) on 1.5% agar plates. Genomic DNA was isolated by using the U.S. Department of Energy Joint Genome Initiative “DNA Isolation Bacterial CTAB Protocol” (http://my.jgi.doe.gov/general/index.html).
Genome Sequencing.
The A. marina genome was sequenced by using a pyrosequencing approach on a Roche-454 GS20 instrument at 454 Life Sciences. Approximately 24-fold sequence coverage was achieved from 1,860,686 short sequence reads. A total of 775 contigs were assembled from these reads representing 7,570,764 nonredundant bases. This assembly was supplemented with reads generated by a traditional Sanger sequencing strategy (50). Briefly, genomic DNA was fragmented by kinetic shearing, and three shotgun libraries were generated: small and medium insert libraries in pOTWI3 (using size fractions of 2–3 and 6–8 kb, respectively) and a large insert fosmid library in pEpiFOS-5 (insert sizes ranging from 28 to 47 kb), which was used as a scaffold. The relative amount of sequence coverage obtained from the small, medium, and large insert libraries was 4.4X, 0.6X, and 0.6X, respectively. A total of 56,419 end sequences derived from these libraries were generated by using dye-terminator chemistry on Applied Biosystems 3730xl automated sequencers. The combined 454 and Sanger sequence datasets were assembled with the program Arachne and finished as described (50).
Phylogenetic Analysis.
Homology matching was performed by using the Basic Local Alignment Search Tool (BLAST) (51), and neighbor-joining (NJ) trees were constructed by using MEGA 4.0 (52). Bootstrapped NJ trees (1,000×) were constructed by using a JTT substitution model. To generate protein families, Markov clustering (53) was performed iteratively on a matrix generated from BLAST e values with an inflation parameter of 2.8.
Annotation.
Initial automated annotation of the genome was performed with the TIGR/JCVI Annotation Engine (www.tigr.org/AnnotationEngine), where it was processed by TIGR's prokaryotic annotation pipeline using gene finding with Glimmer, SignalP predictions, Blast-extend-repraze (BER), HMM, and TMHMM searches. Automatic annotations were created by AutoAnnotate. Manatee (manatee.sourceforge.net) was used to manually review and confirm the annotation of all genes. Pseudogenes contained one or more mutations that would ablate expression; each mutation was confirmed by using the original sequencing data. The circular genome map was created by using the program CGView (54).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Paul Stothard for assistance with the generation of the circular genome and plasmid maps, Brendon Hill at 454 Life Sciences for coordination of 454 sequencing, and Jonathan Eisen for helpful discussions. This work was supported by the U.S. National Science Foundation Phototrophic Prokaryotes Sequencing Project Grant 0412824 and by a Grant-in-Aid for Creative Scientific Research (no. 17GS0314) from the Japanese Society for Promotion of Science. W.D.S. is funded by the Japanese Society for Promotion of Science Postdoctoral Fellowship for Foreign Researchers (no. P07141). M.C. is funded by an Australian Research Council Discovery Project (DP0665169). Partial support for the participation of P.C.C. and L.E.K. in this research was provided by a grant to Washington University from the Howard Hughes Medical Institute through the Undergraduate Biological Sciences Education Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank databases [accession nos. CP000828 (the main chromosome of A. marina) and CP000838, CP000839, CP000840, CP000841, CP000842, CP000843, CP000844, CP000845, and CP000846 (the nine plasmids, pREB1–pREB9)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0709772105/DC1.
References
- 1.Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, et al. Proc Natl Acad Sci USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, et al. DNA Res. 2001;8:205–213. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 3.Miyashita H, Ikemoto H, Kurano N, Miyachi S, Chihara M. J Phycol. 2003;39:1247–1253. [Google Scholar]
- 4.Miyashita H, Adachi K, Kurano N, Ikemoto H, Chihara M, Miyachi S. Plant Cell Physiol. 1997;38:274–281. [Google Scholar]
- 5.Miyashita H, Ikemoto H, Kurano N, Adachi K, Chihara M, Miyachi S. Nature. 1996;383:402. [Google Scholar]
- 6.Marquardt J, Senger H, Miyashita H, Miyachi S, Morschel E. FEBS Lett. 1997;410:428–432. doi: 10.1016/s0014-5793(97)00631-5. [DOI] [PubMed] [Google Scholar]
- 7.Tomo T, Okubo T, Akimoto S, Yokono M, Miyashita H, Tsuchiya T, Noguchi T, Mimuro M. Proc Natl Acad Sci USA. 2007;104:7283–7288. doi: 10.1073/pnas.0701847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Q, Miyashita H, Iwasaki II, Kurano N, Miyachi S, Iwaki M, Itoh S. Proc Natl Acad Sci USA. 1998;95:13319–13323. doi: 10.1073/pnas.95.22.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shevela D, Noring B, Eckert HJ, Messinger J, Renger G. PhysChemChemPhys. 2006;8:3460–3466. doi: 10.1039/b604389e. [DOI] [PubMed] [Google Scholar]
- 10.Kuhl M, Chen M, Ralph PJ, Schreiber U, Larkum AWD. Nature. 2005;433:820. doi: 10.1038/433820a. [DOI] [PubMed] [Google Scholar]
- 11.Ohkubo S, Miyashita H, Murakami A, Takeyama H, Tsuchiya T, Mimuro M. Appl Environ Microbiol. 2006;72:7912–7915. doi: 10.1128/AEM.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami A, Miyashita H, Iseki M, Adachi K, Mimuro M. Science. 2004;303:1633. doi: 10.1126/science.1095459. [DOI] [PubMed] [Google Scholar]
- 13.Miller SR, Augustine S, Le Olson TL, Blankenship RE, Selker J, Wood AM. Proc Natl Acad Sci USA. 2005;102:850–855. doi: 10.1073/pnas.0405667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de los Rios A, Grube M, Sancho LG, Ascaso C. FEMS Microbiol Ecol. 2007;59:386–395. doi: 10.1111/j.1574-6941.2006.00256.x. [DOI] [PubMed] [Google Scholar]
- 15.McNamara C, Perry T, Bearce K, Hernandez-Duque G, Mitchell R. Microb Ecol. 2006;51:51–64. doi: 10.1007/s00248-005-0200-5. [DOI] [PubMed] [Google Scholar]
- 16.Smith MC, Bowman JP, Scott FJ, Line MA. Antarct Sci. 2000;12:177–184. [Google Scholar]
- 17.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 18.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 19.Palenik B, Ren Q, Dupont CL, Myers GS, Heidelberg JF, Badger JH, Madupu R, Nelson WC, Brinkac LM, Dodson RJ, et al. Proc Natl Acad Sci USA. 2006;103:13555–13559. doi: 10.1073/pnas.0602963103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, Lamerdin J, Regala W, Allen EE, McCarren J, et al. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 21.Konstantinidis KT, Tiedje JM. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KC. BioEssays. 2004;26:1322–1326. doi: 10.1002/bies.20109. [DOI] [PubMed] [Google Scholar]
- 23.Norioka N, Hsu MY, Inouye S, Inouye M. J Bacteriol. 1995;177:4179–4182. doi: 10.1128/jb.177.14.4179-4182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham P, Rangarajan S, Woodgate R, Goodman MF. Proc Natl Acad Sci USA. 2001;98:8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross RL, Muller V. FEBS Lett. 2004;576:1–4. doi: 10.1016/j.febslet.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 26.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, Sieber J, Struchtemeyer CG, Bhattacharyya A, Campbell JW, et al. Proc Natl Acad Sci USA. 2007;104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 28.Gloag RS, Ritchie RJ, Chen M, Larkum AWD, Quinnell RG. Biochim Biophys Acta. 2007;1767:127–135. doi: 10.1016/j.bbabio.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Swingley WD, Hohmann-Marriott MF, Le Olson T, Blankenship RE. Appl Environ Microbiol. 2005;71:8606–8610. doi: 10.1128/AEM.71.12.8606-8610.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beale SI. Photosynth Res. 1999;60:43–73. [Google Scholar]
- 31.Tanaka A, Ito H, Tanaka R, Tanaka NK, Yoshida K, Okada K. Proc Natl Acad Sci USA. 1998;95:12719–12723. doi: 10.1073/pnas.95.21.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Layer G, Heinz DW, Jahn D, Schubert WD. Curr Opin Chem Biol. 2004;8:468–476. doi: 10.1016/j.cbpa.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Nagata N, Tanaka R, Satoh S, Tanaka A. Plant Cell. 2005;17:233–240. doi: 10.1105/tpc.104.027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stickforth P, Steiger S, Hess WR, Sandmann G. Arch Microbiol. 2003;179:409–415. doi: 10.1007/s00203-003-0545-4. [DOI] [PubMed] [Google Scholar]
- 35.Maresca JA, Graham JE, Wu M, Eisen JA, Bryant DA. Proc Natl Acad Sci USA. 2007;104:11784–11789. doi: 10.1073/pnas.0702984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunningham FX, Sun ZR, Chamovitz D, Hirschberg J, Gantt E. Plant Cell. 1994;6:1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adir N. Photosynth Res. 2005;85:15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Zhang Y, Blankenship RE. Photosynth Res. 2008;95:147–154. doi: 10.1007/s11120-007-9255-0. [DOI] [PubMed] [Google Scholar]
- 39.Chen M, Bibby TS. Photosynth Res. 2005;86:165–173. doi: 10.1007/s11120-005-1330-9. [DOI] [PubMed] [Google Scholar]
- 40.Bibby TS, Nield J, Chen M, Larkum AWD, Barber J. Proc Natl Acad Sci USA. 2003;100:9050–9054. doi: 10.1073/pnas.1532271100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen M, Quinnell RG, Larkum AW. FEBS Lett. 2002;514:149–152. doi: 10.1016/s0014-5793(02)02315-3. [DOI] [PubMed] [Google Scholar]
- 42.Chen M, Hiller RG, Howe CJ, Larkum AWD. Mol Biol Evol. 2005;22:21–28. doi: 10.1093/molbev/msh250. [DOI] [PubMed] [Google Scholar]
- 43.Michel KP, Pistorius EK. Physiol Plant. 2004;120:36–50. doi: 10.1111/j.0031-9317.2004.0229.x. [DOI] [PubMed] [Google Scholar]
- 44.Garczarek L, Hess WR, Holtzendorff J, van der Staay GWM, Partensky F. Proc Natl Acad Sci USA. 2000;97:4098–4101. doi: 10.1073/pnas.070040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M, Bibby TS, Nield J, Larkum AWD, Barber J. FEBS Lett. 2005;579:1306–1310. doi: 10.1016/j.febslet.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 46.Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 47.Inoue H, Tsuchiya T, Satoh S, Miyashita H, Kaneko T, Tabata S, Tanaka A, Mimuro M. FEBS Lett. 2004;578:275–279. doi: 10.1016/j.febslet.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Itoh S, Mino H, Itoh K, Shigenaga T, Uzumaki T, Iwaki M. Biochemistry. 2007;46:12473–12481. doi: 10.1021/bi7008085. [DOI] [PubMed] [Google Scholar]
- 49.Chen M, Eggink LL, Hoober JK, Larkum AWD. J Am Chem Soc. 2005;127:2052–2053. doi: 10.1021/ja043462b. [DOI] [PubMed] [Google Scholar]
- 50.Swingley WD, Sadekar S, Mastrian SD, Matthies HJ, Hao J, Ramos H, Acharya CR, Conrad AL, Taylor HL, Dejesa LC, et al. J Bacteriol. 2007;189:683–690. doi: 10.1128/JB.01390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Tamura K, Dudley J, Nei M, Kumar S. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 53.Enright AJ, Van Dongen S, Ouzounis CA. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stothard P, Wishart DS. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.