Abstract
Lung cancers caused by activating mutations in the epidermal growth factor receptor (EGFR) are initially responsive to small molecule tyrosine kinase inhibitors (TKIs), but the efficacy of these agents is often limited because of the emergence of drug resistance conferred by a second mutation, T790M. Threonine 790 is the “gatekeeper” residue, an important determinant of inhibitor specificity in the ATP binding pocket. The T790M mutation has been thought to cause resistance by sterically blocking binding of TKIs such as gefitinib and erlotinib, but this explanation is difficult to reconcile with the fact that it remains sensitive to structurally similar irreversible inhibitors. Here, we show by using a direct binding assay that T790M mutants retain low-nanomolar affinity for gefitinib. Furthermore, we show that the T790M mutation activates WT EGFR and that introduction of the T790M mutation increases the ATP affinity of the oncogenic L858R mutant by more than an order of magnitude. The increased ATP affinity is the primary mechanism by which the T790M mutation confers drug resistance. Crystallographic analysis of the T790M mutant shows how it can adapt to accommodate tight binding of diverse inhibitors, including the irreversible inhibitor HKI-272, and also suggests a structural mechanism for catalytic activation. We conclude that the T790M mutation is a “generic” resistance mutation that will reduce the potency of any ATP-competitive kinase inhibitor and that irreversible inhibitors overcome this resistance simply through covalent binding, not as a result of an alternative binding mode.
Keywords: lung cancer, tyrosine kinase, x-ray crystallography
Mutations in the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) have recently been identified as a cause of nonsmall cell lung cancer (1–7). The most common oncogenic mutations are small, in-frame deletions in exon 19 and a point mutation that substitutes Leu-858 with arginine (L858R). These mutations likely cause constitutive activation of the kinase by destabilizing the autoinhibited conformation (8, 9), which is normally maintained in the absence of ligand stimulation. Importantly, the activating mutations have also been found to confer sensitivity to the small molecule tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib (1–3). As first reported by Carey et al. (10) in studies with erlotinib, the mutant kinases bind the inhibitors more tightly than does the WT EGFR and additionally the deletion and L858R mutations markedly decrease the affinity of the kinase for ATP (8, 10), with which the inhibitors compete for binding. These two effects combine to yield the remarkable potency of gefitinib and erlotinib against tumors and cell lines that are “addicted” to the activated EGFR for survival (5, 11, 12).
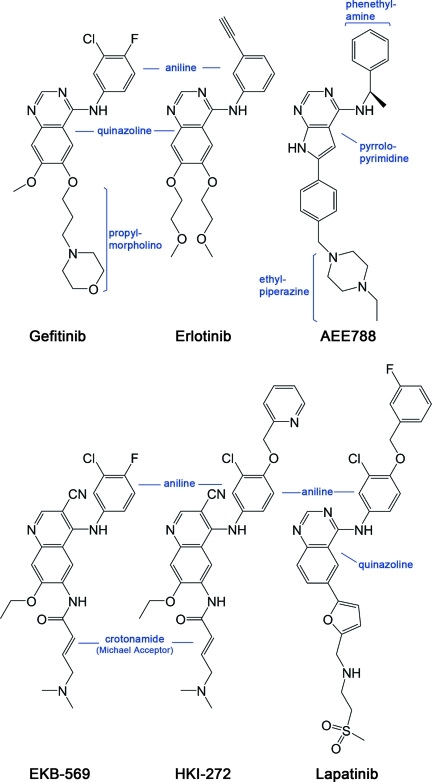
Clinically, the efficacy of these TKIs is often of limited duration because of the emergence of drug resistance conferred by a second mutation: substitution of threonine 790 with methionine (T790M) (13–15). The T790M mutation accounts for about half of all resistance to gefitinib and erlotinib (16, 17). Threonine 790 is the gatekeeper residue in EGFR, so named because its key location at the entrance to a hydrophobic pocket in the back of the ATP binding cleft makes it an important determinant of inhibitor specificity in protein kinases. Substitution of this residue in EGFR with a bulky methionine has been thought to cause resistance by steric interference with binding of TKIs, including gefitinib and erlotinib (13–15). However, the T790M mutant kinase remains sensitive to irreversible inhibitors, including CL-387,785, EKB-569, and HKI-272 (14, 15, 18–20). These compounds closely resemble the reversible anilinoquinazoline inhibitors, but contain a reactive Michael-acceptor group that forms a covalent bond with Cys-797 at the edge of the ATP-binding cleft (Fig. 1). The irreversible inhibitors are designed to target only this cysteine in EGFR because of their specific noncovalent interactions in the ATP binding pocket, which resemble those of reversible anilinoquinazoline compounds. Thus the fact that these irreversible TKIs still inhibit the T790M mutant is at odds with steric hindrance as a mechanism of resistance: the reversible inhibitor gefitinib and the irreversible inhibitor EKB-569 have identical aniline substituents that are expected to bind in the gatekeeper pocket (Fig. 1), so the same steric effects that block gefitinib binding should also prevent the initial binding of EKB-569 (and of the related compound HKI-272).
Fig. 1.
Chemical structures of selected EGFR inhibitors. All compounds are drawn in a consistent orientation and conformation that reflects their approximate binding mode in the EGFR kinase. HKI-272 and EKB-569 are examples of irreversible inhibitors. Lapatinib and HKI-272 are thought to require the inactive conformation of EGFR for binding because of their additional aniline substitutions.
A number of observations indicate that in addition to conferring drug resistance, the gatekeeper mutation may derepress the catalytic activity of EGFR and other kinases. A germ-line T790M mutation has been discovered in a family with a hereditary predisposition to lung cancer, suggesting that this mutation confers a growth advantage in the absence of the selective pressure of TKIs (21). Consistent with this idea, introduction of the T790M in tandem with the L858R mutant in NIH 3T3 cells increases EGFR activity and enhances the transformed phenotype (22). Transgenic mice engineered with lung-specific expression of the T790M mutant develop lung adenocarcinomas (23), albeit with a longer latency than those harboring the L858R or combined L858R and T790M mutations (23, 24). The EGFR T790M mutation was also identified in an untreated case of Barrett's esophagus and the corresponding adenocarcinoma (25). Interestingly, the corresponding mutation in BCR-Abl (T315I) confers resistance to imatinib and other TKIs in the treatment of chronic myelogenous leukemia and has also been found to preexist in untreated CML (26, 27). The equivalent mutation is found in v-Src (T338I) and has long been known to confer transforming activity on c-Src (28). Despite the long history of interest in this key residue in control of tyrosine kinase activity, a structural understanding of its effects is lacking. To better understand its role in inhibitor resistance and kinase deregulation, we have studied the structural and enzymological effects of the T790M mutation in the context of both the WT and the L858R-mutant EGFR kinases.
Results
T790M Mutants Bind Gefitinib with Low Nanomolar Affinity.
We first measured binding of gefitinib to the WT, L858R, T790M, and L858R/T790M mutants by using a direct binding assay in which intrinsic fluorescence of EGFR is quenched by titration with the inhibitor (8). Strikingly, the T790M mutation only modestly affects binding of gefitinib in the context of the L858R mutant (Table 1). The resistant L858R/T790M double-binds gefitinib with Kd = 10.9 nM, which is only ≈4-fold weaker than the exquisitely sensitive L858R mutant (Kd = 2.4 nM). The T790M mutant binds gefitinib with Kd = 4.6 nM, nearly as tightly as the L858R mutant and considerably tighter than the WT kinase. The small difference in gefitinib affinity caused by introduction of the secondary T790M mutation is in stark contrast to the roughly two orders of magnitude differences observed in the sensitivity of cell lines bearing the L858R vs. L858R/T790M or exon 19 deletions with T790M mutations (13–15, 29), and therefore cannot explain the clinically observed drug resistance. We also examined binding of the pyrrolopyrimidine compound AEE788 (Novartis Pharmaceuticals), which binds in a manner similar to gefitinib despite the difference in chemical scaffold (8). The T790M mutant has a more dramatic effect on the affinity for AEE788, but notably the L858R/T790M double mutant retains 18.6 nM affinity for this compound (as compared with Kd = 1.1 nM for the L858R mutant). The larger effect on AEE788 as compared with gefitinib is not unexpected because the phenethylamine substituent on this inhibitor extends further into the hydrophobic pocket that is “guarded” by the gatekeeper residue (8).
Table 1.
Inhibitor dissociation constants for the WT and mutant EGFR kinases
| Kinase |
Kd, nM |
Kd /Km[ATP], ×10−3 |
||
|---|---|---|---|---|
| Gefitinib | AEE788 | Gefitinib | AEE788 | |
| WT | 35.3 ± 0.4 | 5.3 ± 0.3 | 6.8 | 1.0 |
| T790M | 4.6 ± 0.1 | 27.6 ± 0.7 | 0.78 | 4.7 |
| L858R | 2.4 ± 0.1 | 1.1 ± 0.1 | 0.016 | 0.0074 |
| L858R/T790M | 10.9 ± 0.6 | 18.6 ± 0.5 | 1.3 | 2.2 |
The ratio Kd /Km[ATP] provides a relative estimate of inhibitor potency.
Crystal Structures of T790M Mutant.
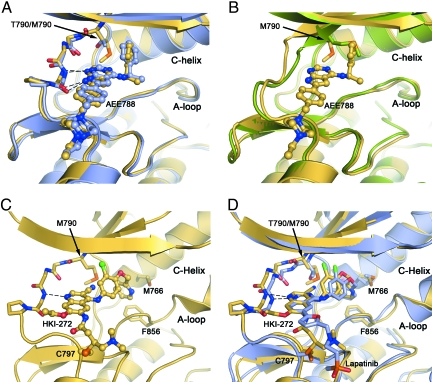
Crystal structures of the T790M mutant show how inhibitors are accommodated in the presence of the gatekeeper mutation, in both the active and inactive conformations of the kinase. We determined structures of the T790M mutant alone and in complex with the irreversible inhibitor HKI-272 in the inactive conformation or in complex with AEE788 in the active conformation [see supporting information (SI) Table 3 for crystallographic statistics]. The structure of the T790M mutant in complex with AEE788 is shown in Fig. 2A. The compound binds in essentially the same manner observed in the WT enzyme, with the pyrrolopyrimidine core making two hydrogen bonds with the hinge region of the kinase and the phenethylamine substituent extending into the gatekeeper hydrophobic pocket. Comparison with the binding of AEE788 to the WT enzyme reveals only a small rotation of the phenethylamine substituent, which is in direct contact with the mutant gatekeeper residue. Comparison of the AEE788 complex with the structure of the T790M mutant in the absence of inhibitor (Apo-T790M) shows that the Met-790 side chain must adopt a different rotamer to accommodate the inhibitor (Fig. 2B).
Fig. 2.
Crystal structures of the EGFR T790M mutant show that inhibitors are readily accommodated in the active and inactive conformations of the kinase. (A) Superposition of EGFR T790M/AEE788 complex (yellow) and WT/AEE788 complex [light blue; drawn from PDB ID code 2J6M (8)]. Dashed lines indicate hydrogen bonds to the kinase hinge region that are preserved in both complexes. The location of the T790M mutation is indicated. (B) Superposition of EGFR T790M/AEE788 complex (yellow) and apo-T790M structure (green). Note the alternate side-chain conformation of Met-790 in the presence of the inhibitor. (C) Crystal structure of HKI-272 in complex with the T790M mutant. The kinase adopts an inactive conformation, with the C-helix displaced. A covalent bond is formed between Cys-797 and the crotonamide Michael acceptor of HKI-272. (D) The structure of the T790M mutant in complex with HKI-272 (yellow) is superimposed on the structure of the WT EGFR kinase in complex with Lapatinib [light blue; drawn from PDB ID code 1XKK (32)]. In both structures, the kinase adopts the same inactive conformation and the inhibitors bind in a similar manner, with a single hydrogen bond to the hinge (dashed lines) and with their aniline substituents extending into the enlarged hydrophobic pocket that is characteristic of the inactive conformation.
The irreversible inhibitor HKI-272 is a 4-(arylamino)quinoline-3-carbonitrile compound and a potent inhibitor of both EGFR and ErbB2 kinases (14, 30). In complex with HKI-272, the EGFR kinase adopts an inactive conformation in which the regulatory C-helix is displaced from its active position (Fig. 2C). The enlarged hydrophobic pocket created by the outward rotation of the C-helix appears to be required to accommodate the bulky aniline substituent found in HKI-272. Both HKI-272 and lapatinib contain additional aromatic groups appended to the aniline ring (a 2-pyridinyl group in HKI-272 and a fluorophenyl group in lapatinib; Fig. 1). Thus it is not surprising that HKI-272, like lapatinib, binds the inactive conformation of the kinase and that the overall binding mode of the two compounds is similar (Fig. 2D). The quinoline core of HKI-272 forms a single hydrogen bond with the hinge region of the kinase in a manner analogous to anilinoquinazoline compounds (31, 32). The 2-pyridinyl group of HKI-272 is surrounded by hydrophobic residues in the expanded pocket, including Met-766 in the C-helix, Phe-856, and Met-790, the mutant gatekeeper residue. The nitrile substituent of HKI-272 also approaches the gatekeeper residue (extending to ≈3 Å from the methionine side chain). In addition to these noncovalent interactions of HKI-272, the expected covalent bond is formed between Cys-797 at the edge of the active site cleft and the crotonamide Michael-acceptor group on the inhibitor, rendering binding irreversible (Fig. 2C). Although the resolution of the structure is modest, electron density for the inhibitor and for the covalent bond is clear (SI Fig. 4).
The structure of the T790M mutant also suggests a possible mechanism of catalytic activation. We hypothesize that the mutation facilitates interconversion between the inactive and active conformations via direct interaction with the Asp-Phe-Gly sequence (DFG motif) at the base of the kinase activation loop (see SI Figs. 5 and 6 and related discussion in SI Text). The mutation may also enhance the stability of the active conformation (relative to the inactive), as it makes favorable hydrophobic interactions with Met-766 and Leu-777 in the active state.
Increased ATP Affinity of the L858R/T790M Mutant Confers Drug Resistance.
The binding data and crystal structures clearly demonstrate that the gatekeeper mutation does not sterically block binding of reversible inhibitors. Why then does the T790M mutation confer resistance? Kinetic characterization of the WT and mutant EGFR kinases reveals a marked decrease in the Michaelis-Menten constant (Km) for ATP in the drug-resistant L858R/T790M mutant as compared with the drug-sensitive L858R mutant (Table 2). As described (8, 10), the L858R mutant activates EGFR, but also reduces the apparent affinity for ATP (Table 2). Strikingly, the T790M mutation restores the ATP affinity to near WT levels in the L858R/T790M double mutant (Km[ATP] = 8.4 μM, as compared with Km[ATP] = 148 μM for the L858R mutant). In isolation, the T790M mutation does not significantly affect ATP affinity. We cannot explain structurally why the T790M mutation increases ATP affinity in the context of the L858R mutant, but not in the context of the WT enzyme.
Table 2.
Enzyme kinetic parameters of WT and mutant EGFR kinases
| Kinase | Km[ATP], μM | kcat, s−1 | kcat/Km[ATP],μM−1·s−1 |
|---|---|---|---|
| WT | 5.2 ± 0.2 | 0.026 | 5.00E-3 |
| T790M | 5.9 ± 0.1 | 0.137 | 2.32E-2 |
| L858R | 148 ± 4 | 1.484 | 1.00E-2 |
| L858R/T790M | 8.4 ± 0.3 | 0.456 | 5.43E-2 |
We also find that the T790M mutation activates the kinase ≈5-fold as compared with the WT enzyme (Table 2); this catalytic activation of the T790M mutant likely explains its presence as a germ-line mutation in a family predisposed to lung cancer (21). Although the L858R/T790M mutant has a modestly decreased kcat relative to the L858R mutant, it is still much more active than the WT enzyme and also exhibits a 5-fold higher kcat/Km[ATP] than the L858R mutant (Table 2).
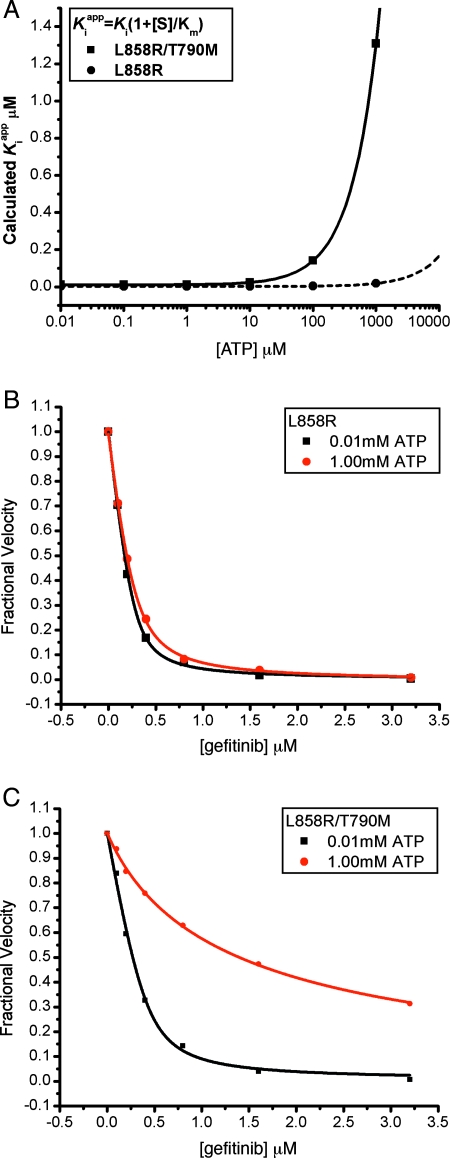
Because TKIs such as gefitinib must compete with ATP for binding to the kinase active site, the enhanced ATP affinity is expected to decrease the apparent inhibitor potency. In the L858R/T790M mutant this “Km effect” combines with the small difference in binding affinity for gefitinib to dramatically decrease inhibitor sensitivity at cellular concentrations of ATP. The expected potency of gefitinib (calculated Kiapp) is plotted as a function of ATP concentration for the L858R and L858R/T790M mutants in Fig. 3A. Whereas the L858R mutant maintains low-nanomolar sensitivity to gefitinib at cellular ATP concentrations (≈1 mM), the L858R/T790M mutant does not. This predicted loss of inhibitor sensitivity in the L858R/T790M mutant is confirmed by direct in vitro measurement of enzyme inhibition by gefitinib at 10 μM vs. 1 mM concentrations of ATP. The L858R/T790M mutant is sensitive to gefitinib at 10 μM ATP, but resistant at 1 mM, whereas the L858R mutant is effectively inhibited even at the higher concentration, which approximates the cellular level of ATP (Fig. 3 B and C). We also observe this effect in other assays using polyE4Y or the signaling adapter Shc as an EGFR substrate and in the presence of either Mn2+ or Mg2+ (data not shown). We conclude that the clinically observed resistance of L858R/T790M mutant stems largely from its enhanced affinity for ATP (as compared with the inhibitor-sensitive L858R mutant) and not from a steric block of inhibitor binding as previously hypothesized.
Fig. 3.
The drug resistance of T790M secondary mutation is manifested only at cellular concentrations of ATP. (A) The calculated Kiapp for the L858R single mutant and L858R/T790M double mutant are plotted versus ATP concentration, using experimentally measured values for the Km[ATP] (Table 2) and setting Ki = Kd as measured (Table 1). Note the expected loss of potency of the double mutant (solid line) as ATP concentrations approach cellular levels (≈1 mM). (B) Inhibition of L858R mutant EGFR kinase by gefitinib in the presence of 10 μM (black squares) or 1.0 mM ATP (red circles). (C) Inhibition of the L858R/T790M double mutant EGFR by gefitinib in the presence of 10 μM (black squares) or 1.0 mM ATP (red circles). In B and C, the in vitro kinase activity of the indicated EGFR mutant was measured in the presence of the indicated concentrations of gefitinib by using an EGFR Tyr-1173 autophosphorylation site peptide (ENAEYLRVA) as substrate.
Discussion
The present work highlights the extent to which the compromised ATP affinity of the EGFR mutants renders them susceptible to inhibition (at least for the L858R, G719S, and exon19 deletions that have been studied to date) (8, 10). The T790M mutation merely restores ATP affinity to the level of the WT kinase. In effect, the diminished ATP affinity of the oncogenic mutants open a “therapeutic window,” which renders them more easily inhibited relative to the WT EGFR and other kinases on which the inhibitors might have activity. The T790M secondary mutation effectively closes this window by restoring ATP affinity to WT levels.
The activating nature of the gatekeeper mutation is not unique to EGFR, as indicated by its effect on Src and its presence in chronic myelogenous leukemia before imatinib treatment. The Km effect may be more idiosyncratic; note that the T790M mutation has little effect on Km in the context of the WT EGFR kinase (Table 2). For the Abl T315I mutant, we measure Km[ATP] = 1.8 μM as compared with Km[ATP] = 6 μM for the WT Abl kinase (data not shown), this change in ATP affinity is expected to have only a modest effect on inhibitor potency. Also, it is clear that the T315I gatekeeper mutation in BCR-Abl directly blocks binding of imatinib and other compounds with a similar binding mode.
Our findings explain the puzzling observation that irreversible anilinoquinazoline inhibitors (and closely related irreversible compounds such as HKI-272) maintain efficacy against the T790M resistance mutant, and additionally they are important for understanding the nature of resistance and possible avenues to the development of more effective drugs. The fact that the T790M substitution confers resistance by increasing the affinity for ATP, rather than by simply sterically interfering with inhibitor binding, means that T790M is a “generic” resistance mutant; it will tend to confer resistance to any ATP competitive inhibitor. To our knowledge, this mechanism of drug resistance, resistance conferred by a mutation that increases affinity for a competing physiologic substrate, has not been previously documented in a clinical context. Interestingly, a distinct, but related, effect has recently been described in a mutant of the mitotic kinesin KSP (also called Eg5). The KSP mutant was discovered in a laboratory screen and confers drug resistance by an allosteric mechanism involving enhanced affinity for ATP (33).
As a class, irreversible inhibitors can overcome T790M resistance through covalent binding; once covalently bound, they are no longer in a competitive, reversible equilibrium with ATP. A number of such compounds are currently in clinical trials in oncology, including HKI-272, but none have yet received approval. One concern with covalent inhibitors is the potential for toxicity caused by off-target effects. At least 10 kinases in addition to EGFR have a reactive cysteine residue in the position equivalent to Cys-797 in EGFR, so it will be important to understand the activity of available irreversible agents against these kinases in particular, which include Tec family kinases, JAK3, and other kinases important for hematopoietic development and immune function. However, our results indicate that irreversible binding is not required for effective inhibition of the T790M mutant. A reversible inhibitor that binds with sufficient affinity to outcompete ATP should work as well. Calculations similar to those shown in Fig. 3A indicate that reversible inhibitors with affinity of ≈200 pM or tighter against the T790M mutant should be effective.
Methods
Protein Preparation and Crystallization.
Constructs spanning residues 696-1022 of the human EGFR and bearing the WT sequence or the T790M and L858R mutations were expressed and purified by using a baculovirus/insect cell system as described (8). Crystals of the T790M mutant were obtained in 0.1 M Hepes (pH 7.5), 21% PEG6000, 0.3 M NaCl, and 5 mM tris(2-carboxyethyl)-phosphine (TCEP), whereas the T790M/HKI-272 complex crystals were made by cocrystallization in 0.1 M Hepes (pH 7.0), 0.2 M Li2SO4, 28% PEG3350, and 5 mM TCEP. T790M/AEE788 complex crystals were made by soaking the apo-T790M crystals in 300 μM AEE788 inhibitor overnight.
Structure Determination and Refinement.
Diffraction data were collected at the Argonne National Laboratory (Argonne, IL) APS ID24 or ID19 beamlines at 100 K. The data were processed with HKL2000 (34). The structures were solved by molecular replacement method with PHASER (35), using the EGFR 696-1022 G719S structure [Protein Data Bank (PDB) ID code 2itn] (8) for apo-T790M and T790M/AEE788 structures and EGFR 696-1022 V948R structure (PDB code 2gs7) (9) for the T790M/HKI-272 structure. CNS/simulated-annealing (36) was then used to obtain less biased 2 Fo − Fc and Fo − Fc maps for manual inspection and adjustment of the model. Repeated rounds of manual refitting and crystallographic refinement were performed by using COOT (37) and refmac5 (38). Inhibitors were modeled into the closely fitting positive Fo − Fc electron density and then included in the following refinement and fitting cycles. Topology and parameter files for the inhibitors were generated by using PRODRG (39).
Enzyme Kinetic Assays, Inhibition Assays, and Data Analysis.
EGFR kinetic parameters were determined in triplicate by using the ATP/NADH coupled assay system in a 96-well format as described (8). The reaction mixture contained 0.5 mg/ml BSA, 2 mM MnCl2, 1 mM phospho(enol) pyruvic acid (PEP; Sigma-Aldrich; catalogue no. P7002), 1 mM TCEP, 0.1 M MOPS 7.5, 5 mM poly-[Glu4Tyr1] peptide (Sigma-Aldrich; catalogue no. P7244), 1/50 of the final reaction mixture volume of pyruvate kinase/lactic dehydrogenase enzymes from rabbit muscle (Sigma-Aldrich; catalogue no. P-0294), 0.5 mM NADH, and 0.5 μM EGFR kinase; ATP at varied concentration was added last to start the reaction. Steady-state initial velocity data were drawn from the slopes of the A340 curves and fit to the Michaelis-Menten equation to determine Vm and Km values. To assure that our derived kcat parameters reflected concentrations of active enzyme, we estimated the active enzyme concentration of every kinase preparation by titration of the samples with the tight binding inhibitor gefitinib or AEE788 (see below).
Inhibition assays were carried out by using the same kinetic assay method, with 10 mM MgCl2 and 1.25 mM EGFR autophosphorylation site peptide (ENAEYLRVA) as the phospho-acceptor substrate. The ATP concentration was fixed at 10 μM or 1 mM, and the indicated concentrations of the inhibitors were added before the addition of ATP.
Binding Constant Assay and Calculation of the Kiapp Values.
The equilibrium fluorescence quenching method was used to obtain the binding constant and estimate the active enzyme concentration as described (8). Fluorescence measurements were carried out in a nitrogen-sparged buffer containing 20 mM Tris, 0.5% glycerol, 250 mM NaCl, and 1 mM TCEP. The obtained Kd values and Km,ATP were used to calculate the Kiapp values using the following equation (40):
assuming that the Kd values obtained in the binding assays are equal to Ki under the condition of the above kinetic assays.
Supplementary Material
ACKNOWLEDGMENTS.
We thank G. Caravotti (Novartis) and S. Rabindran (Wyeth Pharmaceuticals) for the compounds AEE788 and HKI-272, respectively; R. Copeland and C. Walsh for comments on the manuscript; and Y. Li and F. Poy for technical assistance. This work was supported by National Institutes of Health Grants CA080942 (to M.J.E.) and CA116020 (to M.M.). M.J.E. is the recipient of a Scholar Award form the Leukemia and Lymphoma Society.
Footnotes
Conflict of interest statement: M.J.E. and M.M. are consultants for and receive research funding from Novartis Institutes for Biomedical Research.
This article is a PNAS Direct Submission.
Data deposition: The crystallographic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2JIT, 2JIU, and 2JIV).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709662105/DC1.
References
- 1.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson BE, Janne PA. Epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Cancer Res. 2005;65:7525–7529. doi: 10.1158/0008-5472.CAN-05-1257. [DOI] [PubMed] [Google Scholar]
- 5.Gazdar AF, Shigematsu H, Herz J, Minna JD. Mutations and addiction to EGFR: The Achilles “heal” of lung cancers? Trends Mol Med. 2004;10:481–486. doi: 10.1016/j.molmed.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer: Search and destroy. Eur J Cancer. 2006;42:17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 8.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Carey KD, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 11.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate antiapoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 12.Sharma SV, et al. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell. 2006;10:425–435. doi: 10.1016/j.ccr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak EL, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 16.Kosaka T, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 17.Balak MN, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 18.Greulich H, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter TA, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sequist LV. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Oncologist. 2007;12:325–330. doi: 10.1634/theoncologist.12-3-325. [DOI] [PubMed] [Google Scholar]
- 21.Bell DW, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 22.Godin-Heymann N, et al. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–7326. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regales L, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS ONE. 2007;2:e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Kwak EL, et al. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res. 2006;12:4283–4287. doi: 10.1158/1078-0432.CCR-06-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 27.Roche-Lestienne C, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 28.Kato JY, et al. Amino acid substitutions sufficient to convert the nontransforming p60c-src protein to a transforming protein. Mol Cell Biol. 1986;6:4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuza Y, et al. Allele-dependent variation in the relative cellular potency of distinct EGFR inhibitors. Cancer Biol Ther. 2007;6:661–667. doi: 10.4161/cbt.6.5.4003. [DOI] [PubMed] [Google Scholar]
- 30.Tsou HR, et al. Optimization of 6,7-disubstituted-4-(arylamino)quinoline-3-carbonitriles as orally active, irreversible inhibitors of human epidermal growth factor receptor-2 kinase activity. J Med Chem. 2005;48:1107–1131. doi: 10.1021/jm040159c. [DOI] [PubMed] [Google Scholar]
- 31.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 32.Wood ER, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 33.Luo L, et al. ATP-competitive inhibitors of the mitotic kinesin KSP that function via an allosteric mechanism. Nat Chem Biol. 2007;3:722–726. doi: 10.1038/nchembio.2007.34. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski ZM, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Schuttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 40.Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd Ed. New York: Wiley; 2000. pp. 305–317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.