Abstract
Acquisition of additional genetic and/or epigenetic abnormalities other than the BCR/ABL fusion gene is believed to cause disease progression in chronic myeloid leukemia (CML) from chronic phase to blast crisis (BC). To gain insights into the underlying mechanisms of progression to BC, we screened DNA samples from CML patients during blast transformation for mutations in a number of transcription factor genes that are critical for myeloid–lymphoid development. In 85 cases of CML blast transformation, we identified two new mutations in the coding region of GATA-2, a negative regulator of hematopoietic stem/progenitor cell differentiation. A L359V substitution within zinc finger domain (ZF) 2 of GATA-2 was found in eight cases with myelomonoblastic features, whereas an in-frame deletion of 6 aa (Δ341–346) spanning the C-terminal border of ZF1 was detected in one patient at myeloid BC with eosinophilia. Further studies indicated that L359V not only increased transactivation activity of GATA-2 but also enhanced its inhibitory effects on the activity of PU.1, a major regulator of myelopoiesis. Consistent with the myelomonoblastic features of CML transformation with the GATA-2 L359V mutant, transduction of the GATA-2 L359V mutant into HL-60 cells or BCR/ABL-harboring murine cells disturbed myelomonocytic differentiation/proliferation in vitro and in vivo, respectively. These data strongly suggest that GATA-2 mutations may play a role in acute myeloid transformation in a subset of CML patients.
Keywords: blast crisis, chronic phase, genetic alteration, transcriptional regulation
Chronic myeloid leukemia (CML), a malignant clonal disorder of hematopoietic stem/progenitor cells, accounts for ≈15% of all adult leukemias. Typically the disease develops in three phases. An initial chronic phase (CP) is characterized by massive expansion of the granulocytes lasting ≈3–6 years. The disease then progresses, often through an accelerated phase (AP), to a terminal blast crisis (BC) that manifested by a block of cell differentiation resulting in accumulation of myeloid or lymphoid blasts in peripheral blood and/or bone marrow (BM) (1). Treatments including allogeneic hematopoietic stem cell transplantation and the administration of tyrosine kinase inhibitors such as Gleevec can slow down disease progression and greatly improve clinical outcome, but the advent of blast transformation (AP/BC) remains a challenge for physicians.
The discovery of the hallmark chromosomal translocation t(9;22) (Ph chromosome) and subsequent cloning of the BCR/ABL fusion gene have opened a new era in understanding CML. The fusion of BCR sequences adds new regulatory domains/motifs to ABL and increases its tyrosine kinase activity. Numerous studies have demonstrated that BCR/ABL is the major cause of CML, but the molecular mechanisms underlying BC are yet to be clearly defined (2). In the initial CP the granulocytic lineage is expanded, but cells retain their ability to differentiate. Progression to BC is characterized by a dramatic decrease in differentiation capability (3, 4). Additional genetic events have been reported to be associated with CML progression, including duplication of the Ph chromosome, trisomy 8, isochromosome i(17q), and mutations in tumor suppressor genes such as INK4A/ARF, p53, and Rb (5, 6). Mutation of the p53 was detected in up to 30% of CML myeloid BC (7), whereas ≈50% of the patients with lymphoid BC presented a homozygous deletion at the INK4A/ARF gene locus on chromosome 9, which is also found in many acute lymphocytic leukemia patients (8). Chromosomal translocations that are otherwise associated with de novo acute myeloid leukemia (AML), e.g., t(7;11)(p15;p15) generating NUP98/HOXA9 and t(3;21)(q26;q22) generating AML1/EVI, were reported in isolated cases and could be directly implicated in CML disease progression. Coexpression of AML1/EVI or NUP98/HOXA9 with BCR/ABL has been shown to induce a myeloblastic leukemia in mice that resembles CML in blast phase (9, 10).
A number of transcription factors (TFs) have been shown to be important for normal hematopoiesis. Mutations in these TFs may impair normal hematopoietic cell differentiation and survival and therefore collaborate with deregulated tyrosine kinases in leukemogenesis (11). Because CML AP/BC is characterized by a block of cell differentiation, we hypothesized that alterations in certain hematopoiesis-regulating TFs may play critical roles in CML blast transformation. In a screen of 85 CML cases during AP/BC, we discovered two new mutations in GATA-2, which might be involved in acute myeloid transformation in CML.
Results
Identification of GATA-2 Mutations in Myeloid Transformation of CML.
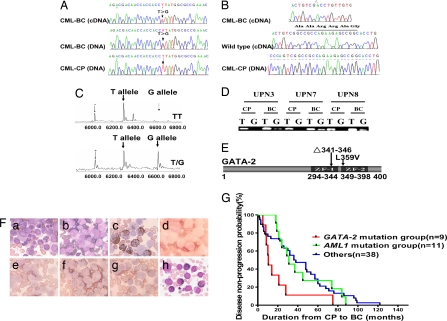
A total of 85 unselected CML patients with blast transformation (28 cases at AP and 57 at BC) were screened for genetic alterations in the protein-coding and/or promoter regions of 11 hematopoietic TFs or two key signal transducers, including Notch1, PU.1, C/EBPα, GATA-1, GATA-2, GATA-3, AML1, CBFβ, MYB, ICSBP, p53, N-Ras, and K-Ras [supporting information (SI) Table 1]. In line with previous reports (12, 13), mutations in p53 (3/85) gene were detected in addition to a number of SNPs in C/EBPα, GATA-3, PU.1, and ICSBP genes. AML1 was found mutated in 11 of 85 cases. In addition, by sequencing both cDNA and genomic DNA samples two new mutations were detected in the GATA-2 gene. One T-to-G substitution was identified in eight cases (9.41%) that did not have any of the mutations mentioned above (SI Table 2). This mutation was located within exon 5 of GATA-2 (nucleotide 1075 of the GATA-2 ORF sequence), which resulted in L359V change at ZF2 domain (Fig. 1 A and E). A small deletion in the GATA-2 gene was detected in one case (UPN6), causing a deletion of amino acids 341–346 (Δ341–346) across the border of ZF1, which is immediately upstream of the GATA-2 L359V mutation (Fig. 1B). The G/T heterozygosity of GATA-2 at nucleotide 1075 in eight patients was confirmed by MALDI-TOF MS, and the highly sensitive allele-specific PCR revealed that over the disease course this mutation existed only at AP and BC, but not at CP (Fig. 1 C and D). Leukemic BM cells from eight cases with L359V and those with the deletion Δ341–346 were myeloblast and/or monoblast in morphology (Fig. 1F). We then screened the BM samples from 200 normal subjects and 233 patients, the latter including 159 cases of primary AML, 30 cases of primary acute lymphocytic leukemia, 25 cases of myeloproliferative disorder, and 19 cases of myelodysplastic syndrome, and we did not detect GATA-2 mutations in any of these cases, rendering the possibility that these two mutations could belong to functionally silent SNPs extremely unlikely. Interestingly, the Wright's staining of BM samples with the GATA-2 Δ341–346 deletion also revealed an increased number of eosinophilic cells (26.5%) (Fig. 1Fh). The 11 patients with AML1 mutations were characterized by acute myeloblastic transformation without involvement of the monocytic lineage. Clinically, the cases with GATA-2 mutations showed a very poor prognosis in terms of disease progression, whereas the AML1 mutation group (n = 11), like the remaining cases (n = 38) whose detailed disease progression history records were available, had a more favorable prognosis (P < 0.05) (Fig. 1G).
Fig. 1.
GATA-2 mutations in myeloid blast transformation of CML. (A) DNA sequencing analyses detect a recurrent T/G heterozygosity at 1,075 nt (coding region sequence) of GATA-2 exon 5 in both cDNA and genomic DNA samples isolated from BM cells of CML patients during AP/BC. (B) An 18-bp deletion of the GATA-2 gene from nucleotide 1021 to nucleotide 1038 specifically detected at BC phase in a BM sample of patient UPN6. (C) MALDI-TOF MS confirms a GATA-2 T-to-G mutation within BM cells at BC phase. (D) The detection of the nucleotide 1075 T-to-G mutation in BM samples of three patients by allele-specific PCR at AP/BC but not at CP. (E) The GATA-2 gene deletion results in a deletion of amino acids 341–346 (Δ341–346) around the ZF1 domain border. L359V substitution is within ZF2 domain. (F) Morphological and histochemical examinations of BM samples from patient UPN3 with L359V or from patient UPN6 with Δ341–346. (a) Wright's staining of a BM cellular smear from patient UPN3 at CP. (b–g) Examination on the BM samples from UPN3 at BC. (b) Wright's staining. (c) Myeloperoxidase staining, a specific marker of myeloid cells. Note the presence of strong positive, weak positive, or even negative contingents, suggesting the identities of myeloblasts and monoblasts. (d) Periodic acid Schiff staining. (e) Naphthol AS-chloracetate esterase staining, a marker of granulocytes. (f) Naphthol AS-D acetate esterase (NAS-DAE) staining. (g) Inhibition of NAS-DAE staining by sodium fluoride, a test to distinguish granulocytes from monocytes. (h) Wright's staining of BM samples from UPN 6 at BC shows the presence of eosinophilia. (G) Disease progression of CML patients with GATA-2 mutation compared with those without GATA-2 mutation.
Functional Consequences of GATA-2 Mutations in CML Acute Myeloid Transformation.
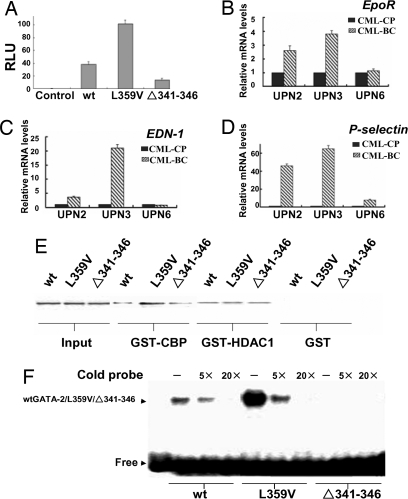
Both GATA-2 mutations were located within or close to the N terminus of the ZF2 domain, which is responsible for DNA binding, self-association, and heterodimerization with the PU.1 protein (14, 15). To explore the functional aspects of the two GATA-2 mutants, we tested their transcription-activating properties on a classic GATA-2 response element-coupled luciferase reporter. The same amounts of plasmids encoding WT GATA-2, GATA-2 L359V, or Δ341–346 were cotransfected with the reporter into 293T cells. Interestingly, compared with wtGATA-2, the GATA-2 L359V mutant exhibited a 3-fold higher activation potential, whereas the activity of Δ341–346 GATA-2 was only half of its WT counterpart (Fig. 2A), suggesting that L359V is a gain-of-function mutation for transactivation. EpoR, EDN-1, and P-selectin are well known GATA-2 target genes (16); therefore, we have examined the expression of these genes in the primary BM samples isolated from patients with GATA-2 mutations (two cases with L359V and one with the deletion) at both CP and AP/BC. Interestingly, EpoR and EDN-1 mRNA levels were elevated in two patients with L359V at BC (Fig. 2 B and C), but not in the case with Δ341–346 deletion. However, compared with CP P-selectin was increased in all three cases at BC (Fig. 2D). These data strongly suggested that the L359V and Δ341–346 mutations confer potentiated transcriptional properties onto the wtGATA-2 protein.
Fig. 2.
GATA-2 L359V has enhanced transcriptional activity on target genes. (A) Transfection assays were conducted in 293T cells using a GATA-2 response element-coupled luciferase reporter, with wtGATA-2-, GATA-2 L359V-, or Δ341–346 mutant-expressing plasmid. (B–D) Examination of mRNA levels of EpoR, EDN-1, and P-selectin using real-time RT-PCR in BM samples isolated at either the CP or BC stage of indicated patients (n = 3). (E) Comparison of the binding affinities of three different GATA-2 proteins with CBP or HDAC1 in a GST pull-down assay. (F) Gel-shift assay on the specific binding of the three GATA-2 proteins with a DNA probe containing a classic GATA-2 response element.
The gain of function in transcriptional activity of TFs could be attributed to enhanced interactions with coactivators, diminished association with corepressors, and/or enhanced binding to cognate DNA target sites. As shown in Fig. 2E, wtGATA-2 and the two GATA-2 mutants exhibited nearly identical binding to corepressor histone deacetylase 1 (HDAC1), but GATA-2 L359V mutant showed an increased binding affinity to the coactivator CREB-binding protein (CBP). L359V mutant protein also appeared to have a higher affinity for the cognate DNA binding site of GATA-2 (Fig. 2F). Therefore, GATA-2 L359V may recruit coactivators more effectively, thus resulting in an increased transactivation of its target genes.
GATA-2 Mutants Interfere with PU.1 Function.
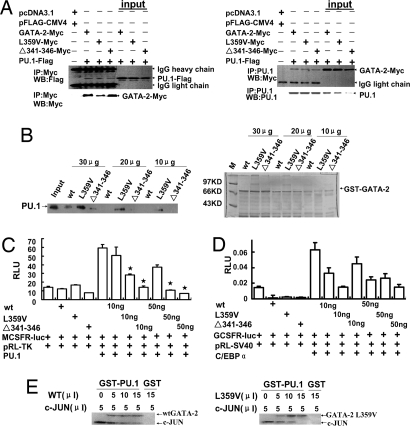
In addition to regulating erythropoiesis, both GATA-1 and GATA-2 play negative regulatory roles in early myelopoiesis by directly inhibiting the activity of PU.1 via a physical interaction of its ZF2 domain with the PU.1 molecule (14, 17). We therefore address whether the two GATA-2 mutants have altered binding properties to PU.1. As shown in Fig. 3A, like wtGATA-2, the two MYC-tagged GATA-2 mutants retained their abilities to physically associate with PU.1, and by using two-way immunoprecipitation assays all three proteins were found in the coprecipitates containing PU.1. A GST pull-down assay demonstrated that the GATA-2 L359V mutant, but not Δ341–346, enhanced binding to PU.1 (Fig. 3B); however, both mutants inhibited ability of PU.1 to activate the M-CSFR promoter-based luciferase reporter more effectively than the wtGATA-2 (Fig. 3C). On the other hand, compared with the WT protein, two GATA-2 mutants did not affect the activity of C/EBPα (Fig. 3D). To further understand the molecular mechanisms, we measured the competitive PU.1-binding affinities of two GATA-2 proteins with c-JUN, a cofactor required for PU.1 transactivation (Fig. 3E). Consistent with the increased inhibitory effect on the expression of PU.1 target genes, GATA-2 L359V decreased the binding of c-JUN with the PU.1 protein to a greater extent than the wtGATA-2.
Fig. 3.
GATA-2 mutants exhibit enhanced inhibition on the transcriptional activity of PU.1. (A) GATA-2 mutants physically associate with PU.1 in vivo. Flag-tagged PU.1 cDNA plus one of three c-Myc-tagged GATA-2 cDNAs were cotransfected into Cos-7 cells. Left shows that the immunoprecipitation of whole-cell lysates by anti-Myc antibodies coprecipitates the PU.1 protein. Right shows that, in a reverse way, PU.1 antibody-mediated precipitates contain three GATA-2 proteins, as probed by an anti-c-Myc antibody. (B Left) Different doses of GST-wtGATA-2, GST-GATA-2 L359V, or GST-Δ341–346 mutant and GST were used to pull down the lysates of the PU.1-transfected Cos-7 cells. (B Right) Coomassie blue staining confirms the quantification of GST fusion proteins by Bradford assay. (C) The M-CSFR promoter-coupled luciferase reporter plasmid was cotransfected into Cos-7 cells, with different doses of wtGATA-2 or GATA-2 mutant expression plasmids as indicated. The relative luciferase units are expressed as mean ± SD. *, P < 0.05. (D) G-CSFR promoter-coupled luciferase reporter was cotransfected with different doses of plasmids as shown in C. (E) GATA-2 L359V competes with c-JUN on binding to PU.1. Combinations of different amounts of 35S-labeled GATA-2 and c-JUN as indicated were incubated with the GST-PU.1 protein. Pull-down efficiencies of GATA-2 and c-JUN by PU.1 were examined by autoradiography.
Enhanced Activity of GATA-2 Mutants Interferes with Myelomonocytic Differentiation/Proliferation.
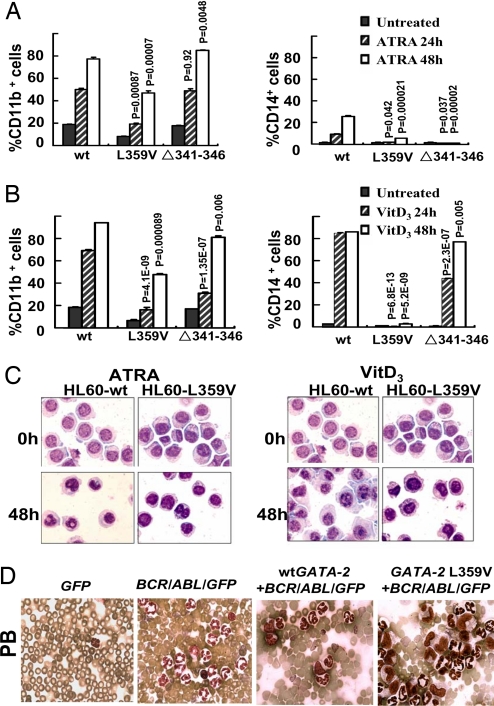
As first step we chose an in vitro differentiation model of myeloid leukemia to examine the functional consequences of the two GATA-2 mutants on granulocytic and monocytic differentiation programs. 1,25-(OH)2-vitamin D3 (VitD3) induced a clear monocytic differentiation of wtGATA-2 transfected HL-60 cells as reflected by expression of monocyte surface markers CD14 and CD11b as well as morphological appearance. However, HL-60 cells with GATA-2 L359V, or to a lesser extent Δ341–346, were refractory to differentiation induction by VitD3 (Fig. 4 B and C). The cells expressing the GATA-2 mutants also displayed diminished up-regulation of myeloid differentiation antigens CD11b and CD14 by all-trans-retinoic acid (ATRA) as compared with those expressing the wtGATA-2 (Fig. 4A); therefore, interference of the GATA-2 L359V and Δ341–346 with the myelomonocytic differentiation program of HL60 cells is likely due to attenuating effects on PU.1 function.
Fig. 4.
Enhanced activities of GATA-2 interfere with myelomonocytic differentiation/proliferation. wtGATA-2- and GATA-2 mutant-expressing plasmids were stably transfected into HL-60 myeloid leukemia cells. One set of representative results from four independent transfection experiments is shown. (A) CD11b (Left) or CD14 (Right) levels were monitored along with 1 × 10−6 M ATRA-induced granulocytic differentiation of indicated HL-60 cells by flow cytometry (means ± SD). P values indicate the comparisons between GATA-2 mutants and wtGATA-2. (B) CD11b and CD14 levels were analyzed at 24 or 48 h after treatment with 1 × 10−7 M VitD3 as in A. (C) Wright's staining of indicated HL-60 subclones before and after 48 h of treatment with ATRA or VitD3. (D) GATA-2 L359V+BCR/ABL/GFP induced a myelomonocytic leukemia-like disease in transduced cell-transplanted mice. Morphological pictures of peripheral blood were taken from deceased recipients under a ×100/1.0 oil-immersion objective lens.
5-FU-treated BM cells were cotransduced with wtGATA-2 or GATA-2 L359V cDNA and BCR/ABL (p210) and then transplanted into syngeneic recipients (SI Fig. 5A). At 3–5 weeks after BM transplantation, BCR/ABL induced a lethal granulocytosis in mice as expected (20/20 in three separate experiments), with a mean survival time of 28.6 days (SI Fig. 5 B and C). Interestingly, in contrast to granulocytic cell predominance in BCR/ABL mice, monocytic cells (Gr-1low/− and F4/80+) are the majority of GFP+ cells (an average of 80% at 19 days after BM transplantation) in GATA-2 L359V+BCR/ABL/GFP mice (SI Fig. 5C and data not shown). The fraction of monocytic cells was also increased in wtGATA-2+BCR/ABL/GFP mice, albeit to a lesser extent (an average of 50% at 19 days after BM transplantation) (SI Fig. 5C and data not shown). This result indicates that GATA-2 overexpression alters myelomonocytic differentiation of BCR/ABL-harboring hematopoietic stem/progenitors and that this activity of GATA-2 is dramatically increased by the L359V mutation.
Similar to the previously reported negative effects of wtGATA-2 overexpression on normal hematopoiesis, the overexpression of either of the two GATA-2 cDNAs (WT or L359V) inhibited proliferation of BCR/ABL-harboring hematopoietic stem/progenitors, resulting in the rescue of 80–90% lethal effects of BCR/ABL-induced granulocytosis (SI Fig. 5B). A total of 19.2% (5/26) of mice transduced with wtGATA-2+BCR/ABL/GFP and 12% (3/25) of mice transduced with GATA-2 L359V+BCR/ABL/GFP did develop a fatal disease for, on average, 38.6 and 41.3 days, respectively. At the time of death, 20–30% monocyte-like cells in peripheral blood and BM were detected in both wtGATA-2+BCR/ABL/GFP and GATA-2 L359V+BCR/ABL/GFP mice compared with 3–4% in BCR/ABL alone mice (Fig. 4D and SI Fig. 5B). However, although the percentage of monocytic cells is about the same as in the diseased wtGATA-2+BCR/ABL/GFP and GATA-2 L359V+BCR/ABL/GFP mice, pathological examination revealed that the monocytic cells in wtGATA-2+BCR/ABL/GFP mice are morphologically normal, whereas monocytic cells in GATA-2 L359V+BCR/ABL/GFP mice have an atypical morphology, indicating incomplete differentiation. This result suggests that GATA-2 L359V not only has gain of function in altering myelomonocytic differentiation of BCR/ABL-harboring hematopoietic stem/progenitors, but also disturbs full differentiation of monocytic cells. Similar observations were made in two repeated experiments.
Discussion
CML has long been considered a unique model for studying the multistep processes of leukemogenesis. More than 95% of CML cases are associated with the presence of the Ph chromosome and the BCR/ABL fusion gene (18). Numerous studies show that BCR/ABL endows the malignant cells with survival and/or growth advantages via multiple pathways such as RAS (19), PI-3K/Akt (20), STAT5 (21), and cell cycle deregulation (22). However, cellular differentiation programs do not appear to be major targets of BCR/ABL, because at CP the Ph chromosome exists in most of the differentiated hematopoietic cells. Therefore, a differentiation arrest leading to the accumulation of immature hematopoietic cells at AP/BC is believed to arise from the occurrence of new genetic or molecular abnormalities in the affected clone.
Among several reported molecular events associated with CML acute transformation, p53, INK4A/ARF, and RB abnormalities mainly deregulate the cell cycle (7, 8). Lineage commitment and differentiation abilities of p53−/− progenitors were not affected, suggesting that loss of p53 per se does not cause differentiation arrest. AML-related fusion genes NUP98/HOXA9 and AML1/EVI could cooperate with BCR/ABL to induce CML BC in animal models, but only AML1/EVI inhibited cell differentiation (23). The exact mechanisms by which these genetic events lead to differentiation block remain to be determined.
Evidence from both human and animal studies suggests that molecular defects of essential signaling molecules like protein tyrosine kinases usually lead to a CML-like phenotype, whereas involvement of TFs mainly results in myelodysplastic syndrome (24). For a full-blown acute leukemia phenotype to occur, both events may be required. Based on this rationale, we hypothesized that involvement of TFs should be critical in CML acute transformation. In line with this hypothesis, we have identified specifically in CML-AP/BC mutations in GATA-2 and AML1, two TFs essential for early hematopoiesis and stem cell self-renewal. Interestingly, despite the fact that the GATA-2 mutant with a Δ341–346 deletion behaved slightly differently from that with L359V, all of the eight cases with GATA-2 L359V displayed differentiation arrest at myeloblast and/or monoblast stages, and the Δ341–346 deletion was associated with myeloblastic crisis with eosinophilia. At the cellular level, GATA-2 mutations disturbed VitD3- or ATRA-induced myelomonocytic differentiation of HL-60 cells, a scenario consistent with the lineage identity of blast cells found in patients. Likewise, when introduced into BCR/ABL-harboring hematopoietic cells, GATA-2 L359V also reprogrammed the myelomonocytic differentiation of transduced myeloid progenitors in vivo. This gain of function of GATA-2 L359V may underline the blast crisis with the myelomonocytic phenotype observed in patients. The unfavorable outcome of patients with the GATA-2 L359V mutation as compared with the AML1 mutation group and the remaining patients observed here suggests that this mutation may also serve as a useful prognostic marker.
Our data also suggest that GATA-2 mutations might mediate CML transformation through two different molecular mechanisms. First, GATA-2 L359V is a gain-of-function mutation leading to enhanced DNA binding and coactivator recruitment when compared with WT protein. In particular, the L359V mutant showed enhanced transactivation of target genes in vivo. Second, both GATA-2 mutations repressed the transcription activation function of myeloid master regulator PU.1 via aberrant protein–protein interaction. It has been previously reported that PU.1 and C/EBPα are two major TFs for myeloid cell differentiation. PU.1 regulates both granulocytic and monocytic maturation, whereas C/EBPα mainly regulates granulocyte maturation (25, 26). Partial inactivation of PU.1 in adult mice led to the development of myeloid leukemia (27), whereas mice with targeted disruption of C/EBPα demonstrated a selective block of granulocyte differentiation (28). Clinically, heterozygous PU.1 or C/EBPα mutations were associated with AML, and PU.1 and C/EBPα are down-regulated or inactivated in certain AML patients (29, 30). We have demonstrated that GATA-2 L359V possessed a higher propensity to bind PU.1 and consequently exerted a stronger repressing effect on PU.1 transactivation. The GATA-2 protein has two zinc fingers, ZF1 (aa294–344) and ZF2 (aa349–398). The ETS domain of PU.1 can bind to either GATA-2 ZF2 or c-JUN, but the functional consequences are opposite: GATA-2 binding represses whereas c-JUN binding enhances the transactivation functions of PU.1 (14, 31). Our results show that both GATA-2 L359V and Δ341–346 mutants repress PU.1 activity more effectively than wtGATA-2, although with distinct molecular mechanisms. The two GATA-2 mutants did not show enhanced repressing effects on C/EBPα, most likely because of the fact that interaction between GATA-2 and C/EBP is mainly mediated by a region from amino acid 377 to amino acid 415 of GATA-2 that is probably not affected by observed mutations (32).
It is of interest to know at which stage of CML the mutations in TFs arise in hematopoietic cells. Our observation that mutation of GATA-2 was detected only at AP/BC but not at CP by using a highly sensitive allele-specific PCR seems to be in favor of the view that the additional mutations in CML AP/BC represent secondary genetic events in BCR/ABL oncogene-containing cells. It has been shown that BCR/ABL proteins may translocate from the cytoplasm to the nucleus where they interfere with DNA repair by interacting with effectors of the ataxia telangiectasia and rad 3-related (ATR) protein, a phenomenon that may underlie the genomic instability of the CML clone (33). BCR/ABL also down-regulates DNA repair protein DNA-PKcs (34) and BRCA1, the latter being shown to play a major role in the maintenance of genome integrity (35) in response to DNA damage (36, 37). The mutator phenotype of BCR/ABL is also associated with enhanced expression of DNA polymerase β (38). All of these events facilitate the generation of secondary genetic abnormalities that channel the disease toward acute transformation. For those patients without detectable additional genetic or molecular abnormalities at AP/BC, consideration should be given to alternate mechanisms, for example, epigenetic deregulation or posttranslational modification of important hematopoietic TFs (39, 40).
Materials and Methods
Patients.
All 85 CML patients in this study were diagnosed and followed at Shanghai Rui Jin Hospital, Renji Hospital, Xinhua Hospital, Huadong Hospital, Tongji Hospital, Changhai Hospital, Shanghai First People's Hospital, and Shanghai Tenth People's Hospital during the period of April 1996 to December 2004. BM samples were obtained with informed consent, and then DNA and RNA were extracted by using standard protocols. Detailed clinical data with regard to the disease progression were available among nine patients with the GATA-2 gene mutation, 11 patients with AML1 gene abnormality, and 38 patients among the remaining cases.
DNA Sequencing, MALDI-TOF MS, and TrueSNP Allele-Specific PCR.
PCRs were carried out by using standard protocols. PCR products were sequenced by ABI 3700 Automatic Sequencer (PE Biosystems). Sequencing was performed in triplicate, and mutations were double-checked at both genomic DNA and cDNA samples. In addition, heterozygous mutations were confirmed by MALDI-TOF MS (Sequenom). TrueSNP allele-specific PCR primers were synthesized by Proligo. The forward primers are 5′-GCAAATTGTCAGACGACAACCACCACC+TTA-3′ (WT allele primer) and 5′-CAAATTGTCAGACGACAACCACCACC+G-3′ (T-to-G mutation allele primer).
Plasmid Constructions.
cDNAs containing full-length wtGATA-2, GATA-2 L359V, and Δ341–346 mutants were cloned by RT-PCR from patients' BM samples. EcoRI and XhoI were used for cloning into the pcDNA3.1/Myc-His(−)B vector (Invitrogen) or pGEX-4T-2 (Amersham Pharmacia). PU.1 and C/EBPα cDNA were inserted into the pFLAG-CMV4 vector (Sigma) via HindIII and BamHI. The M-CSF receptor promoter sequence (−416 to 124) was cloned by PCR into the pGL4.15 vector (Promega), which was designated as pGL4.15-M-CSF-R-Luc. The G-CSF receptor promoter sequence (−1097 to 194) was cloned into pGL3 (Promega) via KpnI and BglII sites and designated as pGL3-G-CSF-R-Luc. A NcoI-MluI-NotI-NcoI adaptor was cloned into the NcoI site of the retroviral vector MSCV-IRES-GFP (a kind gift from Warren S. Pear, University of Pennsylvania, Philadelphia). wtGATA-2 or GATA-2 L359V cDNA was cloned to the XhoI and EcoRI sites of the modified MSCV-IRES-GFP vector. BCR/ABL cDNA was cloned to the MluI and NotI sites of modified MSCV-IRES-GFP. SI Fig. 5A shows a schematic diagram of all retroviral constructs.
Cell Culture and Transient Transfection Assay.
293T and Cos-7 cells were grown in DMEM supplemented with 10% FBS (HyClone). wtGATA-2-, GATA-2 L359V-, or Δ341–346-encoding plasmids were cotransfected with CD34×2-Luc reporter plasmid (kindly provided by Tariq Enver, Weatherall Institute of Molecular Medicine, Oxford, U.K.), or an internal control pRL-SV40 plasmid (Promega) into 293T or Cos-7 cells using SuperFect transfection reagent (Qiagen). Luciferase assays on pGL4.15-M-CSF-R-Luc or pGL3-G-CSF-R-Luc were performed by the same method. Luciferase activity was measured in a Luminometer with the Dual-Luciferase Reporter Assay System (Promega).
Drug-Induced Cell Differentiation.
VitD3- and ATRA-induced cell differentiation experiments were carried out as previously described (41).
Gel-Shift Assay.
wtGATA-2, GATA-2 L359V, and Δ341–346 mutant proteins were generated in vitro by using a TNT-coupled rabbit reticulocyte lysate system (Promega). The GATA-2 response element probe (5′-cacttgataacagaaagtgataactct-3′) was used in this experiment. The experiment was performed in accordance with previously described protocol (42).
Quantitative Real-Time RT-PCR.
After reverse transcription, real-time PCR was performed by using a SYBR Green PCR Master Mix (Applied Biosystems). The 18S rRNA was coamplified as an endogenous control.
In Vitro Protein/Protein Binding Assay.
The in vitro interactions between different GATA-2 and CBP or HDAC1 proteins were measured with previously described procedures (42). Examination on the interaction between different GATA-2 proteins with PU.1 was performed with the same protocol except that the purified GST-wtGATA-2, GST-GATA-2L359V, and GST-Δ341–346 GATA-2 proteins were incubated with the whole-cell lysates of pFLAG-CMV4-PU.1 transfected Cos7 cells. [35S]Methionine-labeled GATA-2 and c-JUN were in vitro translated by using a TNT-coupled rabbit reticulocyte lysate system (Promega). Different amounts of GATA-2 and c-JUN were incubated with GST-PU.1. A GST pull-down experiment was performed as described above.
Coimmunoprecipitation.
A total of 10 μg of different GATA-2-expressing plasmids was cotransfected with 10 μg of PU.1-expressing plasmid into Cos-7 cells. pcDNA3.1/MYC-His(−)B vector and pFLAG-CMV4 vector were used as transfection mocks. Whole-cell lysates after 48 h were extracted with RIPA buffer (Sigma). A total of 500 μg of proteins from each lysate was immunoprecipitated by either mouse anti-MYC (Santa Cruz Biotechnology) or rabbit anti-PU.1 (Santa Cruz Biotechnology) antibody at 4°C overnight and probed with mouse anti-Flag M2 monoclonal antibody (Sigma) or mouse anti-MYC antibody (Santa Cruz Biotechnology) using a standard protocol.
Viral Preparations and BM Reconstitution Assay.
Retroviral supernatants were generated, and BM transplantation was performed as described (43). All of the retroviral titers for the mouse experiments were matched to ≈40–45% GFP+ NIH 3T3 cells. Total cell lysates from infected NIH 3T3 cells were processed for Western blot as described (44). The following primary antibodies were used: anti-ABL3, anti-GATA-2 (Santa Cruz Biotechnology), and anti-dynamin (Affinity Bioreagents).
Hematopathologic Analysis and Flow Cytometry.
Smears and cytospin were stained with the Hema 3 stain set (Fisher) for routine cell morphology. Peripheral blood or single-cell suspensions of murine tissues were prepared and stained with phycoerythrin- or allophycocyanin-conjugated mouse antibodies (BD Pharmingen) for flow cytometry as previously described (10).
Supplementary Material
ACKNOWLEDGMENTS.
We thank all members of the Shanghai Institute of Hematology for their support and Dr. Jiang-Xiang Liu for his constructive discussion. This work was supported in part by the Chinese National High Tech Program (Grant 863:2006AA02A405), the Chinese National Key Program for Basic Research (Grant 973:2004CB518600), the Key Disciplines Program of Shanghai Municipal Education Commission (Grant Y0201), the Innovation Group of the National Natural Science Foundation of China (Grant 30521003), the National Natural Science Foundation of China for Young Scholars (Grant 30500298), the Shanghai Municipal Commission for Science and Technology (Grant 06DZ22021), the Samuel Waxman Cancer Research Foundation Laboratory, and the National Heart, Lung, and Blood Institute (Grant HL083515 to R.-B.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711824105/DC1.
References
- 1.Kantarjian HM, Talpaz M, Giles F, O'Brien S, Cortes J. New insights into the pathophysiology of chronic myeloid leukemia and imatinib resistance. Ann Intern Med. 2006;145:913–923. doi: 10.7326/0003-4819-145-12-200612190-00008. [DOI] [PubMed] [Google Scholar]
- 2.Ramaraj P, et al. Effect of mutational inactivation of tyrosine kinase activity on BCR/ABL-induced abnormalities in cell growth and adhesion in human hematopoietic progenitors. Cancer Res. 2004;64:5322–5331. doi: 10.1158/0008-5472.CAN-03-3656. [DOI] [PubMed] [Google Scholar]
- 3.Jaiswal S, et al. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci USA. 2003;100:10002–10007. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman JM, Melo JV. Chronic myeloid leukemia: Advances in biology and new approaches to treatment. N Engl J Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 5.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 6.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 7.Feinstein E, et al. p53 in chronic myelogenous leukemia in acute phase. Proc Natl Acad Sci USA. 1991;88:6293–6297. doi: 10.1073/pnas.88.14.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sill H, Goldman JM, Cross NC. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood. 1995;85:2013–2016. [PubMed] [Google Scholar]
- 9.Dash AB, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci USA. 2002;99:7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuenco GM, Ren R. Cooperation of BCR-ABL and AML1/MDS1/EVI1 in blocking myeloid differentiation and rapid induction of an acute myelogenous leukemia. Oncogene. 2001;20:8236–8248. doi: 10.1038/sj.onc.1205095. [DOI] [PubMed] [Google Scholar]
- 11.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 12.Ahuja H, Bar-Eli M, Advani SH, Benchimol S, Cline MJ. Alterations in the p53 gene and the clonal evolution of the blast crisis of chronic myelocytic leukemia. Proc Natl Acad Sci USA. 1989;86:6783–6787. doi: 10.1073/pnas.86.17.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steer EJ, Goldman JM, Cross NC. Mutations of the transcription factor AML1/CBFA2 are uncommon in blastic transformation of chronic myeloid leukaemia. Leukemia. 2001;15:476–477. doi: 10.1038/sj.leu.2402062. [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikonomi P, et al. Levels of GATA-1/GATA-2 transcription factors modulate expression of embryonic and fetal hemoglobins. Gene. 2000;261:277–287. doi: 10.1016/s0378-1119(00)00510-2. [DOI] [PubMed] [Google Scholar]
- 17.Walsh JC, et al. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 18.Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- 19.Goga A, McLaughlin J, Afar DE, Saffran DC, Witte ON. Alternative signals to RAS for hematopoietic transformation by the BCR-ABL oncogene. Cell. 1995;82:981–988. doi: 10.1016/0092-8674(95)90277-5. [DOI] [PubMed] [Google Scholar]
- 20.Skorski T, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye D, Wolff N, Li L, Zhang S, Ilaria RL., Jr STAT5 signaling is required for the efficient induction and maintenance of CML in mice. Blood. 2006;107:4917–4925. doi: 10.1182/blood-2005-10-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonuleit T, et al. Bcr-Abl kinase down-regulates cyclin-dependent kinase inhibitor p27 in human and murine cell lines. Blood. 2000;96:1933–1939. [PubMed] [Google Scholar]
- 23.Mitani K. Molecular mechanisms of leukemogenesis by AML1/EVI-1. Oncogene. 2004;23:4263–4269. doi: 10.1038/sj.onc.1207777. [DOI] [PubMed] [Google Scholar]
- 24.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer Cell. 2002;1:417–420. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 25.Koschmieder S, Rosenbauer F, Steidl U, Owens BM, Tenen DG. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int J Hematol. 2005;81:368–377. doi: 10.1532/ijh97.05051. [DOI] [PubMed] [Google Scholar]
- 26.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 27.Metcalf D, et al. Inactivation of PU.1 in adult mice leads to the development of myeloid leukemia. Proc Natl Acad Sci USA. 2006;103:1486–1491. doi: 10.1073/pnas.0510616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang DE, et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pabst T, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 30.Mueller BU, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100:998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- 31.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 32.Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol Cell Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dierov J, Dierova R, Carroll M. BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell. 2004;5:275–285. doi: 10.1016/s1535-6108(04)00056-x. [DOI] [PubMed] [Google Scholar]
- 34.Deutsch E, et al. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood. 2001;97:2084–2090. doi: 10.1182/blood.v97.7.2084. [DOI] [PubMed] [Google Scholar]
- 35.Deutsch E, et al. Down-regulation of BRCA1 in BCR-ABL-expressing hematopoietic cells. Blood. 2003;101:4583–4588. doi: 10.1182/blood-2002-10-3011. [DOI] [PubMed] [Google Scholar]
- 36.Skorski T. BCR/ABL regulates response to DNA damage: The role in resistance to genotoxic treatment and in genomic instability. Oncogene. 2002;21:8591–8604. doi: 10.1038/sj.onc.1206087. [DOI] [PubMed] [Google Scholar]
- 37.Nieborowska-Skorska M, et al. ATR-Chk1 axis protects BCR/ABL leukemia cells from the lethal effect of DNA double-strand breaks. Cell Cycle. 2006;5:994–1000. doi: 10.4161/cc.5.9.2722. [DOI] [PubMed] [Google Scholar]
- 38.Canitrot Y, et al. Mutator phenotype of BCR-ABL transfected Ba/F3 cell lines and its association with enhanced expression of DNA polymerase beta. Oncogene. 1999;18:2676–2680. doi: 10.1038/sj.onc.1202619. [DOI] [PubMed] [Google Scholar]
- 39.Yang MY, Liu TC, Chang JG, Lin PM, Lin SF. JunB gene expression is inactivated by methylation in chronic myeloid leukemia. Blood. 2003;101:3205–3211. doi: 10.1182/blood-2002-05-1598. [DOI] [PubMed] [Google Scholar]
- 40.Perrotti D, et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 41.Bunce CM, Wallington LA, Harrison P, Williams GR, Brown G. Treatment of HL60 cells with various combinations of retinoids and 1α,25 dihydroxyvitamin D3 results in differentiation towards neutrophils or monocytes or a failure to differentiate and apoptosis. Leukemia. 1995;9:410–418. [PubMed] [Google Scholar]
- 42.Dong S, et al. Amino-terminal protein-protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-alpha fusion protein. Proc Natl Acad Sci USA. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gross AW, Zhang X, Ren R. Bcr-Abl with an SH3 deletion retains the ability to induce a myeloproliferative disease in mice, yet c-Abl activated by an SH3 deletion induces only lymphoid malignancy. Mol Cell Biol. 1999;19:6918–6928. doi: 10.1128/mcb.19.10.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parikh C, Subrahmanyam R, Ren R. Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood. 2006;108:2349–2357. doi: 10.1182/blood-2004-08-009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.