Abstract
The recent discovery of ammonia-oxidizing archaea (AOA) dramatically changed our perception of the diversity and evolutionary history of microbes involved in nitrification. In this study, a moderately thermophilic (46°C) ammonia-oxidizing enrichment culture, which had been seeded with biomass from a hot spring, was screened for ammonia oxidizers. Although gene sequences for crenarchaeotal 16S rRNA and two subunits of the ammonia monooxygenase (amoA and amoB) were detected via PCR, no hints for known ammonia-oxidizing bacteria were obtained. Comparative sequence analyses of these gene fragments demonstrated the presence of a single operational taxonomic unit and thus enabled the assignment of the amoA and amoB sequences to the respective 16S rRNA phylotype, which belongs to the widely distributed group I.1b (soil group) of the Crenarchaeota. Catalyzed reporter deposition (CARD)–FISH combined with microautoradiography (MAR) demonstrated metabolic activity of this archaeon in the presence of ammonium. This finding was corroborated by the detection of amoA gene transcripts in the enrichment. CARD-FISH/MAR showed that the moderately thermophilic AOA is highly active at 0.14 and 0.79 mM ammonium and is partially inhibited by a concentration of 3.08 mM. The enriched AOA, which is provisionally classified as “Candidatus Nitrososphaera gargensis,” is the first described thermophilic ammonia oxidizer and the first member of the crenarchaeotal group I.1b for which ammonium oxidation has been verified on a cellular level. Its preference for thermophilic conditions reinvigorates the debate on the thermophilic ancestry of AOA.
Keywords: ammonia oxidation, archaea, nitrification, thermophile, amoA
Nitrification, the successive microbial oxidation of ammonia via nitrite to nitrate, is a crucial step in the biogeochemical nitrogen cycle, and ammonia-oxidizing microorganisms catalyze the first, rate-limiting step of this process. Until recently, the microbiology of ammonia oxidation was thought to be well understood. Aerobic, chemolithoautotrophic bacteria within the Beta- and Gammaproteobacteria were the only known ammonia-oxidizing microorganisms (1, 2). However, in the last few years, this understanding has been radically changed, first, by the discovery that ammonium can also be oxidized anaerobically by a clade of deep branching planctomycetes (3, 4), and later by the equally surprising cultivation of ammonia-oxidizing archaea (AOA) belonging to the Crenarchaeota (5). Since then, AOA were found to outnumber ammonia-oxidizing bacteria (AOB) in several terrestrial and marine systems, including different soils (6), the North Sea and Atlantic Ocean (7), the Pacific Ocean (8), and the Black Sea (9). Furthermore, molecular analyses demonstrated that AOA also occur in association with marine sponges (10–13), and amoA sequences related to recognized AOA were retrieved in numerous studies from a wide variety of other habitats (7–9, 14–18), including two moderately thermophilic sites with a temperature below 50°C (19, 20). The latter findings raise the question of whether ammonia oxidation takes place at elevated temperatures and in particular whether AOA thrive under these conditions. This scenario seems theoretically possible because ammonium was detected in concentrations between 2.5 μM and 46.7 mM in thermophilic and hyperthermophilic environments (21, 22) and can be formed there biotically (23) or abiotically (24–26). However, this question cannot be answered by mere gene fragment retrieval. Direct evidence for autotrophic ammonia oxidation in these systems via process measurements, in situ demonstration of metabolic activity of thermophilic AOA or AOB, or isolation of these organisms is still lacking.
Recently, ammonia-oxidizing enrichment cultures were established from microbial mats of the Siberian Garga hot spring, and the temperature optimum for ammonia oxidation of these consortia was estimated to be 50°C (27). Here, one of these ammonia-oxidizing enrichment cultures, which had been maintained for 6 years at 46°C (termed Ga9.2a in the original publication), was screened for ammonia-oxidizing microorganisms with a set of molecular tools. We demonstrate the presence and metabolic activity of AOA in this moderately thermophilic enrichment and provide initial insights into their ecophysiology.
Results
The ammonia-oxidizing enrichment culture was grown aerobically (5.1–5.6 mg of O2 liter−1; measured in five different replicate flasks at 46°C) and mediated the near stoichiometric conversion of ammonium to nitrite without any detectable nitrate production [supporting information (SI) Fig. 4].
Screening for Known Beta- and Gammaproteobacterial AOB and Nitrite Oxidizers.
Initially, the bacterial diversity of the enrichment was monitored by PCR amplification of 16S rRNA gene sequences by using the general bacterial primers 616V and 630R. A total of 42 clones were screened by restriction fragment length polymorphism (RFLP) and assigned to five different patterns; 18 clones representing all RFLP types were sequenced and phylogenetically analyzed (SI Fig. 5). All clones formed a monophyletic group within the Betaproteobacteria, but no 16S rRNA gene sequences related to known AOB or nitrite oxidizers were found. Furthermore, no amplicon was obtained with a 16S rRNA gene-based PCR assay for betaproteobacterial AOB by using primers βamoF and Nso1225R. A combination of these primers with primers targeting the domain Bacteria (616V or 630R) also did not yield any PCR product. The enrichment was also screened for amoA genes of beta- and gammaproteobacterial AOB, but although the applied assays together target all known AOB, no PCR product was observed. Identical results were observed with the addition of PCR-enhancing substances, suggesting that the negative results were not caused by PCR inhibition. This finding was further confirmed by the addition of suitable control DNA to the enrichment biomass before PCR, which always led to successful amplification (data not shown). Consistent with the PCR results, no FISH signals were detectable after hybridization of the formaldehyde-fixed biomass with a suite of AOB- and nitrite oxidizer-specific probes (SI Table 1).
Detection of AOA.
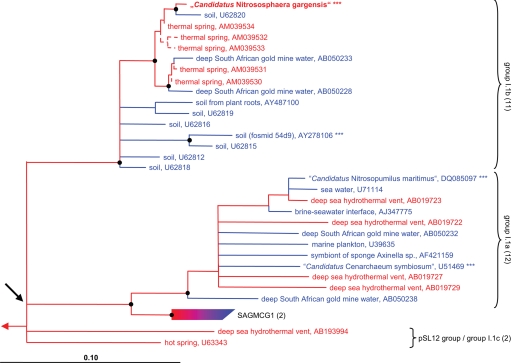
Archaeal 16S rRNA genes were PCR-amplified by using two different sets of primers, cloned, RFLP-screened, and sequenced. In total, 46 clones representing all RFLP types were partly or fully sequenced. All 46 sequences were related at >99.1% similarity to each other and assigned to a monophyletic cluster within Crenarchaeota group I.1b (soil group; Fig. 1) that also included partial 16S rRNA gene sequences recently retrieved from a subsurface radioactive thermal spring (20) and a Wisconsin soil (28). In addition, crenarchaeotal amo genes were PCR-amplified from the enrichment, and 19 and 7 clones were sequenced for amoA and amoB, respectively. All cloned amoA and amoB sequences showed similarities of >99.2% and >99.3% to each other, respectively. No amplification product for amoC could be obtained. Consistent with the 16S rRNA tree topology, the AmoA and AmoB sequences retrieved from the enrichment formed a monophyletic branch within the crenarchaeotal I.1b (soil) cluster (Fig. 2A and SI Fig. 6).
Fig. 1.
16S rRNA-based phylogenetic consensus tree of putative AOA within the Crenarchaeota, showing the positioning of the moderately thermophilic AOA “Candidatus Nitrososphaera gargensis.” For this analysis, only sequences longer than 1,300 nt were considered; 1,264 nt positions that were conserved in at least 50% of all archaea in the data set were used for phylogeny inference. The 16S rRNA consensus tree was constructed from a maximum-likelihood tree, and all nodes that were not supported by TreePuzzle and maximum parsimony were collapsed. In addition, following Robertson et al. (67), all nodes with <70% parsimony bootstrap support (calculated without conservation filters by using 100 iterations) were collapsed. Sequences that have been obtained from (hyper)thermophilic organisms or environments are labeled in red, and those from mesophilic organisms or habitats are depicted in blue. Transits from red to blue indicate presence of both (hyper)thermophilic and mesophilic organisms within a group. Black dots indicate bootstrap support >90%. Dashed lines denote short sequences (<1,300 nt), which were added to the tree without changing the overall tree topology. Asterisks mark organisms for which the presence of an amoA gene has been demonstrated. Numbers in parentheses give the number of sequences within a group that were used for phylogenetic analyses. The scale bar represents 10% estimated sequence divergence. According to SI Fig. 6, the arrow shows the last common ancestor of current AOA that definitely possessed amoA and amoB genes.
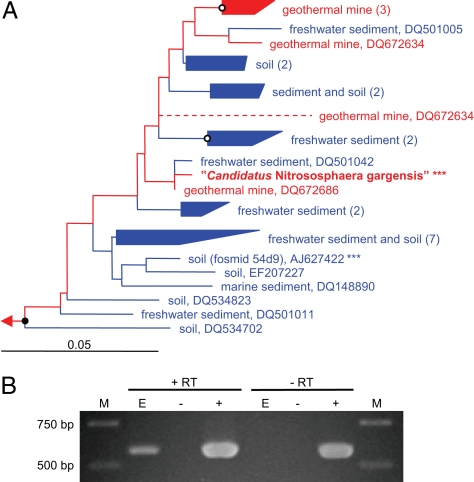
Fig. 2.
Phylogenetic and transcriptional analysis of amoA of “Candidatus Nitrososphaera gargensis.” (A) AmoA evolutionary distance (Fitch) tree showing the phylogenetic positioning of the thermophilic AOA studied (in bold) within the I.1b (soil) cluster. For phylogenetic analyses, 188 aa positions were considered. Sequences obtained from thermophilic environments are labeled in red, whereas those from mesophilic habitats are shown in blue. White and black dots indicate bootstrap (100 iterations) support of >70% and >90%, respectively. Dashed lines indicate short sequences that were added after construction of the tree. Asterisks mark organisms for which the 16S rRNA gene is known. Numbers in parentheses give the number of sequences within a group. The scale bar represents 5% estimated sequence divergence. A more complete AmoA tree as well as an AmoB phylogenetic tree are available for download (SI Fig. 6). (B) Crenarchaeotal amoA mRNA detection in the enrichment, E. +RT, mRNA detection via reverse transcription PCR; −RT, PCR control for DNA contamination in the RNA extract; +, positive control, with use of a cloned amoA gene fragment; −, negative control without nucleic acids; M, size marker.
To directly link the retrieved crenarchaeotal 16S rRNA gene sequences with single microbial cells in the enrichment, catalyzed reporter deposition (CARD)–FISH with the newly designed clone-specific probe RHGA702 was applied. This probe hybridized exclusively to small cocci of 0.9 ± 0.3 μm (n = 22) in diameter, occurring in irregular-shaped microcolonies of 10–50 μm in diameter (Fig. 3). Not all cells of such microcolonies showed a signal after hybridization. Identical results were observed in parallel control experiments with the crenarchaeotal probe Cren512 and the general archaeal probe Arch915. The relative abundance of the crenarchaeotes in the enrichment was determined by an indirect CARD-FISH assay to be 50.4% ± 12.5 (SD) (for details, see SI Results).
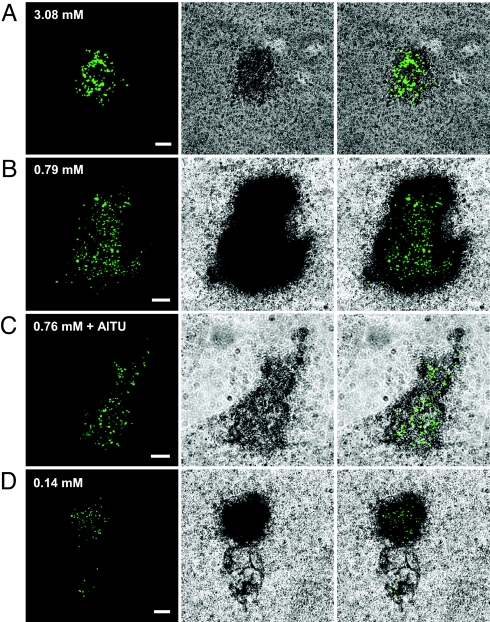
Fig. 3.
Combination of CARD-FISH and MAR for visualization of actively metabolizing AOA in the thermophilic enrichment. CARD-FISH/MAR results after 12-h incubation with radioactive bicarbonate and ammonium concentrations of 3.08 mM (A), 0.79 mM (B), 0.76 mM plus addition of the inhibitor allyl-thiourea (AlTU) (C), and 0.14 mM (D) are shown. (Scale bars: 10 μm.) Without addition of ammonium to the medium, neither CARD-FISH nor MAR signals could be observed. Experimental conditions (except for the ammonium concentrations and addition of AlTU) and microscopic settings were exactly the same for all experiments shown.
Activity of AOA in the Enrichment.
Transcription of crenarchaeotal amoA was demonstrated with biomass from the enrichment (grown in batch culture with 2.0 mM ammonium in the medium) via RT-PCR by using primers specifically targeting the obtained amoA sequences (amo16F/amo586R) (Fig. 2B). Cloning and sequencing of the RT-PCR product resulted in amoA sequences that were identical to amoA sequences obtained by PCR amplification from the enrichment.
Ammonia-oxidizing activity of the detected crenarchaeote was demonstrated by CARD-FISH/microautoradiography (MAR). The enriched biomass was preincubated at 46°C for 3 h with different ammonium concentrations without addition of labeled bicarbonate to allow adaptation of the cells to the incubation conditions. After this step, the ammonium concentration was determined, radioactive bicarbonate was added as sole external carbon source, and the biomass was incubated at 46°C for 12 h. For all ammonium concentrations measured (0.14 mM, 0.79 mM, and 3.08 mM, after preincubation) MAR signals coincided with the AOA microcolonies, but bicarbonate fixation was much lower in the presence of 3 mM ammonium than at lower ammonium concentrations (Fig. 3). For all experiments, chemical analyses showed that 12–22% of the ammonium was removed during the 12-h incubation step, and no cells other than AOA identified by the specific FISH probe showed a positive MAR signal (SI Fig. 7). It should, however, be noted that because the maximum exposure time examined was 15 days, unknown (bacterial) ammonia oxidizers with a very weak activity might have been overlooked. Neither CARD-FISH nor MAR signals were detected for AOA microcolonies in control experiments without added ammonium or with UV-sterilized biomass (data not shown). The absence of CARD-FISH signals without ammonium in the medium suggests that the AOA either reduce their cellular ribosome content in response to starvation and/or that, under these conditions, changes in the cell envelope composition occurred, which negatively affected probe permeability. Addition of 100 μM of the inhibitor allyl-thiourea (AlTU) in the presence of 0.76 mM ammonium resulted in strongly decreased MAR signals of the AOA microcolonies (Fig. 3C).
Discussion
This study demonstrates that an ammonia-oxidizing enrichment grown at 46°C contains a crenarchaeote that, according to its 16S rRNA gene, is a member of the group I.1b. This group consists mainly of sequences retrieved from soil and also includes a 16S rRNA gene sequence from a fosmid clone that also carries an archaeal amo-operon (29, 30). As only one archaeal 16S rRNA phylotype (by using a threshold of 97% sequence similarity) was found in the enrichment, the retrieved archaeal amoA and amoB gene fragments most likely also originated from this phylotype. In contrast to previous studies, which only reported the presence of AOA-related 16S rRNA or crenarchaeotal amoA gene fragments in thermophilic environments (19, 20, 31–33), we specifically identified the respective organism by CARD-FISH with 16S rRNA-targeted probes as cocci in irregularly shaped aggregates. Furthermore, we propose three independent lines of evidence that, taken together, strongly suggest that the detected archaeon is actually an AOA. First, despite the application of several well established screening tools, no recognized bacterial or archaeal ammonia oxidizers could be identified in the ammonia-oxidizing enrichment, although we cannot exclude that the lysis procedure was not suitable for all microorganisms in the enrichment. Second, the crenarchaeote in the enrichment was metabolically active at 46°C, while ammonia was oxidized to nitrite as revealed by detection of its amoA mRNA (Fig. 2B). The presence of amoA mRNA is a strong hint that the AOA contributes to the ammonia oxidation in the enrichment by use of its Amo protein. If the detected AOA lived from substrates other than ammonium (or ammonia), e.g., from excretion products of unknown ammonia oxidizers in the enrichment, detection of amoA mRNA would have been very unlikely because transcription of crenarchaeotal and bacterial amoA has been shown to be induced by ammonia (30, 34–36). Finally, incorporation of bicarbonate into single archaeal cells as monitored by CARD-FISH/MAR was observed in the presence of ammonium (Fig. 3) but was absent in medium lacking ammonium.
CARD-FISH/MAR experiments were also applied to gain insights into the ecophysiology of the AOA, although the very limited amount of available living biomass from the enrichment prevented a more encompassing assessment. Interestingly, the AOA was highly active in the presence of 0.14 mM and 0.79 mM ammonium, whereas partial inhibition was observed with 3.08 mM ammonium (Fig. 3). Inhibition of AOB at such a low ammonium concentration has not been reported, to our knowledge, and some AOB strains grow at more than 200 mM ammonium at a comparable or higher pH (37–39). Apparently, the AOA investigated in this study is even more sensitive to ammonium than members of the Nitrosomonas oligotropha lineage (1, 40), with strain JL-21 as current record holder for ammonium sensitivity whose growth is inhibited at a concentration of 21.4 mM at pH 7.2 (38). The high sensitivity of the AOA to ammonium might be partially explained by the shift of the ammonia/ammonium equilibrium to ammonia at elevated temperature. However, little is known about the mechanism(s) of inhibition. Recent studies showed that ammonia or ammonium is not only taken up passively by AOB via diffusion, but also that at least some AOB possess active uptake systems (41, 42). Putative ammonia or ammonium transporters are also encoded in the genomes of the AOA “Candidatus Cenarchaeum symbiosum” (11) and “Candidatus Nitrosopumilus maritimus” (http://genome.jgi-psf.org/draft_microbes/nitma/nitma.info.html), relatives of the AOA studied here. Such accumulation mechanisms, if not tightly regulated, might, above a certain extracellular substrate concentration, lead to intracellular ammonia/ammonium concentrations that negatively influence the cytoplasmic pH and thus could result in inhibition. However, as little is known about the ammonia oxidation pathways in Archaea (11), other causes for inhibition like the accumulation of toxic intermediates, such as hydroxylamine, cannot be excluded.
Taken together, the CARD-FISH/MAR experiments indicated that the AOA in the enrichment is adapted to moderately low substrate concentrations compatible with the 5.88 μM ammonium measured in the Garga hot spring at the time of sampling (27). Surprisingly, the AOA was not completely inhibited by the addition of 100 μM AlTU (Fig. 3C), a concentration known to completely inhibit ammonia oxidation by AOB (43–45). Potential causes for this residual activity are that (i) during the preincubation, which took place with ammonium but without AlTU, energy storage compounds were built by the AOA, allowing low rates of bicarbonate incorporation in the absence of a functional Amo; and (ii) crenarchaeotal AmoA is not as susceptible to AlTU as its bacterial counterpart, e.g., by having a higher affinity to or not being as dependent on copper as bacterial AmoA (46).
On the basis of the results of this study, we propose, according to Murray and Schleifer (47) and Murray and Stackebrandt (48), provisional classification of the novel archaeon as “Candidatus Nitrososphaera gargensis.” The short description of “Candidatus Nitrososphaera gargensis” is as follows: phylogenetically related to the crenarchaeotal 16S rRNA sequence cluster I1.b (soil group), not isolated; enriched from the Garga hot spring in the Buryat Republic (Russia); cocci have a diameter of 0.9 ± 0.3 μm (but note that cell morphology might be slightly altered because of the CARD-FISH procedure) in irregular shaped microcolonies; basis of assignment: amoA, amoB, and 16S rRNA gene sequences (GenBank accession nos. EU281317, EU281322, and EU281334) and detection by the phylotype-specific oligonucleotide probe RHGA702 (5′-GTG GTC TTC GGT GGA TCA-3′) complementary to helix 24a of the 16S rRNA, aerobic chemolithoautotrophic moderately thermophilic ammonia oxidizer [on the basis of the proposal by Reysenbach and Shock (22) that thermophily starts at 45°C], partially inhibited by ammonium in a concentration of 3.08 mM at pH 7.4 and 46°C.
The discovery of the moderately thermophilic “Candidatus Nitrososphaera gargensis” within the group I.1b is noteworthy as it represents the first organism within this group for which, on a cellular level, ammonia oxidation has been shown. Furthermore, all previously described AOA and AOB are mesophiles. It should be mentioned that enrichment of a thermophilic ammonia oxidizer capable of growth at 55°C was reported from geothermal springs of Kamchatka (Russia), but the obtained cultures were unstable and the putative ammonia oxidizers were not identified beyond a morphological description (49). The discovery of a thermophilic AOA adds to our picture of biogeochemical nitrogen cycling in thermophilic environments for which nitrite oxidation, nitrate ammonification, denitrification, and nitrogen fixation have already been reported (23, 50, 51). Furthermore, a thermophilic origin was hypothesized for anaerobic ammonium oxidation as well (52). The existence of thermophilic AOA is also consistent with the previously postulated idea that the mesophilic crenarchaeotes are descendants of ancestral thermophiles (53) and thus suggests that archaeal ammonia oxidation evolved under thermophilic conditions with the mesophilic lifestyles exemplified by soil or marine AOA likely representing independent, secondary adaptations to lower temperatures. Ammonia oxidation as an ancient form of energy conservation is consistent with the postulated early earth chemically driven nitrogen cycle (54), which provides for the formation of ammonia at high temperatures (24–26).
Materials and Methods
Chemical Analyses of the Enrichment.
The concentration of ammonium was measured by fluorescence detection according to Corbin (55) after precolumn-derivatization with OPA reagent and HPLC separation. Nitrite and nitrate concentrations were determined quantitatively by UV detection after ion-pair chromatography on a Hypersil ODS C18 column (56). The oxygen concentration in enrichment flasks was determined with an electrode (OXI 96; Nova Analytics).
Amplification, Cloning, and Phylogenetic Analyses of 16S rRNA and amo Genes.
PCR amplifications of archaeal and bacterial 16S rRNA and amo genes were performed directly from harvested cells (diluted in 0.9% NaCl) from an ammonia-oxidizing enrichment from the Garga hot spring in the Buryat Republic, Russia (27), by using primers and annealing temperatures as listed in SI Table 2. At the time of analyses, this enrichment had been maintained for 6 years but had not been screened for ammonia oxidizers with nucleic acid-based methods. All PCRs started with an initial heating step at 95°C for 5–7 min to induce cell lysis. PCR-enhancing substances (betaine, bovine serum albumin, dimethyl sulfoxide, or formamide) were added according to the literature (57, 58). Cloning and sequencing were done as described in ref. 59. The diversity of sequences within the clone libraries was screened with RFLP, by using the enzyme MspI (Fermentas Life Sciences Inc.). From each resulting RFLP pattern, at least one representative clone was sequenced. Sequencing was performed by using the BigDye Terminator Cycle Sequencing Kit v3.1 and the ABI 3130xl Sequencer (Applied Biosystems) following the manufacturer's protocols. Obtained sequences were phylogenetically analyzed by using the software programs ARB (60) and PHYLIP (61) with comprehensive databases that contained 16S rRNA and all available archaeal amo sequences, respectively.
RT-PCR of Crenarchaeotal amoA mRNA.
Cells were harvested from the enrichment by centrifugation and were stored in RNAlater (Ambion) at 4°C until further processing. Total RNA was isolated by using TRIzol (Invitrogen) according to the manufacturer's instructions with the following modifications: RNA was precipitated overnight at −20°C in the presence of 5 μg of glycogen (Sigma). After incubation with DNase (Sigma), reverse transcription was carried out by using the Revert Aid First Strand cDNA Kit (Fermentas Inc.) according to the manufacturer's instructions with the clone-specific primer amo586R (5′-AGC AAT GGG AAC TGA CAG-3′), which had been designed by using ARB. cDNA was amplified by using clone-specific primers amo16F (5′-ACG CAC AAC GCA CTA CTT-3′) and amo586R (length of amplificate: 570 bp) with thermal cycling as follows: initial denaturation at 94°C for 4 min followed by 40 cycles of denaturation at 94°C for 40 s, annealing at 60°C for 30 s, and elongation at 72°C for 40 s, followed by a final elongation step at 72°C for 10 min. Specificity of amplification was confirmed by cloning and sequencing of the amplification product. Absence of contaminating DNA in the RNA extract was demonstrated by performing the aforementioned PCR without the initial reverse transcription step.
FISH and CARD-FISH.
To visualize AOB and nitrite oxidizers in the enrichment, FISH was performed as described by Daims et al. (62) by using probes listed in SI Table 1. No unspecific labeling of cells was observed in any sample by using the nonsense probe. After FISH, cells were stained at 4°C with the DNA-binding dye at 1 μg ml−1. Microscopic observation and documentation were accomplished by using a LSM 510 scanning confocal microscope (Zeiss) and the included software.
To visualize AOA in situ, a specific 16S rRNA-targeting FISH-probe was designed by using ARB. Probe RHGA702 (SI Table 1) targeted all crenarchaeotal 16S rRNA sequences obtained from the enrichment but has at least one central mismatch to all other sequences in our ARB database including all published sequences from Crenarchaeota. For more details regarding this probe, please refer to probeBase (63) at http://www.microbial-ecology.net/probebase. HPLC-purified and lyophilized HRP-conjugated oligonucleotides were obtained from Thermo Electron. For CARD-FISH analyses, ethanol-fixed biomass from the enrichment was immobilized on slides and dehydrated by an increasing ethanol series (50%, 80%, and 96% for 3 min each). Afterward, the slide was dipped into 0.2% agarose (in distilled water) and left to dry at 30°C. Then, permeabilization of cell walls was performed by using 10-min incubation with proteinase K (15 μg ml−1; Sigma). This and all other incubation and washing steps were done at room temperature if not stated otherwise. After permeabilization, the slides were washed for 1 min in distilled water. Subsequently, slides were incubated in 0.01 M hydrogen chloride for 20 min to destroy remaining proteinase K. To inactivate endogenous peroxidases, cells were incubated in a solution of 0.15% hydrogen peroxide in methanol for 30 min. Finally, the slides were washed two times in distilled water for 1 min. The biomass was covered with 20 μl of hybridization buffer (64) at a probe concentration of 0.16 ng μl−1 (65). Hybridization was performed at 46°C for 2.5 h by using the probes Arch915, Cren512, and RHGA702 (10% formamide) in separate experiments (SI Table 1). By using a “nonsense” probe (SI Table 1), no unspecific labeling of cells could be observed. After hybridization, slides were washed in prewarmed washing buffer (64) at 48°C for 15 min. Before tyramide signal amplification, the slides were dipped into distilled water (4°C) and washed for 15 min in 1× PBS. Excess liquid was removed by tapping the slides, and the slides were incubated at 46°C for 45 min with substrate mix by using 1/1,000 parts fluorescein-labeled tyramide (65). Subsequently, the slides were washed in 1× PBS for 10 min and distilled water for 1 min, respectively. Finally, the slides were DAPI-stained and evaluated microscopically as described above. The diameter of the AOA was determined quantitatively by using the digital image analysis software daime (66).
Incubation with Radioactive Bicarbonate and MAR.
For bicarbonate uptake experiments, cells were concentrated from the enrichment via centrifugation (10 min at 13,000 × g), washed in growth medium (27) without ammonium, centrifuged again and resuspended in a small volume of this medium. Resuspended cells were kept at 4°C for no longer than 7 h before the incubation experiments. Resuspended biomass was exposed in parallel experiments to different ammonium concentrations by using a stock solution of 10 mM NH4Cl in a total volume of 3 ml of growth medium. Two biological replicates were performed for each of these experiments. Furthermore, control experiments were performed without addition of ammonium as well as with UV-sterilized biomass to check for physiological activity without added substrate and for chemography, respectively. For all experiments, the biomass was incubated in vertically fixed vials under aerobic conditions for 3 h at 46°C and was shaken at 110 rpm in a water bath. After this preincubation step, which was included to allow the cells to re-adapt to elevated temperature conditions, a subsample was taken (to determine the ammonium concentration via ion exchange chromatography with chemically suppressed conductivity detection) and 10 μCi [14C]bicarbonate (Hanke Laboratory Products) was added to each vial. Subsequently, the biomass was incubated for 12 h under the conditions described above. In addition, 100 μM of the inhibitor AlTU (Fluka) was added after the preincubation to one of the vials containing 0.76 mM ammonium after preincubation. After incubation, biomass was fixed with EtOH or formaldehyde as described by Daims et al. (62). CARD-FISH and subsequent DAPI staining were performed as described above. The hybridized samples were dipped in preheated (48°C) LM-1 emulsion (Amersham), exposed for 12–15 days at 4°C in the dark and developed in Kodak D19 (40 g liter−1 of distilled water) before microscopic examination. For each condition and biological replicate, three microautoradiography slides were analyzed and at least 10 AOA microcolonies were examined per slide.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dave Stahl, Christa Schleper, Alex Loy, and Ingo Schmidt for helpful discussions, and Marc Mußmann for advice with CARD-FISH. Eberhard Bock is acknowledged for financial support of the Baikal expedition, and we thank Peter-Georg Jozsa for help with the chemical analyses. Roland Hatzenpichler is recipient of a DOC (Ph.D.) fellowship of the Austrian Academy of Sciences at the Department of Microbial Ecology, University of Vienna, Austria. This work was partly supported by the Deutsche Forschungsgemeinschaft (Grant SP 667/5–1 to E.V.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU281317–EU281336).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708857105/DC1.
References
- 1.Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops HP, Wagner M. Appl Environ Microbiol. 2000;66:5368–5382. doi: 10.1128/aem.66.12.5368-5382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purkhold U, Wagner M, Timmermann G, Pommerening-Röser A, Koops HP. Int J Syst Evol Microbiol. 2003;53:1485–1494. doi: 10.1099/ijs.0.02638-0. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt I, Sliekers O, Schmid M, Cirpus I, Strous M, Bock E, Kuenen JG, Jetten MSM. FEMS Microbiol Ecol. 2002;39:175–181. doi: 10.1111/j.1574-6941.2002.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 4.Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- 5.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 6.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 7.Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, et al. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 9.Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MM. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Preston CM, Wu KY, Molinski TF, DeLong EF. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, de Nys R, Wagner M, Taylor M. Environ Microbiol. 2007 doi: 10.1111/j.1462-2920.2007.01515.x. [DOI] [PubMed] [Google Scholar]
- 14.Beman JM, Francis CA. Appl Environ Microbiol. 2006;72:7767–7777. doi: 10.1128/AEM.00946-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beman JM, Roberts K, Wegley L, Rohwer F, Francis CA. Appl Environ Microbiol. 2007;73:5642–5647. doi: 10.1128/AEM.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, Di H. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 18.Park HD, Wells GF, Bae H, Criddle CS, Francis CA. Appl Environ Microbiol. 2006;72:5643–5647. doi: 10.1128/AEM.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spear JR, Barton HA, Robertson CE, Francis CA, Pace NR. Appl Environ Microbiol. 2007;73:6172–6180. doi: 10.1128/AEM.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weidler GW, Dornmayr-Pfaffenhuemer M, Gerbl FW, Heinen W, Stan-Lotter H. Appl Environ Microbiol. 2007;73:259–270. doi: 10.1128/AEM.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehta MP, Butterfield DA, Baross JA. Appl Environ Microbiol. 2003;69:960–970. doi: 10.1128/AEM.69.2.960-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reysenbach AL, Shock E. Science. 2002;296:1077–1082. doi: 10.1126/science.1072483. [DOI] [PubMed] [Google Scholar]
- 23.Stetter KO. In: The Molecular Origins of Life: Assembling Pieces of the Puzzle. Brack A, editor. Cambridge, UK: Cambridge Univ Press; 1998. pp. 315–335. [Google Scholar]
- 24.Blöchl E, Keller M, Wächtershäuser G, Stetter KO. Proc Natl Acad Sci USA. 1992;89:8117–8120. doi: 10.1073/pnas.89.17.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandes JA, Boctor NZ, Cody GD, Cooper BA, Hazen RM, Yoder HS. Nature. 1998;395:365–367. doi: 10.1038/26450. [DOI] [PubMed] [Google Scholar]
- 26.Brandes JA, Devol AH, Yoshinari T, Jayakumar DA, Naqvi SWA. Limnol Oceanogr. 1998;43:1680–1689. [Google Scholar]
- 27.Lebedeva E, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E. FEMS Microbiol Ecol. 2005;54:297–306. doi: 10.1016/j.femsec.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schleper C, Jurgens G, Jonuscheit M. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 30.Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 31.Ehrhardt CJ, Haymon RM, Lamontagne MG, Holden PA. Environ Microbiol. 2007;9:900–912. doi: 10.1111/j.1462-2920.2006.01211.x. [DOI] [PubMed] [Google Scholar]
- 32.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 33.Takai K, Horikoshi K. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollmann A, Schmidt I, Saunders AM, Nicolaisen MH. Appl Environ Microbiol. 2005;71:1276–1282. doi: 10.1128/AEM.71.3.1276-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebie Y, Miura H, Noda N, Matsumura M, Tsuneda S, Hirata A, Inamori Y. Water Sci Technol. 2002;46:281–288. [PubMed] [Google Scholar]
- 36.SayavedraSoto LA, Hommes NG, Russell SA, Arp DJ. Mol Microbiol. 1996;20:541–548. doi: 10.1046/j.1365-2958.1996.5391062.x. [DOI] [PubMed] [Google Scholar]
- 37.Sato C, Schnoor JL, McDonald DB, Huey J. Appl Environ Microbiol. 1985;49:1101–1107. doi: 10.1128/aem.49.5.1101-1107.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suwa Y, Imamura Y, Suzuki T, Tashiro T, Urushigawa Y. Water Res. 1994;28:1523–1532. [Google Scholar]
- 39.Suwa Y, Sumino T, Noto K. J Gen Appl Microbiol. 1997;43:373–379. doi: 10.2323/jgam.43.373. [DOI] [PubMed] [Google Scholar]
- 40.Koops HP, Pommerening-Röser A. FEMS Microbiol Ecol. 2001;37:1–9. [Google Scholar]
- 41.Schmidt I, Look C, Bock E, Jetten MS. Microbiology. 2004;150:1405–1412. doi: 10.1099/mic.0.26719-0. [DOI] [PubMed] [Google Scholar]
- 42.Weidinger K, Neuhauser B, Gilch S, Ludewig U, Meyer O, Schmidt I. FEMS Microbiol Lett. 2007;273:260–267. doi: 10.1111/j.1574-6968.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 43.Adamczyk J, Hesselsoe M, Iversen N, Horn M, Lehner A, Nielsen PH, Schloter M, Roslev P, Wagner M. Appl Environ Microbiol. 2003;69:6875–6887. doi: 10.1128/AEM.69.11.6875-6887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ginestet P, Audic JM, Urbain V, Block JC. Appl Environ Microbiol. 1998;64:2266–2268. doi: 10.1128/aem.64.6.2266-2268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hooper AB, Terry KR. J Bacteriol. 1973;115:480–485. doi: 10.1128/jb.115.2.480-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bedard C, Knowles R. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray RG, Schleifer KH. Int J Syst Bacteriol. 1994;44:174–176. doi: 10.1099/00207713-44-1-174. [DOI] [PubMed] [Google Scholar]
- 48.Murray RG, Stackebrandt E. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 49.Golovacheva RS. Mikrobiologiia. 1976;45:329–331. [PubMed] [Google Scholar]
- 50.Lebedeva EV, Alawi M, Maixner F, Jozsa P-G, Daims H, Spieck E. Int J Syst Evol Microbiol. 2008;58:242–250. doi: 10.1099/ijs.0.65379-0. [DOI] [PubMed] [Google Scholar]
- 51.Mehta MP, Baross JA. Science. 2006;314:1783–1786. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- 52.Canfield DE, Rosing MT, Bjerrum C. Philos Trans R Soc London B Biol Sci. 2006;361:1819–1834. doi: 10.1098/rstb.2006.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barns SM, Delwiche CF, Palmer JD, Pace NR. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mancinelli RL, McKay CP. Origins Life Evol Biosphere. 1988;18:311–325. doi: 10.1007/BF01808213. [DOI] [PubMed] [Google Scholar]
- 55.Corbin JL. Appl Environ Microbiol. 1984;47:1027–1030. doi: 10.1128/aem.47.5.1027-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meincke M, Bock E, Kastrau D, Kroneck PMH. Arch Microbiol. 1992;158:127–131. [Google Scholar]
- 57.Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovarova M, Draber P. Nucleic Acids Res. 2000;28:E70. doi: 10.1093/nar/28.13.e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Röser A, Koops HP, Wagner M. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Felsenstein J. PHYLIP. Seattle: Dept of Genetics, Univ of Washington; 1993. Version 3.65. [Google Scholar]
- 62.Daims H, Stoecker K, Wagner M. In: Molecular Microbial Ecology. Osborn AM, Smith CJ, editors. Abingdon, UK: BIOS Scientific Publ; 2005. pp. 213–239. [Google Scholar]
- 63.Loy A, Horn M, Wagner M. Nucleic Acids Res. 2003;31:514–516. doi: 10.1093/nar/gkg016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pernthaler A, Pernthaler J, Amann R. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pernthaler A, Pernthaler P, Amann R. In: Molecular Microbial Ecology Manual. Kowalchuk GA, editor. Boston: Kluwer; 2004. pp. 711–726. [Google Scholar]
- 66.Daims H, Lücker S, Wagner M. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- 67.Robertson CE, Harris JK, Spear JR, Pace NR. Curr Opin Microbiol. 2005;8:638–642. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.