Abstract
The trimeric translation initiation factor a/eIF2 of the crenarchaeon Sulfolobus solfataricus is pivotal for binding of initiator tRNA to the ribosome. Here, we present in vitro and in vivo evidence that the a/eIF2 γ-subunit exhibits an additional function with resemblance to the eukaryotic cap-complex. It binds to the 5′-triphosphate end of mRNA and protects the 5′ part from degradation. This unprecedented capacity of the archaeal initiation factor further indicates that 5′ → 3′ directional mRNA decay is a pathway common to all domains of life.
In Escherichia coli, the decay of most RNA transcripts appears to be initiated by 5′ pyrophosphate removal (1), followed by endonucleolytic cleavages that are generated by RNase E (2, 3). The intermediate cleavage products are further degraded by the 3′ → 5′ exonucleases polynucleotide phosphorylase (PNPase), RNase II, and oligoribonuclease, converting the decay intermediates into poly- and mononucleotides (4). At variance with E. coli, Bacillus subtilis possesses a 5′ → 3′ exonuclease activity, which could explain why RNAs in this organism are stabilized for great distances downstream of stable secondary structures or bound ribosomes (5). In eukaryotes, mRNA decay is mainly catalyzed by exonucleases (6). Eukaryotic mRNAs generally have a 7-methylguanosine cap at their 5′ end and a poly(A) tail at their 3′ end. Removal of these terminal modifications is considered rate-limiting for mRNA decay (7). Translation initiation factor eIF4E binds to the 7-methylguanosine cap and thereby protects the cap structure from the decapping enzyme and consequently the mRNA from 5′ → 3′ exonucleolytic decay (7). Different RNases with either endo- or exonuclease activity (8–11) have been inferred or described in Archaea. In S. solfataricus a 3′ → 5′ directional decay by a multisubunit exosome complex has been demonstrated in vitro (10). In addition, the Sulfolobus solfataricus exosome is able to polyadenylate the 3′ end of RNAs in the presence of ADP (12). With the exception of the exosome, no other endo- or exonuclease activities have been described in S. solfataricus. At variance with a previous study wherein the longevity of selected S. solfataricus mRNAs was found to be rather high (13), a recent microarray-based analysis (14) indicated a rather short mRNA half-life, comparable with that of bacterial mRNAs.
Two different mechanisms for translational initiation seem to exist in Sulfolobus (15). One is based on a canonical SD/anti-SD interaction and operates on internal cistrons of polycistronic mRNAs. In contrast, monocistronic mRNAs and proximal genes of polycistronic mRNAs are frequently devoid of a 5′ untranslated region. Decoding of these leaderless mRNAs requires, analogously to Bacteria (16), pairing of the start codon with initiator-tRNA (15). The complexity of archaeal translational initiation seems to be underscored by the presence of a larger-than-bacterial set of factors because Archaea encode ≈10 orthologs of eukaryal and bacterial initiation factors (17). Like its eukaryotic counterpart eIF2 (18), the archaeal factor a/eIF2 consists of three subunits, α, β, and γ, and recruits initiator tRNA (tRNAi) to the 30S ribosomal subunit. In a/eIF2, the α- and γ-subunits are required for tRNAi binding (19, 20), whereas in eIF2, the β- and γ-subunits (18) are responsible for this task.
Some proteins of the translation machinery have additional functions. For instance, E. coli ribosomal protein L4 has a second function in transcription termination, whereas S1 is part of the replicase of the E. coli phage Qβ (21, 22). The ribosomal proteins S16 of E. coli and L32–2 of Schizosaccharomyces pombe display endonuclease activity (23) and act as a transcription activator (24), respectively. Here, we show that the a/eIF2 γ-subunit (a/eIF2-γ) of S. solfataricus has a dual function. Beside its requirement for tRNAi binding to the ribosome, the initiation factor binds to the triphosphorylated 5′ end of mRNA and counteracts 5′ → 3′ directional decay.
Results
a/eIF2(-γ) Binds to the 5′ End of mRNA.
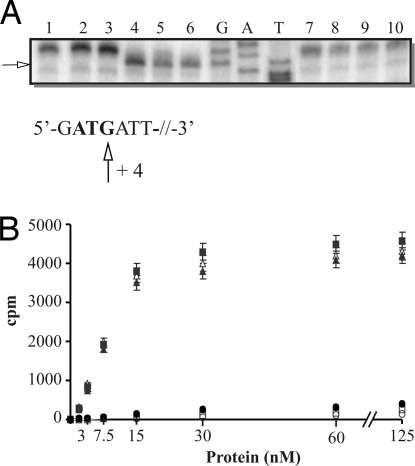
During our efforts to establish an in vitro translation initiation assay for S. solfataricus, we observed that translation initiation factor a/eIF2 binds to the 5′ end of RNA. Primer extension experiments in the presence of a/eIF2 and S. solfataricus 2508sh mRNA revealed an a/eIF2-specific stop signal positioned at nucleotide +4 with respect to the 5′ end of the RNA (Fig. 1A, lane 5). The position of the stop signal was independent of the RNA substrate because a +4 stop signal was also observed with heterologous and other homologous mRNAs including E. coli ompA and λ cI mRNAs as well as S. solfataricus 104 and tfb-1 mRNAs (data not shown). We then asked which individual subunit of a/eIF2 is required for 5′ end binding. As shown in Fig. 1A, lane 4, a/eIF2-γ alone was able to bind to the 5′ end, i.e., block the reverse transcriptase at position +4, whereas the α- and β-subunits failed to do so (Fig. 1A, lanes 2 and 3). Because a/eIF2 and a/eIF2-γ binding was independent of the nature of the mRNAs, the only constant recognition motif for the protein appeared to be the triphosphate group at the 5′ end. Hence, we tested whether a/eIF2 or a/eIF2-γ bind to 5′-dephosphorylated 2508sh mRNA. As shown in Fig. 1A (lanes 7–10), the a/eIF2-dependent stop signal of the reverse transcriptase was not obtained with 5′-dephosphorylated RNA, indicating that the triphosphate group is required for 5′-end binding of a/eIF2-γ.
Fig. 1.
The a/eIF2 γ-subunit binds to the 5′ end of triphosphorylated 2508sh mRNA. (A) Lanes 1–6, primer extension reactions with 2508sh mRNA (0.05 nM) without addition of factors (lane 1), in the presence of 0.1 μM the a/eIF2 α-subunit (lane 2), the a/eIF2 β-subunit (lane 3), the a/eIF2 γ-subunit (lane 4), reconstituted a/eIF2 (lane 5), or cross-linked a/eIF2 (lane 6). Lanes 7–10, primer extension reactions with 5′-dephosphorylated 2508sh mRNA (0.05 nM) without addition of factors (lane 7), in the presence of 0.1 μM concentrations of the a/eIF2 γ-subunit (lane 8), reconstituted a/eIF2 (lane 9), or cross-linked a/eIF2 (lane 10). G, A, and T represent sequencing reactions obtained with 2508sh mRNA containing a 21-nt extension at the 5′ end. The sequence of the 5′-proximal part of 2508sh mRNA is shown below the autoradiograph. The start codon is in bold. The arrow indicates the position of the a/eIF2(-γ)-dependent stop signal. (B) Radiolabeled 2508sh mRNA (1 μM) was incubated with increasing amounts of the a/eIF2 α-subunit (○), the a/eIF2 β-subunit (□), the a/eIF2 γ-subunit (■), with reconstituted a/eIF2 (▴), and with cross-linked a/eIF2 (Δ) at 70°C. Dephosphorylated radiolabeled 2508sh mRNA (1 μM) was incubated with increasing amounts of a/eIF2 (●) at 70°C. Samples were withdrawn after 5 min, and the amount of RNA retained on the filter was determined. The experiment was performed in triplicate. The error bars represent standard deviations.
Next, the binding affinity of a/eIF2 and a/eIF2-γ for 2508sh mRNA was determined by using filter-binding assays. The apparent Kd of a/eIF2 and of cross-linked a/eIF2 for 2508sh mRNA was calculated with ≈10 nM (Fig. 1B). The binding affinity of a/eIF2-γ was indistinguishable from that of a/eIF2 or cross-linked a/eIF2 (Fig. 1B). As expected from the results shown in Fig. 1A, neither the a/eIF2 α- nor the β-subunit displayed any binding activity, and a/eIF2 did not bind to dephosphorylated 2508sh mRNA (Fig. 1B) or to monophosphorylated 2508sh mRNA generated by tobacco acid pyrophospatase (data not shown). The a/eIF2 γ-subunit contains the guanine nucleotide-binding pocket of a/eIF2 (20), and binding of a/eIF2 to tRNAi depends on GTP (19). However, as assessed by filter-binding assays, the affinity for 2508sh mRNA of both a/eIF2 and the a/eIF2 γ-subunit was unaffected in the presence of GTP or GDP (data not shown).
Binding of the a/eIF2 γ-Subunit Protects the 5′ End of mRNA from Degradation in Vitro.
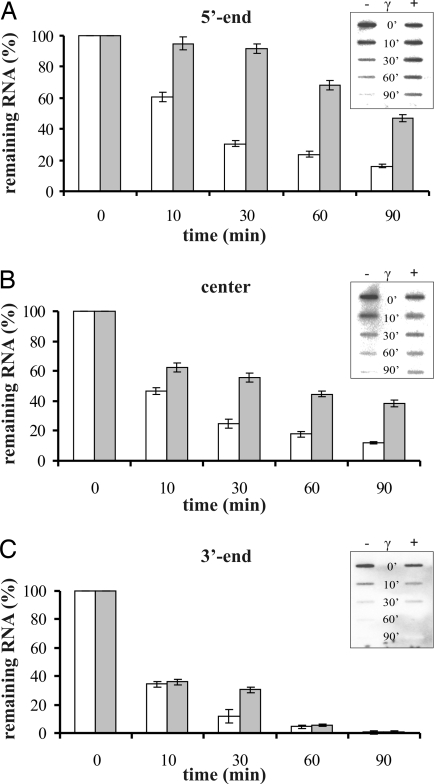
The intact cap-structure at the 5′ end of eukaryotic mRNAs (7) and the 5′ triphosphate group of prokaryotic mRNAs (1) counteract degradation of transcripts. Therefore, we speculated that one possible role for specific binding of a/eIF2-γ to the triphosphorylated 5′ end of mRNAs could be the protection from 5′ → 3′ decay. To test this hypothesis, the a/eIF2 γ-subunit was bound to 5′-triphosphorylated full-length 2508 mRNA, and the segmental stability of the RNA was determined upon incubation with S. solfataricus S100 extracts. Three radioactively labeled DNA oligonucleotides, complementary to the 5′ end, the central region, and the 3′ end of the RNA were used as probes. In the absence of a/eIF2-γ, the signal intensities for the 5′ end decreased to ≈60% and ≈20% after 10 and 60 min (Fig. 2A), respectively. The signal intensities for the 3′ end decreased faster; after 10 and 60 min only ≈35% and ≈5% remained (Fig. 2C), respectively. For mRNA prebound to the a/eIF2 γ-subunit, the signal intensities for the 5′ end decreased only to ≈95% and ≈70% after 10 and 60 min (Fig. 2A), respectively, whereas the decrease in the signal intensities for the 3′ end was comparable to that seen in the absence of the factor (Fig. 2C). The signal intensities for the central part of 2508 mRNA decreased faster than the corresponding signal intensities for the 5′ end and slower than that for the 3′ end (Fig. 2B). Moreover, in agreement with the lack of binding of a/eIF2-γ to dephosphorylated 2508sh mRNA (Fig. 1), the presence of the a/eIF2-γ did not confer protection to the 5′ end of dephosphorylated 2508 mRNA [supporting information (SI) Fig. 5]. In summary, these in vitro studies indicated that the a/eIF2 γ-subunit protects the 5′ end of 2508 mRNA from degradation and that 3′ → 5′ decay is not affected by a/eIF2-γ.
Fig. 2.
The a/eIF2 γ-subunit protects the 5′ end of triphosphorylated 2508 mRNA from degradation in vitro. The 2508 full-length mRNA (0.25 μM) was preincubated in the absence (white bars) or presence (gray bars) of 0.5 μM a/eIF2-γ for 5 min at 70°C. S. solfataricus S100 extract was added, and samples were withdrawn after 0, 10, 30, 60, and 90 min. The remaining RNA was determined with DNA probes complementary to the 5′ end (A), the central part (B), or the 3′ end (C) of 2508 full-length mRNA. An autoradiograph of a corresponding slot blot is shown in the Insets. The signals were visualized by a PhosphorImager and quantified with ImageQuant software. The signal at time 0 was set to 100%. The graphical representations shown are an average of two independent experiments. The error bars represent standard deviations.
Overproduction of the a/eIF2 γ-Subunit Stabilizes S. solfataricus mRNA.
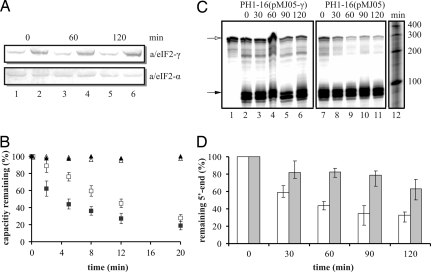
We next tested whether overproduction of a/eIF2-γ would result in stabilization of mRNAs in vivo. The S. solfataricus strain PH1–16 was transformed with plasmid pMJ05-γ, which harbors the a/eIF2-γ gene under transcriptional control of the araS promoter (25). When compared with strain PH1–16 containing the parental vector pMJ05, the levels of a/eIF2-γ were ≈10-fold increased in strain PH1–16(pMJ05-γ) during growth in the presence of arabinose (Fig. 3A). As a measure for the overall stability of bulk mRNA, we first monitored the total protein synthesizing capacity by determining the incorporation of [35S]methionine in both strains at several times after addition of the transcription inhibitor actinomycin D. In contrast to the control strain PH1–16(pMJ05), the total protein synthesizing capacity decreased with a slower rate in strain PH1–16(pMJ05-γ), suggesting that overproduction of a/eIF2-γ resulted in stabilization of bulk mRNA (Fig. 3B).
Fig. 3.
Overproduction of a/eIF2-γ in S. solfataricus stabilizes mRNA. (A) Synthesis of the a/eIF2 α- and γ-subunits in S. solfataricus strains PH1–16(pMJ05) (lanes 1, 3, and 5) and PH1–16(pMJ05-γ) (lanes 2, 4, and 6) as revealed by quantitative immunoblotting. Samples were withdrawn 0, 60, and 120 min after addition of actinomycin D. (B) S. solfataricus strains PH1–16(pMJ05) (■) and PH1–16(pMJ05-γ) (□) were grown in Brock′s medium supplemented with 0.1% tryptone and 0.2% d-arabinose. At time 0, 10 μg/ml actinomycin D was added, and labeling with [35S]methionine was performed at the times indicated. After TCA precipitation, the incorporated radioactivity was determined by using a scintillation counter. The radioactivity incorporated at time 0 was set to 100%. Labeling of aliquots of strains PH1–16(pMJ05) (▴) and PH1–16(pMJ05-γ) (▵) grown in the absence of actinomycin D served as a control. The graphical representation is an average of three independent experiments. The error bars represent standard deviations. (C) As described in Materials and Methods, total RNA was prepared from S. solfataricus strains PH1–16(pMJ05-γ) (lanes 2–6) and PH1–16(pMJ05) (lanes 7–11) at the indicated times after addition of 10 μg/ml actinomycin D. The amount of remaining 5′ end of 2508 mRNA and of 5S rRNA was determined by using an RNase protection assay. The signals of the protected fragments of 2508 mRNA (open arrow) and 5S rRNA (filled arrow) show the expected length of 289 and 90 nt, respectively. Lane 1, protected fragment of 2508 mRNA. Lane 12, RNA ladder (Fermentas). (D) The signals were visualized by a PhosphorImager and quantified with ImageQuant software. The signals at time 0 were set to 100%. The graphical representation (average of three independent experiments) shows the relative amount (normalized to 5S rRNA signals) of remaining 5′ end of 2508 mRNA as a function of time after addition of actinomycin D in strains PH1–16(pMJ05-γ) (gray bars) and PH1–16(pMJ05) (white bars). The error bars represent standard deviations.
To test whether overproduction of a/eIF2-γ would result in stabilization of the 5′-segment of 2508 mRNA in vivo, an RNase protection assay was used. Total RNA was prepared from S. solfataricus strains PH1–16(pMJ05) and PH1–16(pMJ05-γ) at several times after addition of actinomycin D, and the 5′ end of 2508 mRNA was detected with a probe spanning nucleotides 1 to 289 of the mRNA. As shown in Fig. 3 C and D, this experiment corroborated the in vitro studies (Fig. 2), in that a significantly increased stability of the 5′ end of 2508 mRNA was observed in strain PH1–16(pMJ05-γ), i.e., in the presence of increased levels of a/eIF2-γ.
Hierarchy of mRNA and Met-tRNAi Binding to Free and Ribosome-Bound a/eIF2.
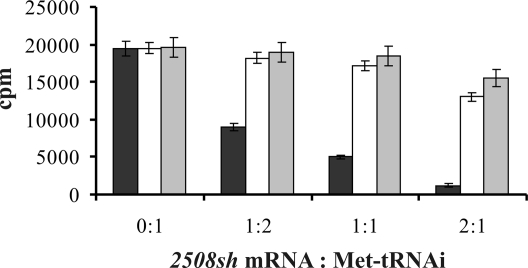
The apparent Kd of free a/eIF2 for Met-tRNAi in the presence of GTP was determined with ≈150 nM (SI Fig. 6), which is ≈15-fold lower than that of a/eIF2 or a/eIF2-γ for the 5′-triphosphorylated end of mRNA (Fig. 1B). Accordingly, the addition of increasing amounts of 2508sh mRNA competed with [35S]Met-tRNAi for binding to free a/eIF2 (Fig. 4). In light of the function of the eukaryotic ortholog of a/eIF2, which is believed to serve as a shuttle for Met-tRNAi to the 40S subunit (26), these observations were surprising because preferential binding of a/eIF2 to mRNA would interfere with its pivotal function in translation initiation (19, 20), i.e., with Met-tRNAi binding to the ribosome. To address this paradox, we asked whether ribosome-bound a/eIF2 displays an increased affinity for Met-tRNAi, which would parallel the mode of action of bacterial IF2 (27). In the presence of ribosomes, the binding affinity of a/eIF2 for Met-tRNAi was ≈3-fold increased (SI Fig. 6). Consequently, 2508sh mRNA competed much less efficiently with Met-tRNAi binding to a/eIF2 in the presence of ribosomes (Fig. 4). Translation initiation factor aIF1 is known to stimulate binding of Met-tRNAi to ribosomes in the presence of GTP and a/eIF2 (28). When compared with free a/eIF2, we observed a ≈5-fold increase in the binding affinity of a/eIF2 for Met-tRNAi in the presence of ribosomes and aIF1 (SI Fig. 6), and 2508sh mRNA no longer competed with Met-tRNAi binding under these conditions (Fig. 4). These experiments suggested that, in the presence of all initiation factors, 30S-bound a/eIF2 preferentially binds Met-tRNAi and thereby performs its task in translation initiation
Fig. 4.
Effect of S. solfataricus ribosomes and aIF1 on tRNAi and mRNA binding to a/eIF2. The reconstituted a/eIF2 trimer (0.5 μM) was incubated in the presence of GTP (1 mM) at 70°C for 5 min with [35S]Met-tRNAi (0.25 μM). Then different concentrations of 2508sh mRNA (0, 0.125, 0.25, and 0.5 μM) were added. The molar ratios of 2508sh mRNA:[35S]Met-tRNAi are given below the graph. The experiments were performed in the absence (black bars) and presence (white bars) of S. solfataricus ribosomes as well as in the presence of S. solfataricus ribosomes and aIF1 (gray bars). The amount of [35S]Met-tRNAi retained by a/eIF2 was determined by filtration through nitrocellulose filters. The graphical representation shown is an average of three independent experiments. Error bars represent standard deviations.
Discussion
Based on the crystal structures of the S. solfataricus a/eIF2-αγ heterodimer (29) and a/eIF2-γ (30), two models for Met-tRNAi binding have been put forward. One model specifies that the α-subunit is not directly involved in Met-tRNAi binding and that Met-tRNAi binding is achieved by the γ-subunit through a methionine-binding pocket and interactions with the terminal A76 and the T-stem of Met-tRNAi (29). The other model (30) deviates from the first in that Met-tRNAi forms extensive contacts with both the α- and γ-subunit and thus supports the observation that the a/eIF2-αγ heterodimer is necessary and sufficient for the stable interaction with tRNAi (19, 20). The competition experiment shown in Fig. 4 implied an overlap of the binding sites for Met-tRNAi and the 5′-triphosphate end of mRNA on a/eIF2. However, because an a/eIF2-γ mutant protein defective in Met-tRNAi binding (20) retained the same affinity for the 5′ end of mRNAs (data not shown), there is either only a partial overlap of the binding sites for either substrate, or alternatively, binding of a/eIF2-γ to the 5′ end of mRNAs interferes with GTP-dependent structural rearrangements in the protein required for binding of Met-tRNAi (29, 30).
Although the level of a/eIF2 was ≈10-fold enhanced in strain PH1–16(pMJ05-γ) when compared with the control strain PH1–16(pMJ05), total protein synthesis in strain PH1–16(pMJ05-γ) was not increased before addition of actinomycin D (Fig. 3 A and B). Hence, the slower decrease in the protein synthesizing capacity after inhibition of transcription (Fig. 3B) seen in the a/eIF2-γ-overproducing strain cannot be attributed to a stimulating effect of increased a/eIF2-γ levels on protein synthesis. Taken together with the stabilization of the 5′ part of 2508 mRNA upon binding of a/eIF2-γ in vitro (Fig. 2), we therefore conclude that in vivo overproduction of a/eIF2-γ in S. solfataricus stabilizes mRNA, as exemplified by the increased stability of 2508 mRNA (Fig. 3C). The E. coli RNases E/G and the B. subtilis RNases J1 and J2 show 5′ end-dependent cleavage. They bind to the 5′ end of RNA, sense the state of phosphorylation, and preferentially cleave monophosphorylated RNA (2, 5). Similarly, the initial velocity of the 5′ → 3′ exonuclease activity of B. subtilis RNase J1 was found to be increased with a 5′ monophosphorylated substrate (5). Given the modus operandi of these enzymes, it seems reasonable to pursue the idea that binding of a/eIF2 to the 5′-triphosphorylated end of S. solfataricus mRNAs diminishes dephosphorylation (1), and thus subsequent processing by an endo- or exonuclease yet to be identified in this organism. Whatever the nature of the nuclease(s) involved, this study revealed a striking functional parallel between a/eIF2(-γ) and the eukaryotic cap-complex (7) and further suggests that 5′ → 3′ directional mRNA decay is common to all domains of life.
In Bacteria, ribosome-bound IF2 recruits fMet-tRNAfMet (27), whereas in Eukaryotes the a/eIF2·GTP·Met-tRNAi complex is formed in the cytoplasm and then binds to the 40S subunit (26). The binding and competition studies indicated that ribosome-bound a/eIF2 exhibits an increased affinity for Met-tRNAi (Fig. 4), suggesting that Met-tRNAi recruitment parallels the bacterial (27) rather than the eukaryal pathway. It seems conceivable that, under conditions of a feast lifestyle, i.e., when there is a surplus of ribosomes, a/eIF2 fulfills primarily its role in translation initiation. In contrast to energetically favorable chemoorganotrophic growth, Sulfolobus can also thrive in sulfur-rich hot springs wherein it grows chemolithotrophically. Under the latter conditions, the cells grow rather poorly because the free energy available is low. In Bacteria and Archaea, the levels of ribosomes vary with growth conditions (31, 32). We speculate that, under famine conditions or during stringent control (33), when ribosome synthesis is likely to be decreased, a/eIF2-γ counteracts 5′-to-3′ mRNA decay. Preliminary experiments designed to mimic feast and famine conditions seem to be in line with this hypothesis. When compared with logarithmic growth in rich medium, the ratio between a/eIF2-γ and ribosomal protein S21 (taken as an indicator for the 30S concentration) was ≈5-fold increased during stationary phase in minimal medium (SI Fig. 7A). Interestingly, the increased a/eIF2/ribosome ratio in stationary phase was paralleled by an increased in vivo stability of the 5′ segment of 2508 mRNA when compared with logarithmic growth (SI Fig. 7 B and C).
There is another aspect of mRNA stabilization by a/eIF2(-γ) that merits consideration. Using in vitro toeprinting, we—not surprisingly—observed that translation initiation on leaderless S. solfataricus 2508 mRNA was inhibited when a/eIF2-γ was prebound to the mRNA (D.H., unpublished data). Given that leaderless mRNAs are prevalent in S. solfataricus (15) and that Met-tRNAi is unlikely to compete for the factor when bound to the 5′-triphosphorylated end of mRNAs (see Fig. 4), a question that requires further experimentation clearly concerns the recycling of a/eIF2(-γ)-protected mRNAs for translation.
Materials and Methods
Plasmids, Strains, and Growth Conditions.
The construction of plasmid pMJ05-γ is described in (SI Text). Electroporation of S. solfataricus PH1–16 and the isolation of transformants was done as described (34). The S. solfataricus pyrEF mutant PH1–16 was grown at 80°C in Brock's medium with or without 10 μg/ml uracil in the presence of 0.1% tryptone and 0.2% d-arabinose (34).
Preparation of S. solfataricus Ribosomes, S. solfataricus tRNAi, and Translation Initiation Factors.
S. solfataricus ribosomes were prepared from frozen cells as described (35). Initiator tRNA from S. solfataricus was transcribed and charged with cold methionine or [35S]methionine (Amersham Pharmacia Biotech) as described (19). All three subunits from a/eIF2 and N-terminal His-tagged aIF1 were prepared as described (19, 28). Cross-linked a/eIF2 was prepared as follows. The reconstituted a/eIF2 trimer was incubated at room temperature in the presence of 5% formaldehyde for 1 h. The cross-linked protein was purified from a native gel by using the ElutaTube protein extraction kit (Fermentas) as specified by the manufacturer.
RNA Preparation.
The bicistronic 2508 mRNA (1,507 nt) from S. solfataricus, encoding an acetyl-CoA-acetyltransferase (SSO2508), was prepared as follows. PCR templates for the generation of 2508 full-length mRNA and for a shorter variant (2508sh mRNA; nucleotides 1–135 of 2508 mRNA) were prepared by using S. solfataricus chromosomal DNA as template and the following oligonucleotides: 5′-AGATAATACGACTCACTATAGATGATTGTAGGATTTGCCGGAAAACT-3′ (2508_FP), 5′-GTTAGCCATTATCCCATCGACGTCAGCG-3′ (2508sh_RP) and 5′-ATATTGTGAAATGATTAAAAGTATTTAAAAGTTA-3′ (2508fl_RP). The mRNAs were synthesized in vitro by using the PCR templates and T7 RNA polymerase (Fermentas) and then gel-purified.
Dephosphorylation of 2508sh and full-length 2508 mRNA was performed with calf intestine alkaline phosphatase (Fermentas). The reactions were carried out two times to increase the yield of dephosphorylated mRNA. To ensure dephosphorylation, the phosphatase-treated RNAs were rephosphorylated with [γ-32P]ATP (Amersham Pharmacia Biotech). As judged by this assay, >95% of the RNA(s) were dephosphorylated.
Filter-Binding Assays.
Various concentrations (0–500 nM) of the factors (a/eIF2, a/eIF2-γ or aIF1) used in the respective assays were incubated for 5 min at 70°C in incubation buffer [18 mM MgCl2, 20 mM Tris·HCl (pH 7), 10 mM KCl] in the presence or absence of 1 mM GTP or GDP and/or in the presence or absence of 1 μM S. solfataricus ribosomes. Internally labeled 2508sh mRNA or [35S]Met-tRNAi was added to the reaction(s), and incubation was prolonged for 5 min at 70°C before the mixture was added to a filtration apparatus equipped with 0.45-μm nitrocellulose filters (Millipore). After washing, the radioactivity retained on the filter was measured by using a scintillation counter. The filter-binding experiments were carried out in triplicate, and the results were averaged. Rate constants measured at variable protein concentrations were fitted to binding curves from which the dissociation constant of the studied protein-RNA complexes was deduced.
Primer Extension Analysis.
The [32P]-5′ end-labeled oligonucleotide 2508tp (5′-TGCCTCATCAGTGACCTCTTTCAATAACTC-3′; complementary to nucleotides +55 to +84 of 2508sh mRNA) was annealed to 5′-triphosphorylated or 5′-dephosphorylated 2508sh mRNA and used to prime cDNA synthesis by reverse transcriptase. Then 0.1 μM concentrations of the individual a/eIF2 subunits, the a/eIF2 trimer, or cross-linked a/eIF2 were added to 2508sh mRNA (0.05 nM) and incubated for 5 min at 70°C in incubation buffer. The reverse-transcriptase reaction was carried out following standard procedures.
In Vitro Protection of mRNA from Degradation.
Full-length 2508 mRNA (0.25 μM) was first incubated with or without 0.5 μM a/eIF2 γ-subunit at 70°C for 5 min in incubation buffer. S100 extracts, prepared from S. solfataricus (35), were preincubated for 1 h at 70°C to degrade most of the endogenous RNAs. The S100 extracts were added to 2508 mRNA, and incubation was continued for 0–90 min at 70°C. The samples were extracted with phenol/chloroform, and the RNA was resuspended in 10 mM Tris·HCl (pH 7). The segmental stability of 2508 mRNA was analyzed by using a Schleicher & Schuell SRC 072/0 Minifold II Slot Blot apparatus. After loading of the RNA samples, the slots were washed with 10 volumes of incubation buffer. Then [32P]-5′ end-labeled DNA probes complementary to the 5′ end (5′-CTTTCTCATAGTTTTTATATAGTTTTCCGGCAAATCCTAC-3′; nucleotides +6 to +47), the central part (5′-ACATCTTCGACTGTAATAGGATTTCTGAACAGAGCTTTAGG-3′; nucleotides +535 to +575); and to the 3′ end (5′-AGTTATGCAAATGTGTTACAGTCTTAAACAGGGGATATTT-3′; nucleotides +1468 to +1507) of full-length 2508 mRNA were added. The signal intensities obtained with the radioactively labeled probes were quantified by a PhosphorImager.
Quantitative Immunoblotting.
Strain S. solfataricus PH1–16-harboring plasmid pMJ05 or pMJ05-γ was grown in Brock's medium in the presence of 0.2% d-arabinose to an OD600 of 0.4. Then, 10 μg/ml actinomycin D was added, and the cells were harvested before addition and 60 and 120 min after addition of the transcription inhibitor. Samples were separated on a 12% SDS/polyacrylamide gel and Western blotting was carried out by using rabbit anti-a/eIF2-α and anti-a/eIF2-γ-subunit antibodies (Pineda) following standard procedures (see SI Fig. 7A).
Determination of Total Protein-Synthesizing Capacity.
Strain S. solfataricus PH1–16-harboring plasmid pMJ05 or pMJ05-γ was grown in Brock's medium in the presence of 0.2% d-arabinose. At an OD600 of 0.4, actinomycin D was added to a final concentration of 10 μg/ml. Samples of 1 ml of culture were withdrawn before and at different times (see Fig. 3B) after addition of actinomycin D. Pulse-labeling was performed by addition of 1 μl of [35S]methionine (10 μCi/ml; Amersham Pharmacia Biotech) for 2 min at 75°C, and then cold methionine (10 mM final concentration) was added. Upon precipitation with 10% TCA, the pellets were washed with cold acetone (90%) and resuspended in water, and the incorporated radioactivity was determined by scintillation counting.
RNase Protection Assay.
Total RNA from S. solfataricus strains PH1–16(pMJ05) and PH1–16(pMJ05-γ) was prepared before and at different times (see Fig. 3C) after addition of actinomycin D as described (36). PCR templates for the generation of 2508 and 5S riboprobes were prepared by using S. solfataricus chromosomal DNA as template and the following oligonucleotides: 5′-GGAGGTAACCTATATATAACGGGTATC-3′ (5S_FP), 5′-TAATACGACTCACTATAGGGCTTAGTGGGGCT. GCGGATCCTCACG-3′ (5S_RP), 5′-CGTGTTTCTCTGCTAACTAGTCGC-3′ (2508_5_FP), and 5′-TAATACGACTCACTATAGGGCTTGTAAGCCCTATATATCATTGAC-3′ (2508_5_RP). The RNase Protection Assay was performed by using the RPA III kit (Ambion) according to the manufacturer's instructions. Samples were separated on 8% polyacrylamide-8M urea gels, and the protected fragments were visualized by using a PhosphorImager.
Supplementary Material
ACKNOWLEDGMENTS:
We are grateful to Drs. M. Garber, O. Nikonov, E. Stolboushkina, and S. Nikonov for comments on the manuscript and Dr. C. Schleper and S. Albers (University of Bergen, Bergen, Norway) for providing materials. This work was supported by Austrian Academic Exchange Service Travel Grant I.3/04 and Austrian Science Fund (FWF) Grant P-15334 (to U.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708894105/DC1.
References
- 1.Celesnik H, Deana A, Belasco J-G. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callaghan A-J, et al. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 3.Carpousis A-J. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher M-P. Promiscuous exoribonucleases of Escherichia coli. J Bacteriol. 1993;175:4577–4583. doi: 10.1128/jb.175.15.4577-4583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathy N, et al. 5′-to-3′ exoribonuclease activity in Bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 6.Newbury S-F. Control of mRNA stability in Eukaryotes. Biochem Soc Trans. 2006;34:30–34. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- 7.Tourriere H, Chebli K, Tazi J. mRNA degradation machines in eukaryotic cells. Biochimie. 2002;84:821–837. doi: 10.1016/s0300-9084(02)01445-1. [DOI] [PubMed] [Google Scholar]
- 8.Buttner K, Wenig K, Hopfner K-P. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Even S, et al. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzetti B, Sohlberg B, Zaccai G, von Gabain A. Biochemical and serological evidence for an RNase E-like activity in halophilic Archaea. J Bacteriol. 1997;179:1180–1185. doi: 10.1128/jb.179.4.1180-1185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portnoy V, et al. RNA polyadenylation in Archaea: Not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus. EMBO Rep. 2005;6:1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bini E, Dikshit V, Dirksen K, Drozda M, Blum P. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA. 2002;8:1129–1136. doi: 10.1017/s1355838202021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson A-F, et al. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 2006;7:R99. doi: 10.1186/gb-2006-7-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benelli D, Maone E, Londei P. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol Microbiol. 2003;50:635–643. doi: 10.1046/j.1365-2958.2003.03721.x. [DOI] [PubMed] [Google Scholar]
- 16.Grill S, Gualerzi C-O, Londei P, Bläsi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: Evolutionary implications for translation. EMBO J. 2000;19:4101–4110. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell S-D, Jackson S-P. Transcription and translation in Archaea: A mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 18.Kimball S-R. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–29. doi: 10.1016/s1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 19.Pedulla N, et al. The archaeal eIF2 homologue: Functional properties of an ancient translation initiation factor. Nucleic Acids Res. 2005;33:1804–1812. doi: 10.1093/nar/gki321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yatime L, Schmitt E, Blanquet S, Mechulam Y. Functional molecular mapping of archaeal translation initiation factor 2. J Biol Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- 21.Squires C-L, Zaporojets D. Proteins shared by the transcription and translation machines. Annu Rev Microbiol. 2000;54:775–798. doi: 10.1146/annurev.micro.54.1.775. [DOI] [PubMed] [Google Scholar]
- 22.Wool I-G. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 23.Oberto J, et al. The Escherichia coli ribosomal protein S16 is an endonuclease. Mol Microbiol. 1996;19:1319–1330. doi: 10.1111/j.1365-2958.1996.tb02476.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Yuan S, Jiang S. The ribosomal protein L32–2 (RPL32–2) of S. pombe exhibits a novel extraribosomal function by acting as a potential transcriptional regulator. FEBS Lett. 2006;580:1827–1832. doi: 10.1016/j.febslet.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 25.Albers S-V, et al. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapp L-D, Lorsch J-R. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 27.Boelens R, Gualerzi C-O. Structure and function of bacterial initiation factors. Curr Protein Pept Sci. 2002;3:107–119. doi: 10.2174/1389203023380765. [DOI] [PubMed] [Google Scholar]
- 28.Hasenöhrl D, Benelli D, Barbazza A, Londei P, Bläsi U. Sulfolobus solfataricus translation initiation factor 1 stimulates translation initiation complex formation. RNA. 2006;12:674–682. doi: 10.1261/rna.2289306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structural switch of the gamma subunit in an archaeal aIF2 alpha gamma heterodimer. Structure (London) 2006;14:119–128. doi: 10.1016/j.str.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Nikonov O, et al. New insights into the interactions of the translation initiation factor 2 from Archaea with guanine nucleotides and initiator tRNA. J Mol Biol. 2007;373:328–336. doi: 10.1016/j.jmb.2007.07.048. [DOI] [PubMed] [Google Scholar]
- 31.Gourse R-L, Gaal T, Bartlett M-S, Appleman J-A, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 32.Nercessian D, Conde R-D. Control of ribosome turnover during growth of the haloalkaliphilic archaeon Natronococcus occultus. Res Microbiol. 2006;157:625–628. doi: 10.1016/j.resmic.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Cellini A, et al. Stringent control in the archaeal genus Sulfolobus. Res Microbiol. 2004;155:98–104. doi: 10.1016/j.resmic.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Jonuscheit M, Martusewitsch E, Stedman K-M, Schleper C. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol. 2003;48:1241–1252. doi: 10.1046/j.1365-2958.2003.03509.x. [DOI] [PubMed] [Google Scholar]
- 35.Londei P, Altamura S, Cammarano P, Petrucci L. Differential features of ribosomes and of poly(U)-programmed cell-free systems derived from sulphur-dependent archaebacterial species. Eur J Biochem. 1986;157:455–462. doi: 10.1111/j.1432-1033.1986.tb09689.x. [DOI] [PubMed] [Google Scholar]
- 36.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.