Abstract
Sexual reproduction requires the specification of cells with distinct fates in plants and animals. The EMS1 (also known as EXS) leucine-rich repeat receptor-like kinase (LRR-RLK) and TPD1 small protein play key roles in regulating somatic and reproductive cell fate determination in Arabidopsis anthers. Here, we show that ectopic expression of TPD1 causes abnormal differentiation of somatic and reproductive cells in anthers. In addition, ectopic TPD1 activity requires functional EMS1. Yeast two-hybrid, pull-down, and coimmunoprecipitation analyses further demonstrate that TPD1 interacts with EMS1 in vitro and in vivo. Moreover, TPD1 induces EMS1 phosphorylation in planta. Thus, our results suggest that TPD1 serves as a ligand for the EMS1 receptor kinase to signal cell fate determination during plant sexual reproduction.
Keywords: Arabidopsis, sexual reproduction, anther, ligand, signal transduction
Cell fate determination is a critical process in development of all multicellular organisms. A fundamental feature of sexual reproduction in both plants and animals is the specification of distinct types of cells that give rise to eggs and sperm. In animals, primordial germ cells are determined and segregated from their somatic neighbors very early during embryonic development. In contrast, plant sexual reproduction takes place postembryonically (1). In flowering plants, after the switch from vegetative growth to reproductive development, reproductive cells are specified from somatic cells in flowers. So far, little is known about the molecular mechanisms underlying cell fate determination during sexual reproduction in plants (1–3).
Anthers are the male parts of flowers that bear pollen for producing sperm (4, 5). A mature anther is usually a four-lobed structure. Each lobe contains five types of highly specialized cells, which are the epidermis, endothecium, middle layer, tapetum, and microsporocytes (pollen mother cells). Microsporocytes are reproductive cells that generate pollen via meiosis, whereas somatic cells, particularly the tapetum, are required for normal development and the release of pollen. An abnormal tapetum has been shown to cause male sterility in plants. Anther development involves cell division, cell differentiation, and cell death, resulting in the specification of both reproductive and somatic cells in the same organ. Recently the anther has emerged as a prime model system for the study of cell fate determination and receptor-linked signaling, in addition to its central importance to plant breeding and reproduction (2, 3, 5–14).
In Arabidopsis the EXCESS MICROSPOROCYTES1 (EMS1, also known as EXTRA SPOROGENOUS CELLS, EXS) and TAPETUM DETERMINANT1 (TPD1) genes play key roles in anther cell fate determination (2, 7, 15). The ems1 mutant anthers lack the tapetum but produce more microsporocytes at the expense of tapetal cells, suggesting that there is a tradeoff between somatic and reproductive cells (2). The tpd1 mutant has a phenotype indistinguishable from that of ems1 (7). The EMS1 gene encodes a leucine-rich repeat receptor-like kinase (LRR-RLK), whereas TPD1 encodes a small, putatively secreted protein. Therefore, we hypothesize that TPD1 may act as a key signaling molecule, possibly a ligand, in the EMS1-mediated signal transduction pathway.
In Arabidopsis, >600 genes encode RLKs, representing 2.5% of the total genes (16, 17). This number is almost double in the rice genome (18). The LRR-RLKs, with 223 members in Arabidopsis (www4.ncsu.edu/∼sclouse/Clouse2010.htm), form the largest family of RLKs. Molecular genetic studies show that LRR-RLKs are involved in a wide range of plant growth and development processes (17, 19), including stem cell maintenance (20, 21), cell fate determination and patterning (2, 8–10, 12, 15, 22, 23), steroid hormone signaling (24–28), organ size and shape regulation (29–31), organ abscission (32), defense responses (33–35), plant transpiration (36), and nodulation (37, 38). However, most signaling molecules, including ligands, remain unidentified even in the known LRR-RLK-linked signal transduction pathways. Here, we report that the ectopic expression of TPD1 causes abnormal differentiation of somatic and reproductive cells in anthers. Genetic studies demonstrate that TPD1 signaling requires functional EMS1. Furthermore, yeast two-hybrid, pull-down, and coimmunoprecipitation experiments show that TPD1 interacts with EMS1 in vitro and in vivo. In addition, we found that TPD1 binding activates phosphorylation of the EMS1 kinase. Thus, interaction between EMS1 and TPD1 is required for the specification of somatic and reproductive cells in Arabidopsis anthers, which indicates that TPD1 serves as a ligand for the EMS1 receptor kinase.
Results
Ectopic Expression of TPD1 Causes Abnormal Differentiation of Somatic and Reproductive Cells in Anthers.
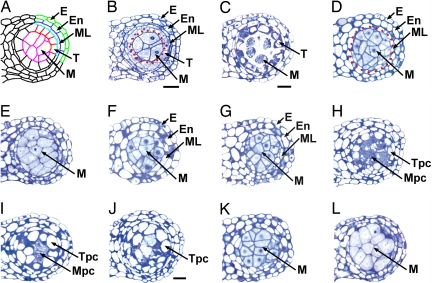
In wild-type anthers at stage 5 of development, each lobe comprises five types of differentiated cells: the epidermis, endothecium, middle layer, tapetum, and microsporocytes (Fig. 1A). Microsporocytes are located at the center of each lobe, surrounded by a tapetum layer (Fig. 1B). At stage 6, tapetal cells are partially vacuolated, and microsporocytes are separated from each other (Fig. 1C). The ems1 anther at stage 5 completely lacks tapetum but produces excess microsporocytes (Fig. 1D). At stage 6, microsporocytes are abnormally enlarged and still attached (Fig. 1E). The tpd1 anther phenotype [see supporting information (SI) Text] is similar to that of ems1 (Fig. 1F). Consistent with previous findings (7), our results show that the ems1 tpd1 double-mutant anther exhibits the identical phenotype to the ems1 and tpd1 single mutants (Fig. 1G), which suggests that TPD1 and EMS1 function in the same genetic pathway.
Fig. 1.
Ectopic expression of TPD1 causes abnormal anther cell differentiation, and TPD1 signaling requires EMS1. Semithin sections show one lobe from anthers. (Scale bars: 20 μm.) B, D, F–H, and K; C, E, I, and L have the same magnification. (A) Diagram of a mature wild-type anther shows epidermis (E), endothecium (En), middle layer (ML), tapetum (T), and microsporocytes (M). (B) A wild-type anther at stage 5. Enclosed by red dotted line are microsporocytes. (C) A wild-type anther at stage 6 shows strongly stained tapetal layer and isolated microsporocytes. (D) An ems1 anther at stage 5 lacks the tapetum layer but has excess microsporocytes (enclosed by red dotted line). (E) An ems1 anther at stage 6 lacks the tapetum layer. Microsporocytes are abnormally enlarged and not isolated. (F) A tpd1 anther at stage 5 has the same phenotype as that of ems1 (D). (G) An ems1 tpd1 double-mutant anther at stage 5 shows the same phenotypes as those of ems1 (D) and tpd1 (F). (H–J) Anthers of CaMV35S::TPD1 transgenic plants. (H) A stage-5 anther shows vacuolated cells in place of a normal tapetum (tapetum-positioned cells, Tpc) and degenerating cells instead of normal microsporocytes (microsporocyte-positioned cells, Mpc). (I) A stage-6 anther exhibits completely vacuolated Tpc and degenerating Mpc. (J) An anther after stage 6 shows the mix of completely vacuolated Tpc and Mpc. (K and L) Anthers from CaMV35S::TPD1 ems1 plants at stage 5 (K) and 6 (L) exhibit identical phenotypes to ems1 (D and E).
We then tested whether ectopic expression of TPD1 affects anther cell differentiation by expressing TPD1 with the cauliflower mosaic virus CaMV35S promoter (CaMV35S). Both the TPD1 gene and TPD1 protein levels were elevated in CaMV35S::TPD1 transgenic plants (data not shown). Seventy-seven percent of the 581 CaMV35S::TPD1 transgenic lines studied produced wider and shorter seed pods than wild type (SI Fig. 5), which is in agreement with the findings of Yang et al. (39). Fifty-two percent of lines exhibited the greatly reduced fertility. Moreover, anthers in 25% of the lines had no or few viable pollen grains (SI Fig. 6). Our analysis of semithin sections revealed abnormal cell differentiation in these anthers. Well differentiated tapetum layers were not detected. Instead, cells in the position of a tapetum would normally be located are highly vacuolated (Fig. 1 H–J). Microsporocyte-like cells also appear to have degenerated (Fig. 1 I and J). High-resolution confocal microscopy confirmed that the tapetal cell and microsporocyte differentiation are impaired in these anthers. In wild-type anthers, tapetal cells typically contain two separated nuclei that result from endomitosis (9, 40) (SI Fig. 7A). However, in CaMV35S::TPD1 anthers, we did not observe two nuclei in the tapetum-positioned cells (SI Fig. 7B), suggesting that the differentiation of tapetum is not complete. In wild-type anthers, the tapetum is established at stage 5 (Fig. 1B) and begins to degenerate at stage 10. Yang et al. (39) found that formation of tapetum is normal in the TPD1 ectopic expression plants but that degeneration of tapetal cells is delayed. In addition, microsporocytes do not degenerate. Our results, which show more severe defects than the previous findings, indicate that ectopic TPD1 signaling affects tapetal cell fate determination and inhibits microsporocyte development.
TPD1 Signaling Depends on Functional EMS1.
To test whether TPD1 signaling requires functional EMS1, we introduced the CaMV35S::TPD1 transgene into the ems1 mutant. The resulting CaMV35S::TPD1 ems1 transgenic plants exhibit the typical ems1 phenotype in seed pod size and fertility (SI Fig. 5). Furthermore, anthers in CaMV35S::TPD1 ems1 plants at stages 5 and 6 (Fig. 1 K and L) have the same phenotypes as ems1 (Fig. 1 D and E). Thus, TPD1 signaling requires the presence of EMS1, and the phenotypes observed in CaMV35S::TPD1 transgenic plants are the result of ectopic TPD1 signaling.
TPD1 Biochemically Interacts with EMS1 in Yeast Cells.
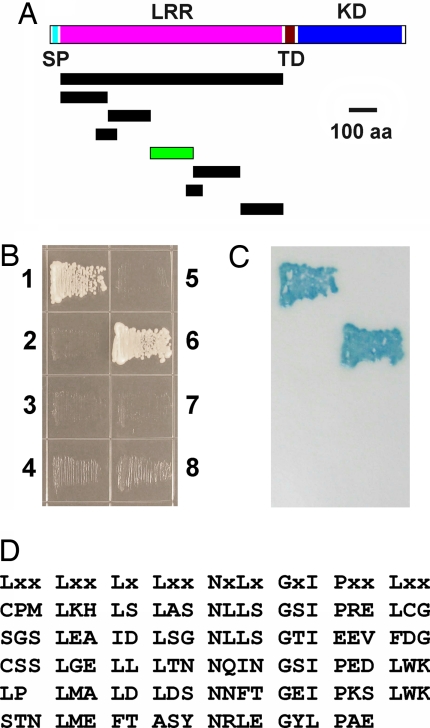
To test whether TPD1 interacts with EMS1, we conducted yeast two-hybrid experiments that have been used to identify the protein ligand LAT52 for the LePRK2 LRR-RLK in tomato (41). Initially, we did not detect any interaction between EMS1 and TPD1 using the entire EMS1 LRR domain (Fig. 2 A and B). LePRK2 contains 5 LRRs with a low potential for glycosylation, whereas EMS1 has 29 LRRs with 11 potential N-linked glycosylation sites that may have interfered with protein interaction (42). Therefore, we cloned a series of shortened cDNAs that encode truncated EMS1 LRRs (Fig. 2A). We found that one small fragment interacts with TPD1 (Fig. 2 A–C, SI Fig. 8). We named this small fragment the TPD1-interacting region (TIR). The TIR contains four typical LRRs and one LRR with low similarity to the typical LRR (Fig. 2D). Therefore, our results indicate that TPD1 biochemically interacts with EMS1.
Fig. 2.
TPD1 interacts with EMS1 in yeast two-hybrid assays. (A) Diagram shows constructs for mapping the TPD1 interacting region (TIR) of EMS1. Bar with green color represents the TIR. (B) Yeast grown on synthetic complete (SC) medium without Leu, Trp, and His. (1) Positive control. (2) Negative control. (3) The entire EMS1 LRR domain and TPD1. (4) The entire EMS1 LRR domain. (5) TPD1 alone. (6) TIR and TPD1. (7) TIR alone. (8) Non-TIR and TPD1. (C) Verification of the interaction by filter lift assay showing blue color of yeast. (D) The amino acid sequence of TIR. The consensus LRR sequence is shown at the top.
TPD1 Directly Interacts with EMS1.
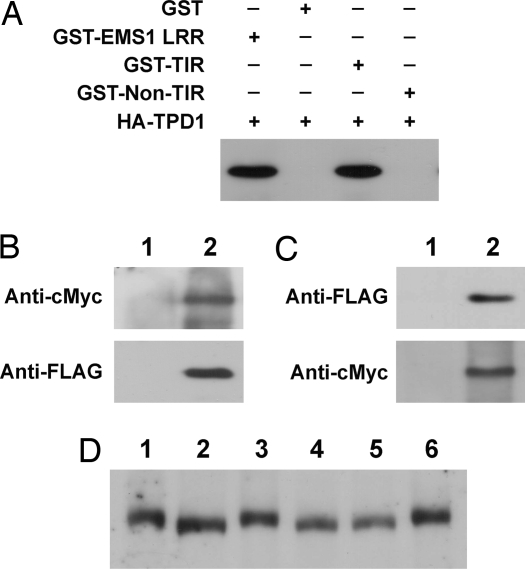
Direct interaction between TPD1 and EMS1 was further confirmed by GST pull-down experiments. We produced fusion proteins of GST to the entire EMS1 LRR domain, TIR and non-TIR (SI Figs. 9 and 10). The TPD1 protein was fused to three HA tags. Instead of GST and non-TIR, GST-EMS1 LRR and GST-TIR were able to pull down HA-TPD1 from crude protein extracts (Fig. 3A). Thus, our results strongly indicate that TPD1 directly interacts with EMS1 in a specific LRR extracellular region, which is TIR.
Fig. 3.
In vitro and in vivo interaction between TPD1 and EMS1 and TPD1 induces the phosphorylation of EMS1. (A) TPD1 interacts with EMS1 in GST pull-down assay. Three micrograms of GST and GST fusion proteins were used to pull down the same amount of crude protein (200 μg) extracts containing HA-TPD1, respectively. (B and C) TPD1 interacts with EMS1 in planta in coimmunoprecipitation assay. (B) EMS1-cMyc (Upper) and TPD1-FLAG (Lower) were detected in EMS1::EMS1-cMyc TPD1::TPD1-FLAG double-transgenic plants, respectively, by Western blot. (1) Wild type. (2) EMS1::EMS1-cMyc TPD1::TPD1-FLAG double-transgenic plant. (C) TPD1-FLAG was detected when proteins from EMS1::EMS1-cMyc TPD1::TPD1-FLAG double-transgenic plants were immunoprecipitated with an anti-cMyc antibody (Upper). EMS1-cMyc was also detected when the same proteins were immunoprecipitated with an anti-FLAG antibody (Lower). (1) TPD1::TPD1-FLAG (Upper) and EMS1::EMS1-cMyc (Lower) single-transgenic plants. (2) EMS1::EMS1-cMyc TPD1::TPD1-FLAG double-transgenic plant. (D) TPD1 binding induces EMS1 phosphorylation. Lanes 1, 3, 4, and 6: EMS1::EMS1-cMyc transgenic plants; lanes 2 and 5: EMS1::EMS1-cMyc tpd1 transgenic plants, in which TPD1 is not present. Proteins in lanes 4 and 5 were treated with calf intestinal alkaline phosphatase (CIP). EMS1-cMyc protein from EMS1::EMS1-cMyc tpd1 plants (lane 2) migrated faster than those from EMS1::EMS1-cMyc plants (lanes 1 and 3). A shift in mobility of EMS1-cMyc did not occur (lane 4) after the CIP treatment (lanes 5 and 6 are controls).
TPD1 Interacts with EMS1 in Planta.
To determine whether TPD1 interacts with EMS1 in vivo, we performed coimmunoprecipitation experiments. EMS1::EMS1-cMyc and TPD1::TPD1-FLAG transgenic plants were created to express EMS1-cMyc and TPD1-FLAG proteins by their native promoters. Both transgenes complemented phenotypes of their corresponding mutants. EMS1-cMyc and TPD1-FLAG proteins were also detected in double-transgenic plants (Fig. 3B). Membrane protein extracts from young inflorescences of double-transgenic plants were immunoprecipitated with an anti-cMyc antibody. We detected TPD1-FLAG by Western blot with an anti-FLAG antibody (Fig. 3C Upper). In the reciprocal coimmunoprecipitation experiment, we also detected EMS1-cMyc from the anti-FLAG immunoprecipitates (Fig. 3C Lower). In conclusion, our results strongly indicate that TPD1 interacts with EMS1 in planta.
TPD1 Induces the Phosphorylation of EMS1.
We investigated whether the binding of TPD1 to EMS1 triggers a downstream signaling event. EMS1 is predicted to be a receptor-like serine/threonine kinase. Based on the well established paradigm for mammalian receptor kinase action, ligand binding can elicit receptor kinase phosphorylation. Such phosphorylation often causes a change in mobility in SDS/PAGE (43). We generated EMS1::EMS1-cMyc tpd1 plants and found that EMS1-cMyc proteins from EMS1::EMS1-cMyc tpd1 plants (Fig. 3D, lane 2) migrated faster in SDS/PAGE than those from EMS1::EMS1-cMyc plants (Fig. 3D, lanes 1 and 3). To test whether the mobility shift was caused by phosphorylation, we treated proteins with calf intestinal alkaline phosphatase (CIP). This treatment shifted the slower migrating band (Fig. 3D, lane 4) to the same position as the faster band (Fig. 3D, lane 5, lane 6 is the control). Therefore, our results suggest that TPD1 binding leads to EMS1 phosphorylation.
In summary, our studies indicate that interaction between TPD1 and EMS1 is required for cell fate determination during plant sexual reproduction. Furthermore, TPD1 interacts with EMS1 in vitro and in vivo, and TPD1 binding activates EMS1 phosphorylation. Thus, we propose that the TPD1 small protein serves as a ligand for the EMS1 receptor kinase.
Discussion
Our results indicate that TPD1 is a ligand for the EMS1 LRR-RLK. So far, studies have identified only a few ligands for some LRR-RLKs with known functions. Steroid brassinolide is the ligand for the Arabidopsis BRI1, BRL1, and BRL3 LRR-RLKs (24, 43, 44). Small peptides are found to serve as ligands for LRR-RLKs, such as PSK for carrot and Arabidopsis PSKRs (45, 46), flg22 for Arabidopsis FLS2 (33), and EF-Tu for Arabidopsis EFR (35). CLV3, a small secreted protein (47), acts as the ligand of CLV1 (48, 49). Recent studies show that the major active domain of CLV3 is a 12- to 15-aa peptide (50–53). Genetic evidence supports the idea that the EPF1 small secreted peptide acts as a ligand for ERECTA family receptor kinases (54). Small protein LAT52 is a protein ligand for the tomato LRR-RLK, LePRK2 (41). TPD1 is a small protein with 176 aa that contains a predicted N-terminal-secreted signal peptide, followed by a predicted cleavage site between the 21st and 22nd amino acid residues (www.cbs.dtu.dk/services/SignalP). Our results provide several lines of evidence to strongly support the idea that TPD1 serves as a ligand of EMS1. Determination of whether secretion of TPD1 is required for its function remains for future study as well as identification of additional signaling molecules in the EMS1 signal transduction pathway.
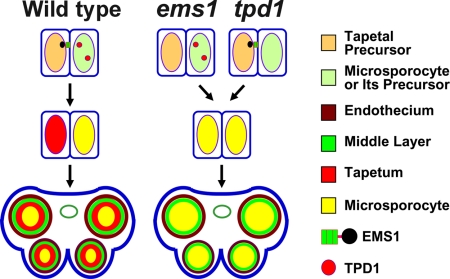
Very little is known about the signaling mechanisms governing cell fate determination during reproduction in plants and in many other organisms. Among LRR-RLKs with known functions, EMS1 specifically directs the determination of somatic and reproductive cell fates (2). The EMS1 gene is predominantly expressed in the tapetum, whereas TPD1 expression is restricted to microsporocytes (2, 7). Therefore, we hypothesize that TPD1 could be secreted from microsporocytes or their precursors and then bind to EMS1 localized on the surface of tapetum precursors (Fig. 4). Signals relayed via EMS1, in turn, direct tapetum formation. In the absence of EMS1-TPD1 signaling, genes promoting microsporocyte differentiation might be persistently expressed in tapetum precursors, resulting in tapetal precursors adopting a microsporocyte fate (Fig. 4). It is also possible that tapetal cells inhibit further proliferation of microsporocytes. Our studies will provide insights into molecular mechanisms of cell fate determination during sexual reproduction in plants and other organisms.
Fig. 4.
A model for EMS1-TPD1 signaling in anther cell fate determination. In the wild-type anther, TDP1 small protein is secreted from microsporocytes or their precursors and then bind to EMS1 receptor kinases that are localized to tapetal precursors. EMS1-TPD1 signaling ensures specification of tapetal cell fate by activating the downstream signaling cascade. In the absence of TPD1 ligand or EMS1 receptor in the tpd1 or ems1 mutant, signals directing tapetum differentiation are blocked. Consequently, tapetal precursors adopt a microsporocyte fate, resulting in the formation of excess microsporocytes.
Abnormal differentiation of the tapetum and microsporocytes in CaMV35S::TPD1 anthers could possibly be due to a negative feedback regulation between the EMS1-TPD1 signal transduction pathway and its target genes, such as SPL/NZZ (55, 56). The spl/nzz mutant fails to produce microsporocytes and anther wall layers, and SPL/NZZ is expressed in both parietal and sporogenous cells, indicating that SPL/NZZ promotes anther cell differentiation, including the tapetum during early stages (55, 56). The fact that both ems1 and tpd1 anthers form excess microsporocytes suggests that EMS1-TPD1 signaling may repress microsporocyte formation (2, 7). Our unpublished data (G.J. and D.Z.) show that SPL/NZZ is epistatic to EMS1/EXS. The expression of EMS1/EXS was significantly increased after the SPL/NZZ function was induced by dexamethasone treatment. Therefore, SPL/NZZ possibly controls the expression of genes required for differentiation of both microsporocyte and tapetum, for example EMS1/EXS.
On the other hand, EMS1-TPD1 signaling may direct tapetum formation by repressing genes required for microsporocyte differentiation in tapetum precursors. For example, without EMS1-TPD1 signaling, SPL/NZZ expression might be persistent in tapetum precursors (i.e., parietal cells), which reverses their cell fate and results in the formation of excess microsporocytes. However, ectopic EMS1-TPD1 signaling might repress the expression of SPL/NZZ, which could affect parietal and sporogenous cell differentiation and subsequently cause aberrant differentiation of tapetum and microsporocytes. It is also possible that functions of other genes, such as DYT1 and AMS, are affected by ectopic expression of TPD1 (11, 57). In the shoot apical meristem (SAM), the CLV3 ligand interacts with its CLV1/CLV2 receptor complex to elicit a negative signal that represses the WUS expression (48, 58). Ectopic expression of CLV3 causes down-regulation of WUS, leading to termination of the SAM (48, 59). The abnormal tapetal layer in some CaMV35S::TPD1 anthers might possess characteristics of outer layers. In this case, other signaling pathways, like the one mediated by RPK2, could be affected (12). TPD1 might also restrict its own expression domain. It will be necessary to perform more experiments to test these hypotheses.
The anther primordium (stage 1) consists of L1, L2 and L3 three layers of cells from outer to inner (2). L1 cells form the epidermis, whereas vascular tissues are derived from L3 cells. The L2 cells, which give rise to the majority of anther cells, first differentiate into archesporial cells at stage 2, then develop into primary sporogenous cells and primary parietal cells at stage 3. The primary sporogenous cells eventually become microsporocytes by stage 5. The primary parietal cells divide into secondary parietal cells at stage 4, and subsequently develop into endothecium, middle layer and tapetum by stage 5.
So far, six LRR-RLKs have been identified that play important roles in signaling anther cell differentiation. Without detectable defects before stage 4, the serk1 serk2 double-mutant anther has a phenotype same as that of ems1/exs (2, 8, 9). Similar to BRI1 and BAK1 (also known as SERK3) (26, 27), EMS1/EXS and SERK1/2 could be involved in the same signaling pathway possibly by heterodimerization to control anther cell fate determination after the formation of sporogenous and parietal cells. Mutations in the RPK2 gene result in failure of middle layer formation and abnormal degeneration of the tapetum after meiosis (12). Therefore, RPK2 signaling might function early to control middle layer determination and later to maintain the tapetum. RPK2 and EMS1/EXS1 might also elicit antagonistic signals to determine the middle layer or tapetum from parietal cells. The bam1 bam2 mutant, which has a more severe phenotype than ems1/exs, serk1 serk2, and rpk2 mutants, lacks the endothecium, middle layer, and tapetum, but produces extra microsporocytes (10). Thus, BAM1/2 might signal differentiation of archesporial cells, which functions earlier than EMS1/EXS, SERK1/2 and RPK2 do. It will be interesting to determine how these LRR-RLK-linked signaling pathways coordinate to regulate anther cell differentiation.
EMS1-TPD1 signaling appears to be a conserved mechanism for anther cell differentiation, because an apparent ortholog to EMS1 exists in rice and possibly in maize (6, 60). We also found several genes that encode proteins similar to TPD1 in the rice genome (D.Z., unpublished data). Eudicots and monocots diverged at least 120 million years ago and together represent >90% of angiosperms, so the presence of the EMS1-TPD1 signal transduction pathway in both suggests that this pathway might have existed in early angiosperms. Land plants produce sporangia that vary in the complexity of structures, such as the tapetum. Ferns, which are nonflowering plants, form a simple sporangium with an outer wall and a tapetum. Thus, studies of EMS1-TPD1 signaling in other vascular plants will shed light on evolution of male reproductive structures in plants.
Materials and Methods
Plant Materials and Growth Condition.
Arabidopsis thaliana plants were of the Landsberg erecta (Ler) ecotype and were grown in Metro-Mix 360 soil (Sun-Gro Horticulture) in growth chambers under a 16-h light/8-h dark photoperiod at 22°C and 50% humidity.
Microscopy.
Semithin sections were made according to the previous procedure (2). Images were photographed with an Olympus BX51 microscope by using an Olympus DP 70 digital camera.
Yeast Two-Hybrid Assay.
In this study, all DNA and cDNA fragments for cloning were PCR-amplified by the Phusion High-fidelity DNA polymerase (SI Table 1) (New England Biolabs). The resulting constructs were verified by sequencing.
The TPD1 cDNA was cloned into pAD-GAL4 vector (HybriZAP 2.1 Two-Hybrid System; Stratagene). cDNAs encoding the entire EMS1 LRR domain and shortened EMS1 LRRs were cloned into the pBD-GAL4 vector (Fig. 2A). Yeasts containing a combination of bait and prey constructs were grown on synthetic complete (SC) medium without Leu, Trp, His, and SC medium with a series of concentrations (5, 10, and 30 mM) of 3-Amino-1,2,4-Triazole (3AT). The filter lift assay was performed according to the manufacturer's manual. Positive and negative controls were provided by the manufacturer.
Pull-Down Assay.
EMS1 cDNA fragments encoding the EMS1 LRR domain, TPD1-interacting region (TIR), and a non-TIR were PCR-amplified, respectively (SI Fig. 9 and SI Table 1), and then cloned into pGEX-4T-2 vector (Amersham) to produce GST fusion proteins (SI Fig. 10). The TPD1 cDNA was cloned into pWS93 vector to produce HA-TPD1 proteins in yeast. Three μg each of GST and GST fusion proteins bound to Glutathione-Sepharose-4B beads (Amersham) were incubated with 200 μg of crude protein extract overnight at 4°C. After washing three times, the pull-down mixtures were analyzed by Western blot using an anti-HA antibody (Roche).
Generation of Transgenic Plants.
The Gateway binary vectors were used to generate transgenic plants. The 1740-bp EMS1 promoter and full-length EMS1 cDNA were PCR-amplified (SI Table 1), and then cloned into the pENTR TOPO vector (Invitrogen). The EMS1::EMS1 fragment was cloned into the Gateway binary vector pGWB16 with cMyc tag by an LR reaction (Invitrogen). The 2,770-bp TPD1 promoter was PCR-amplified (SI Table 1) and then cloned into a vector harboring the TPD1 cDNA. Similarly, the TPD1::TPD1 fragment was cloned the Gateway binary pGWB10 vector with the FLAG tag. To ectopically express TPD1, the TPD1 cDNA was cloned into the Gateway vector pGWB2 with a CaMV35S promoter.
The resulting constructs were introduced into Agrobacterium strain GV3101. After plant transformation, transformants were screened with 50 mg/liter kanamycin and hygromycin. Forty individual EMS1::EMS1-cMyc and TPD1::TPD1-FLAG transgenic lines were crossed to their corresponding mutants to examine for complementation of the phenotype. EMS1::EMS1-cMyc and TPD1::TPD1-FLAG plants were then crossed to produce double-transgenic plants. Forty CaMV35S::TPD1 transgenic lines that occasionally produced a few pollen grains were also crossed to the ems1 mutant.
Coimmunoprecipitation.
Young inflorescences harvested from wild-type, single- and double-transgenic plants were ground into fine powder by using liquid N2 and then in cold grinding buffer [20 mM Tris·HCl (pH 8.8), 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 100 μM Na3VO4, 20% glycerol, and protease inhibitor cocktails]. Samples were spun at 6,000 × g for 15 min at 4°C, and supernatants were further spun at 100,000 × g for 40 min at 4°C to pellet membrane fractions (26, 61). The pellets were sonicated and resuspended in membrane protein extraction buffer [10 mM Tris·HCl (pH 7.3), 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 100 μM Na3VO4, 10% glycerol, 2% Triton X-100, and protease inhibitors]. After centrifugation at 100,000 × g for 25 min at 4°C, supernatants were incubated with prewashed anti-FLAG M2 affinity gel (Sigma–Aldrich) overnight at 4°C. After three washes, the eluted immunoprecipitates were used in Western blots to detect EMS1-cMyc (Anti-cMyc; Roche). The reciprocal coimmunoprecipitation was performed by using the Profound c-Myc Tag IP/Co-IP kit (Pierce).
Kinase Phosphorylation Assay.
Proteins extracted from EMS1::EMS1-cMyc and EMS1::EMS1-cMyc tpd1 plants were separated on a 5% SDS/PAGE gel (43), followed by Western blotting to detect EMS1-cMyc proteins. To confirm whether the mobility shift was caused by phosphorylation, proteins were treated with 10 units of calf intestinal alkaline phosphatase (New England Biolabs) at 37°C for 60 min (61).
Supplementary Material
ACKNOWLEDGMENTS.
We thank S. Forst, S. Kuchin, Y. Sun, X. Wang, and C. Zeng for technical assistance; D. Heathcote, J. Li, and Z. Wang for critical comments on this manuscript; M. Chen and T. Schuck for plant care; and T. Nakagawa for offering Gateway binary vectors. D.Z. particularly thanks H. Ma for his advice and subcontracting the Department of Energy (DOE) grant. This work was supported by DOE Grant DE-FG02-02ER15332 (to H.M. and D.Z.), and by a grant from the Research Growth Initiative (RGI) at the University of Wisconsin-Milwaukee (to D.Z.) D.Z. gratefully acknowledges the support of the Shaw Scientist Award from the Greater Milwaukee Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708795105/DC1.
References
- 1.Walbot V, Evans MM. Unique features of the plant life cycle and their consequences. Nat Rev Genet. 2003;4:369–379. doi: 10.1038/nrg1064. [DOI] [PubMed] [Google Scholar]
- 2.Zhao DZ, Wang GF, Speal B, Ma H. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002;16:2021–2031. doi: 10.1101/gad.997902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg RB, Beals TP, Sanders PM. Anther development: Basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. Plant Cell. 2004;16(Suppl):S46–S60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nonomura K, et al. The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell. 2003;15:1728–1739. doi: 10.1105/tpc.012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SL, et al. Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell. 2003;15:2792–2804. doi: 10.1105/tpc.016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell. 2006;18:1667–1680. doi: 10.1105/tpc.105.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, et al. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno S, et al. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2007;50:751–766. doi: 10.1111/j.1365-313X.2007.03083.x. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Duncan D, Morrow DJ, Fernandes J, Walbot V. Transcriptome profiling of maize anthers using genetic ablation to analyze pre-meiotic and tapetal cell types. Plant J. 2007;50:637–648. doi: 10.1111/j.1365-313X.2007.03074.x. [DOI] [PubMed] [Google Scholar]
- 14.Wijeratne AJ, et al. Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J. 2007;52:14–29. doi: 10.1111/j.1365-313X.2007.03217.x. [DOI] [PubMed] [Google Scholar]
- 15.Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol. 2002;12:1718–1727. doi: 10.1016/s0960-9822(02)01151-x. [DOI] [PubMed] [Google Scholar]
- 16.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torii KU. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol. 2004;234:1–46. doi: 10.1016/S0074-7696(04)34001-5. [DOI] [PubMed] [Google Scholar]
- 18.Shiu SH, et al. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy DR, Chory J. Conservation and innovation in plant signaling pathways. Cell. 2000;103:201–209. doi: 10.1016/s0092-8674(00)00113-6. [DOI] [PubMed] [Google Scholar]
- 20.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 21.DeYoung BJ, et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 22.Clay NK, Nelson T. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell. 2002;14:2707–2722. doi: 10.1105/tpc.005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- 24.Cano-Delgado A, et al. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 26.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 27.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhou A, Wang H, Walker JC, Li J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004;40:399–409. doi: 10.1111/j.1365-313X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- 29.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- 31.Torii KU, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Gomez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 34.Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 35.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 36.Masle J, Gilmore SR, Farquhar GD. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature. 2005;436:866–870. doi: 10.1038/nature03835. [DOI] [PubMed] [Google Scholar]
- 37.Endre G, et al. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- 38.Searle IR, et al. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- 39.Yang SL, et al. Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS. Plant Physiol. 2005;139:186–191. doi: 10.1104/pp.105.063529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss H, Maluszynska J. Molecular cytogenetic analysis of polyploidization in the anther tapetum of diploid and autotetraploid Arabidopsis thaliana plants. Ann Bot. 2001;87:729–735. [Google Scholar]
- 41.Tang W, Ezcurra I, Muschietti J, McCormick S. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell. 2002;14:2277–2287. doi: 10.1105/tpc.003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Hoorn RA, et al. Structure–function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell. 2005;17:1000–1015. doi: 10.1105/tpc.104.028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 44.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 45.Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- 46.Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006;142:45–53. doi: 10.1104/pp.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 48.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 49.Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell. 2002;14:969–977. doi: 10.1105/tpc.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiers M, et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito Y, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 52.Kondo T, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 53.Ni J, Clark SE. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 2006;140:726–733. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiefthaler U, et al. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorensen AM, et al. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003;33:413–423. doi: 10.1046/j.1365-313x.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- 58.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 59.Lenhard M, Laux T. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development. 2003;130:3163–3173. doi: 10.1242/dev.00525. [DOI] [PubMed] [Google Scholar]
- 60.Sheridan WF, Golubeva EA, Abrhamova LI, Golubovskaya IN. The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics. 1999;153:933–941. doi: 10.1093/genetics/153.2.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.