Abstract
Cytokinesis partitions the cell by a cleavage furrow in animals but by a new cross wall in plants. How this new wall assembles at the molecular level and connects with the mother cell wall remains unclear. A lethal Arabidopsis embryogenesis mutant designated root-, shoot-, hypocotyl-defective (rsh) provides some clues: RSH encodes extensin AtEXT3, a structural glycoprotein located in the nascent cross wall or “cell plate” and also in mature cell walls. Here we report that electron micrographs of rsh mutant cells lacking RSH extensin correspond to a wall phenotype typified by incomplete cross wall assembly. Biochemical characterization of the purified RSH glycoprotein isolated from wild-type Arabidopsis cell cultures confirmed its identity as AtEXT3: a (hydroxy)proline-rich glyco protein comprising 11 identical amphiphilic peptide repeats with a 28-residue periodicity: SOOOOKKHYVYKSOOOOVKHYSOOOVYH (O = Hyp), each repeat containing a hydrophobic isodityrosine cross-link motif (YVY, underlined). Atomic force microscopy of RSH glycoprotein imaged its propensity for self-assembly into a dendritic scaffold. Extensin peroxidase catalyzed in vitro formation of insoluble RSH gels with concomitant tyrosine cross-linking, hence this likelihood in muro. We conclude that self-assembling amphiphiles of lysine-rich RSH extensin form positively charged scaffolds in the cell plate. These react with negatively charged pectin to create an extensin pectate coacervate that may template further orderly deposition of the new cross wall at cytokinesis.
Keywords: cytokinesis, (hydroxy)proline-rich glycoprotein, nanotechnology
Plant cells are hydraulic machines. They generate a hydrostatic pressure that drives cell expansion, rate-limited by a thin “primary” cell wall. This extracellular matrix has dynamic properties and structural integrity, hence an organelle with its own unique proteome. Mechanically it is an elastomer–plastomer composite comprising three interpenetrating networks, two of structural polysaccharides and one of structural glycoproteins (1). Each network is internally cross-linked: cellulose microfibrils by xyloglucans, pectins by calcium salt bridges and apiosyl borate esters, and structural (hydroxy)proline-rich glycoproteins (HRGPs), notably the extensins, by tyrosine derivatives (2, 3).
The integrity of an extracellular matrix depends crucially on correct self-assembly of its individual components. Mutants with defective glycosaminoglycan proteoglycans and hydroxyproline (Hyp)-rich collagens are often lethal in animals (4). Likewise, defects in the cell wall or extracellular matrix of green plants (5) and related algae (6) also involve Hyp-rich proteins (7), the extensins (8). These are highly periodic amphiphilic glycoproteins defined by their conserved hydrophilic and hydrophobic regions—short rigid blocks of contiguous O-glycosylated Hyp residues (9), typically hydrophilic Ser-(Hyp)4 motifs. In plants these are interspersed with hydrophobic motifs. Typically these are (i) YXY motifs shown in vitro to form isodityrosine (YXY) (Idt) and to be involved in intermolecular cross-linking via di-Idt formation (3, 10) and (ii) putative intermolecular VYK cross-linking motifs (11). Extensins are secreted as ≈80- to 100-nm rod-like monomers (12) with limited flexibility in an extended polyproline II (three residues per turn) helical conformation further stabilized by their glycomodules. Extensin peroxidase catalyzes polymerization of extensin monomers (11) via di-Idt and pulcherosine (a tetramer and trimer of tyrosine, respectively) to yield cross-linked networks in vitro and in muro (13, 14).
The Arabidopsis genome encodes 20 closely homologous extensin polypeptides [supporting information (SI) Table 4] with distinct patterns of expression at the organ/tissue level (15). However, lethal knockout of a single extensin gene yielded a root-, shoot-, hypocotyl-defective (rsh) seedling (5) where wild-type RSH corresponded to AtEXT3 (Fig. 1) and localized by ImmunoGold labeling to the cell plate and mother wall junctions (5). Here we further identified this lesion at the electron microscope level as defective cell wall assembly that could be ascribed to the absence of AtEXT3. As an extended amphiphilic cell wall protein with Idt motifs, AtEXT3 has evident potential for self-recognition, cross-linking, and scaffold formation, hence the imperative to isolate, purify, and characterize the glycoprotein itself. Thus, subsequent biochemical analysis of AtEXT3 and imaging by atomic force microscopy (AFM) presented here confirmed its propensity for self-recognition and further polymerization. We propose that a positively charged extensin scaffold reacts with acidic pectin to form extensin pectate, which further templates orderly assembly of a nascent cell wall. This work forces us to change the focus on extensin, from its role exclusively in the cessation of growth (16, 17) to an essential role in the initiation of growth.
Fig. 1.
Wild-type, nonhydroxylated AtEXT3 (pre-RSH) deduced from genome sequence. The signal peptide (sp) cleavage site is marked by an asterisk, and the major RSH repeat module (28 aa) is in parentheses. RSH has 60 Tyr residues and 16 YXY (Idt) motifs (underlined).
Results
The rsh Mutant Has Defective Cell Walls.
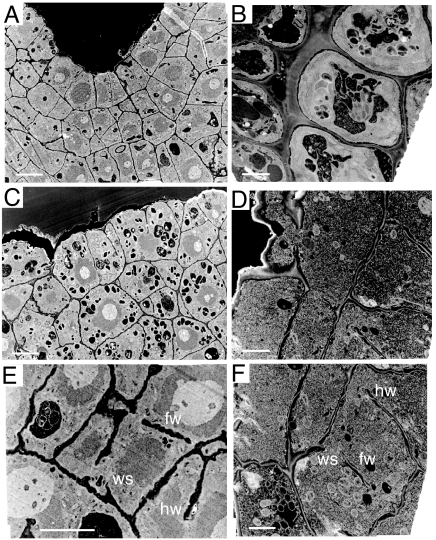
At the electron microscope level, the plane of cell division in the embryo and abortive seedling was aberrant resulting in abnormally shaped cells with incomplete walls (Fig. 2), i.e., “floating walls,” which were unconnected to side walls, “hanging walls,” which were connected at one end only, and “wall stubs,” which were short wall protrusions jutting from the mother cell wall. The newly forming cross wall frequently remained incomplete in the homozygous rsh mutant, apparently unable to connect or remain connected with the mother cell wall at cytokinesis and unable to produce normal walls in nondividing root cells.
Fig. 2.
Electron micrographs showing defective cell walls in the rsh/rsh mutant. (A, C, and E) Images of heart-stage embryo sections. (B, D, and F) Images of 3-day-old root transverse sections. The mutant (C–F) compared with the wild type (A and B) has incomplete cell walls. fw, floating walls; hw, hanging walls; ws, wall stubs. [Scale bars: 10 μm (A, C, and E) and 4 μm (B, D, and F).]
Purification of RSH and Its Identification as AtEXT3.
Although extensins are generally firmly bound as an insoluble wall network, small amounts of soluble monomeric precursors are, to various degrees, salt-elutable. Thus, high salt rapidly desorbed weakly bound HRGPs from the walls of intact Arabidopsis cell cultures. The major soluble component was consistent with the presence of lysine-rich positively charged extensin monomers. These basic macromolecules interact via ion pairs or salt bridges with pectic carboxylate groups (18), thus forming extensin pectate. Sizing by Superose-6 gel filtration (Fig. 3A) and further purification by cation-exchange chromatography yielded a single fraction identified as a monomeric extensin by its size, amino acid, and sugar composition. Subsequent alkaline hydrolysis yielded small Hyp-arabinooligosaccharides (Table 1) characteristic of extensins (19) and consistent with the predictions of the Hyp contiguity hypothesis (20, 21). Deglycosylation of the mature glycoprotein in anhydrous hydrogen fluoride followed by sequencing gave the first 19 residues of the N terminus: NYFYSSOOOOVKHYTOOVK (O = Hyp). This positively identified the glycoprotein as AtEXT3, a HRGP with strict periodicity of 11 identical 28-residue peptide repeats, each with a YVY Idt cross-linking motif. Tryptic degradation yielded two major peptide sequences [SOOOOVK and HYSOOOVY…] predicted from the 28-residue repeat motif, SPPPPKKHYVYKSPPPPVKHYSPPPVYH, confirming the identity of the isolated glycoprotein as AtEXT3 and identical to RSH (5).
Fig. 3.
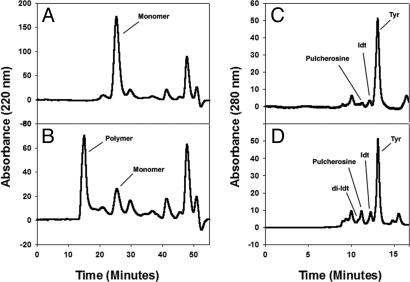
Cross-linking of purified RSH with extensin peroxidase produces di-Idt and pulcherosine. (A) Time 0 control. (B) Five-minute reaction. By 5 min most RSH monomers and oligomers were converted to the large-molecular-weight polymer that voided the column. One nanogram of enzyme was used in each reaction. (C) Acid hydrolysate of RSH monomers separated on a hydroxyethylaspartamide column in size-exclusion mode. (D) Acid hydrolysate of cross-linked RSH. Note that di-Idt appears not to resolve completely from the pulcherosine peak.
Table 1.
Hyp glycoside profile of RSH glycoprotein
| Hyp glycoside* | Mol% of total Hyp |
|---|---|
| Hyp-Ara4 | 33 |
| Hyp-Ara3 | 31 |
| Hyp-Ara2 | 10 |
| Hyp-Ara | 9 |
| Nonglycosylated Hyp | 17 |
*All Hyp glycosides are arabinooligosaccharides (see Materials and Methods).
RSH Forms Tyrosine Cross-Links in Vitro.
Extensin peroxidase catalyzed cross-linking of purified extensin monomers (Fig. 3A) in vitro judging from rapid multimer formation assayed by gel filtration (Fig. 3B). Increased reaction times beyond 5 min formed a gel that remained insoluble even after deglycosylation; on acid hydrolysis this multimer yielded Idt and pulcherosine as major products with only a trace of di-Idt (Fig. 3D and Table 2). A perfect parallel alignment (Fig. 4A) of RSH Idt motifs would yield intermolecular di-Idt cross-links exclusively like the cross-linking extensin analog (YK)20 described previously (14). However, 28-residue RSH repeats offset by 12 residues align all RSH tyrosine residues as potential pulcherosine cross-link motifs (Idt with a biphenyl link to lone Y residues of the HYS motif). Thus, pulcherosine in cross-linked RSH indicates a staggered alignment of RSH monomers (Fig. 4B). The absence of pulcherosine from cross-linked (YK)20 (14) simply reflects the absence of pulcherosine cross-link motifs from (YK)20. Unlike (YK)20 a staggered lateral alignment of RSH would allow growth of the polymer scaffold in two dimensions both by linear and lateral addition of monomers as indicated by the following AFM data.
Table 2.
RSH cross-linked in vitro by extensin peroxidase
| Amino acid | Nanomoles of each amino acid in ≈50 μg of cross-linked RSH* | Expressed as nanomoles of Tyr | Expressed as % of total Tyr | Number of Tyr residues involved as Idt, pulcherosine, or di-Idt in a single RSH molecule containing 60 Tyr residues |
|---|---|---|---|---|
| di-Idt | 0.67 | 2.68 | 9.8 | 5.9 |
| Pulcherosine | 3.06 | 9.18 | 33.7 | 20.2† |
| Idt | 4.65 | 9.30 | 34.2 | 20.5 |
| Tyr | 6.00 | 6.00 | 22.1 | 13.3 |
| Totals | 14.38 | 27.20 | 99.8 | 59.9 |
*Approximately 50 μg of cross-linked RSH was hydrolyzed. Data are from Fig. 3D. The non-cross-linked control (Fig. 3C) contained 10.2 nM Tyr, 2.9 nM Idt, and 1.3 nM pulcherosine but no di-Idt. Small amounts of pulcherosine slowly appeared over time in batches of RSH stored frozen.
†A 12-residue offset (Fig. 4B) with 100% cross-link formation involves all 60 Tyr residues as potential pulcherosine cross-links. The data here showed that approximately one-third were converted to covalent pulcherosine cross-links.
Fig. 4.
Alignments of the RSH 28-aa repetitive motif. (A) Head-to-head parallel alignment gives di-Idt motifs exclusively. (B) Staggered alignment gives pulcherosine motifs exclusively. This shows facile generation of pulcherosine cross-linking motifs. Offsetting the 28-residue canonical RSH motif by 12 residues realigns the Ser-Hyp4 glycomodule of one chain with another (underlined), thus switching from potential di-Idt to pulcherosine cross-links. Hydrophilic residues are shown in yellow, potential di-Idt and pulcherosine cross-links (hydrophobic) are in pink, and other hydrophobic residues are in gray. All motifs are presented in the N terminus (head) to C terminus (tail) direction.
A RSH Network Imaged by AFM.
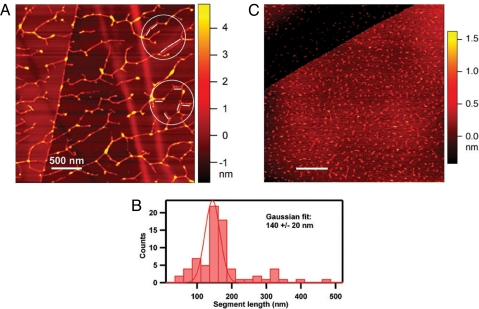
Raster scanning of the surface of samples with an extremely sharp nanoprobe (tip radius < 5 nm) via AFM (22) visualized purified RSH molecules as dendritic structures showing both end-on and lateral adhesion with sites along the main chain (Fig. 5A). AFM molecular height and length measurements (Fig. 5 A and B) of the extended RSH polypeptide showed that the RSH molecules form lateral adhesions of staggered overlapping monomers up to six molecules deep consistent with a role for RSH as an extensin network in muro. Control molecules for AFM included BSA, polyhydroxyproline (Sigma), and the extensin analog (YK)20 containing 20 tandem repeats of the arabinosylated sequence SO4SOSO4YYYK in a polyproline II conformation. None of the AFM controls gave dendritic patterns. Significantly, (YK)20, the control most closely related to AtEXT3 (Fig. 5C), contains 20 tandem repeats of the hexadecapeptide repeat SOOOOSOSOOOOYYYK with a YYY Idt motif but lacks pulcherosine cross-link motifs.
Fig. 5.
Self-assembled RSH network imaged by AFM. (A) A solution of RSH monomer (9 μg/ml water; see Fig. 3A) was dispersed on a highly ordered pyrolytic graphite (HOPG) surface, and the molecular contours were imaged by AFM. The molecular height of the segments, which ranged from 0.6 nm to 5 nm, was estimated by the colors corresponding to the nanometer ruler on the right. The white bars represent 127-nm lengths and correspond to the calculated length of RSH, assuming a strict polyproline II helical secondary structure (three residues per turn; pitch = 9.4 Å; 404 aa). The segments of the dendritic structure, here aligned with the 127-nm rods (circled), likely correspond to a relatively few overlapping RSH molecules aligned to produce rigid segments slightly longer than the RSH molecule, in keeping with its proposed staggered alignment. (B) Exhaustive analysis of all rigid segments in A yielded the histogram. (C) AFM imaging of (YK)20 (10 μg/ml water), an extensin analog produced through synthetic genes and expressed in BY-2 tobacco cells. (YK)20 contains 20 tandem repeats of the arabinosylated extensin sequence SO4SOSO4YYYK, also in a polyproline II helix. (Scale bars: 500 nm.)
RSH Mutant Rescue Requires an Unobstructed C Terminus.
This experiment was designed to test the functionality of RSH when fused to GFP. RSH constructs with their C terminus (but not their N terminus) fused to GFP did not rescue the rsh/rsh mutant, and furthermore it impeded the function of native RSH in wild-type plants (Table 3). This suggests a special role for the C terminus in scaffold formation (see Discussion).
Table 3.
Phenotypes of wild type (RSH/RSH), heterozygotes (RSH/rsh), homozygotes (rsh/rsh), and their transformants
| Transgene used to rescue mutant | Wild-type RSH/RSH | RSH/rsh | rsh/rsh |
|---|---|---|---|
| No transgene | Wild type | Wild type | Embryo lethal |
| RSH | Wild type | Wild type | Wild type |
| RSH::EGFP (C-terminal) | Range* | Range* | Embryo lethal |
| EGFP::RSH (N-terminal) | Wild type | Wild type | Wild type |
*In 30 independent transformants of each, phenotypes ranged from wild type to severely defective embryo mutants; this could be a consequence of competition between monomers of RSH and C-terminal fusions of RSH in polymer formation. Note that all of the N-terminal RSH fusions rescued the mutant.
Discussion
The plant cell wall is a complex and diverse morphogenetic fabric assembled outside the plasma membrane from secreted macromolecular precursors. Currently the rules that govern their self-assembly at the molecular level are unknown. An approach using mutagenesis to disrupt function led to the identification of the recessive embryogenic rsh mutant associated with the knockout of AtEXT3. The mutant had to be propagated as a heterozygote because the mutation is homozygous lethal. Earlier we traced its defective morphology at the light microscope level to an aberrant initial division of the rsh/rsh zygote into a large apical cell and smaller basal cell rather than the small apical cell and larger basal cell of the wild type (5). At the electron microscope level we observed defective wall assembly (Fig. 2). Because this corresponded to a knockout of the AtEXT3 gene, we inferred absence of AtEXT3, i.e., the RSH glycoprotein. We isolated RSH from the wild type and characterized it biochemically, thus identifying it as AtEXT3. Because the cell plate of the wild type contains RSH (5) and the mutant lacks RSH we conclude that defective wall assembly is a direct consequence of RSH absence. RSH is therefore an essential cell wall component entirely consistent with the original conclusion that “[p]rotein containing hydroxyproline is an integral part of the cell wall of actively growing cells” (7). Here we relate the molecular structure of RSH to its self-assembly as a putative molecular scaffold that may template cell plate components in the new cross wall and enable its fusion with the mother cell wall.
Golgi-derived vesicles transport precursors of the cell plate. These vesicles fuse to form a membrane-bound nascent wall that grows outwards from within the cell (23–25). Cell plate components include cellulose (26), xyloglucans, high levels of non esterified polygalacturonic acid (27), arabinogalactan (28), callose (28), arabinofuranose residues typical of extensin (28), and RSH, inferred from expression of RSH–GFP fusions (5); they were all identified by immunocytology. Significantly, two Idt-rich extensins in Catharanthus showed cell cycle-regulated expression, also suggesting that extensins provide material for cell plate formation (29). Thus, HRGP synthesis occurs at an early stage of wall assembly (30). The challenge is to describe how the components of the disk-shaped cell plate undergo orderly self-assembly as it approaches and connects with the mother cell wall. Deposition of callose precedes that of cellulose, which is synthesized during maturation of the cell plate (26) in the final stage of cytokinesis; thus, callose may help in the formation and mechanical stabilization of the growing cell plate (26). Organizational or temporal data for other cell plate components are largely unknown.
Extensins are known as highly insoluble cell wall glycoproteins that form Idt cross-links in vitro. Electron microscopy images of platinum-shaded molecules indicate that these rod-shaped molecules form both intramolecular and short oligo intermolecular cross-links (31). AFM data in this article (Fig. 5) showed that solutions of RSH extensin monomers generated both rope-like and dendritic structures via end-on adhesion and lateral association, consistent with the well known properties of self-assembling amphiphiles in general (32). Thus, RSH protonetworks confirmed its scaffolding propensity. Unlike previously published electron microscopy images, AFM images of RSH showed that all of the available RSH molecules were incorporated into multimeric structures. The extensive dendritic structures observed by AFM could be a consequence of the different technology used compared with that of electron microscopy. Self-assembly of extensin networks has been discussed for many years. Here we outline an assembly paradigm that can be tested experimentally and used for designing novel self-assembling supramolecular structures.
The Extensin HRGP Self-Assembly Paradigm
General rules outlined more than a decade ago (20) identified HRGP putative functional sites involved in molecular alignment for self-recognition and described how monomeric precursors (18) of high periodicity (33) could form extensin networks further stabilized by enzymic cross-linking (11). More recent work identified the crucial role of tyrosine residues as intermolecular cross-links, namely pulcherosine (13) and di-Idt (34), but also as hydrophobic recognition sites; thus, specific tyrosine residues of collagen telopeptides may initiate self-assembly of collagen (35). This may account for the apparent requirement for an unobstructed tyrosine-rich RSH C terminus consistent with its end-on adhesion (Table 3 and Fig. 5). Indeed, a C-terminal tyrosine residue is a notable feature in 18 of the 20 Arabidopsis extensins (SI Table 4).
Earlier proposed features of self-assembling amphiphiles (32) together with the data presented here allow us to propose more specific rules for the initial self-assembly of rod-like AtEXT3 amphiphiles into a positively charged scaffold that templates the in muro deposition of pectin. These rules apply to AtEXT3 and are supported by all of the available data, and they may apply to other extensins, given their highly homologous sequences within (SI Table 4) and between species; these rules involve hydrophobic association, peptide periodicity, covalent cross-link formation, and electrostatic recognition summarized as follows.
A liquid–liquid interface favors self-assembly of highly ordered amphiphilic arrays as flat sheets but not in free solution (32, 36).
Alternating hydrophilic and hydrophobic modules of amphiphilic polypeptides induce like-with-like self-recognition (37).
Strict periodicity aligns extensin monomers (Fig. 4).
Hydrophobic interaction of four tyrosine residues at the RSH C terminus YLYKSPPPPYHY may initiate end-on adhesion (Fig. 5).
Intermolecular covalent tyrosine cross-links stabilize extensin networks (Fig. 3).
Pulcherosine cross-links favor a staggered alignment of RSH monomers (Figs. 3 and 4).
Staggered alignment permits two-dimensional growth of a RSH network.
The acid-base reaction between pectin and extensin (18, 38) forms extensin pectate.
Positively charged extensin scaffolds (Fig. 5) may template orderly deposition of a pectic matrix that is highly organized (39) compared with the disordered pectic gels of jam (27).
Finally, we anticipate that these superabundant, essential HRGPs will not only yield deeper insights into self-assembly processes that underpin plant growth and development but also provide utilitarian benefits based on their future exploitation for nanotechnology (32).
Materials and Methods
Transmission Electron Microscopy.
Immature siliques and 3-day-old seedling root pieces were fixed for 2 h in primary fixative (0.1 M pipes buffer, pH 7.2/4.0% paraformaldehyde/2.5% glutaraldehyde) followed by 2 h in secondary fixative (0.1 M pipes buffer, pH 7.2/1% osmium tetroxide). Samples were washed for 15 min sequentially in 0.1 M pipes buffer (pH 7.2) three times and water twice, followed by dehydration, all at 4°C. After staining [2% uranyl acetate in 0.05 M malate (pH 6.0)] for 2 h, samples were infiltrated with resin (LR White, Medium Grade; London Resin) for 15 h and embedded under long-wave UV light and a stream of nitrogen for a further 15 h, all at 4°C. Sections (80 nm) transferred to formvar-coated nickel grids were examined with a JOEL 100S transmission electron microscope at 80-kV acceleration.
Rescue of the rsh Mutant.
The rsh mutant rescue experiments were done by transforming heterozygous rsh plants (kanamycin-resistant) with the RSH gene fused to GFP, either at the C-terminal coding region of RSH precisely or at the N-terminal coding region of RSH at 5 aa downstream from the signal peptide cleavage site to ensure that a mature GFP–RSH fusion was produced. Green fluorescence in the cell wall in Tris buffer (pH 9.5) and its absence or significant reduction in protoplasts from transformed tissue confirmed the presence of the GFP fusion proteins in the wall. All of the constructs included the entire native RSH regulatory sequences on 6.3 kb of DNA in the EcoRI to XhoI sites of the binary vector SLJ75515 (conferring additional resistance to glufosinate-ammonium for plant selection) (40). The hypervirulent Agrobacterium EHA105 strain (41) was used for plant transformation (42). After T1 seed collection, individual glufosinate-ammonium-resistant T1 plants were selected [glufosinate-ammonium (Sigma 45520, 5 μg per ml)/MS salts (Sigma M5524, 2.2 g/liter)/3% sucrose/0.8% agar), transferred to soil, and self-fertilized to yield T2 seed. Homozygous glufosinate-ammonium-resistant T2 and T3 generation seedlings were scored as rsh homozygous (rsh/rsh), heterozygous (RSH/rsh), or wild-type (RSH/RSH) transformants by germination on plates (as above) in the presence of kanamycin (40 μg/ml). The rsh zygosity of 30 independent transformants for each plasmid construct was examined. Homozygous rsh plants with a restored wild-type phenotype confirmed complementation by the RSH transgene and its tagged fusions. RSH wild-type plants homozygous for the RSH transgene and its tagged fusions were also examined.
RSH Extensin Purification.
After harvesting wild-type Arabidopsis cell cultures by filtration (≈7 days, 20% packed cell volume), the cell pad 200 mM AlCl3 eluate was dialyzed, sized on Superose-6, and then purified via reverse-phase chromatography on a Hamilton PRP-1 preparative column (14).
Amino Acid Analysis.
After acid hydrolysis of peptides and proteins in 6 M HCl at 105°C for 18 h and derivatization with phenylisothiocyanate, amino acids were quantified as their phenylthiocarbamyl derivatives after gradient elution on a C18 column as previously described (14).
Peptide Sequencing.
Peptide sequencing was performed by the Michigan State University Macromolecular Facility (East Lansing, MI).
Sugar Analysis.
After sample hydrolysis for 1 h at 121°C in 2 M trifluoroacetic acid, sugars were quantified as their alditol acetates via gas–liquid chromatography as previously described (14).
Hyp Glycoside Profiles.
Hyp glycosides released by alkaline hydrolysis of RSH were fractionated by cation-exchange chromatography on Chromobeads C and quantified by continuous automated assay of Hyp in the column eluate as previously described (14).
In Vitro Cross-Linking via Extensin Peroxidase.
One nanogram of the extensin peroxidase (pI 4.6) isolated from tomato was used in each mixture containing 60 μg of RSH plus 100 mM H2O2 in 300 μl of 50 mM McIlvaine's buffer, all as previously described (11).
AFM Visualization.
A total of 60 μl of aqueous protein solutions [9 μg/ml RSH (monomeric fraction; see Fig. 3A), 10 μg/ml BSA, 10 μg/ml YK20, and 10 μg/ml polyhydroxyproline (Sigma)] was deposited on graphite for 5 min; the water was drawn off with filter paper, and the graphite surface was rinsed with 100 μl of water. The samples were dried under a stream of nitrogen gas. AFM images were obtained by using an MFP 3D microscope (Asylum Research) in AC mode under ambient conditions. Nanoprobes with a resonance frequency of ≈75 KHz and a spring constant of ≈3.5 N/m (NSC18; Mikromash) were used. The quality factor of probe resonance was electronically enhanced, when necessary, to ensure consistent imaging dynamics of the samples and their graphite background (43).
Supplementary Material
ACKNOWLEDGMENTS.
D.T.A.L. thanks the School of Life Sciences, University of Sussex, for use of their facilities. M.C.C. was supported by the University of Massachusetts and National Science Foundation Grant MCB-0311972, and M.J.K. was supported by Herman Frasch Foundation Grant 526-HF02, National Science Foundation Grant MCB-9874744, and U.S. Department of Agriculture Grant 2002-34490-11919. This work was supported by the Ohio University Biomimetic Nanoscience and Nanotechnology Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711980105/DC1.
References
- 1.Lamport DTA. The protein component of primary cell walls. Adv Bot Res. 1965;2:151–218. [Google Scholar]
- 2.Fry SC. Isodityrosine, a new cross-linking amino acid from plant cell-wall glycoprotein. Biochem J. 1982;204:449–455. doi: 10.1042/bj2040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epstein L, Lamport DTA. An intramolecular linkage involving isodityrosine in extensin. Phytochemistry. 1984;23:1241–1246. [Google Scholar]
- 4.Buehler MJ. Nature designs tough collagen: Explaining the nanostructure of collagen fibrils. Proc Natl Acad Sci USA. 2006;103:12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall Q, Cannon MC. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyams J, Davies DR. Induction and characterization of cell wall mutants of Chlamydomonas reinhardi. Mutat Res. 1972;14:381–389. [Google Scholar]
- 7.Lamport DTA, Northcote DH. Hydroxyproline in primary cell walls of higher plants. Nature. 1960;188:665–666. [Google Scholar]
- 8.Lamport DTA. Oxygen fixation into hydroxyproline of plant cell wall protein. J Biol Chem. 1963;238:1438–1440. [PubMed] [Google Scholar]
- 9.Lamport DTA. In: The Biochemistry of Plants. Preiss J, editor. New York: Academic; 1980. pp. 501–541. [Google Scholar]
- 10.Lamport DTA. In: Biogenesis of Plant Cell Wall Polysaccharides. Loewus FA, editor. New York: Academic; 1973. pp. 149–164. [Google Scholar]
- 11.Schnabelrauch LS, Kieliszewski MJ, Upham BL, Alizedeh H, Lamport DTA. Isolation of pI 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link sites. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- 12.Heckman JW, Terhume BT, Lamport DTA. Characterization of native and modified extensin monomers and oligomers by electron microscoy and gel filtration. Plant Physiol. 1988;86:848–856. doi: 10.1104/pp.86.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brady JD, Sadler IH, Fry SC. Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell walls: Its role in cross-link formation. Phytochemistry. 1998;47:349–353. doi: 10.1016/s0031-9422(97)00592-x. [DOI] [PubMed] [Google Scholar]
- 14.Held MA, Kamyab A, Hare M, Shpak E, Kieliszewski MJ. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem. 2004;279:55474–55482. doi: 10.1074/jbc.M408396200. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleland RE. A possible role for hydroxyproline-containing proteins in the cessation of cell elongation. Plant Physiol. 1967;42:669–671. doi: 10.1104/pp.42.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadava D, Chrispeels MJ. Hydroxyproline-rich cell wall protein (extensin): Role in the cessation of elongation in excised pea epicotyls. Dev Biol. 1973;30:49–55. doi: 10.1016/0012-1606(73)90047-x. [DOI] [PubMed] [Google Scholar]
- 18.Smith JJ, Muldoon EP, Lamport DTA. Isolation of extensin precursors by direct elution of intact tomato cell suspension cultures. Phytochemistry. 1984;23:1233–1239. [Google Scholar]
- 19.Lamport DTA. Hydroxyproline-O-glycosidic linkage of the plant cell wall glycoprotein extensin. Nature. 1967;216:1322–1324. [Google Scholar]
- 20.Kieliszewski MJ, Lamport DTA. Extensin: Repetitive motifs, functional sites, posttranslational codes and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 21.Shpak E, Leykam JF, Kieliszewski MJ. Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc Natl Acad Sci USA. 1999;96:14736–14741. doi: 10.1073/pnas.96.26.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller DJ, Janovjak H, Lehto T, Kuerschner L, Anderson K. Observing structure, function and assembly of single proteins by AFM. Prog Biophys Mol Biol. 2002;79:1–43. doi: 10.1016/s0079-6107(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 23.Verma DPS. Cytokinesis and building of the cell plate in plants. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:751–784. doi: 10.1146/annurev.arplant.52.1.751. [DOI] [PubMed] [Google Scholar]
- 24.Mayer U, Juergens G. Cytokinesis: Lines of division taking shape. Curr Opin Plant Biol. 2004;7:599–604. doi: 10.1016/j.pbi.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Segui-Simarro JM, Austin JR, II, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuels AL, Giddings TH, Jr, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts K. Structures at the plant cell surface. Curr Opin Cell Biol. 1990;2:920–928. doi: 10.1016/0955-0674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- 28.Northcote DH, Davey R, Lay J. Use of antisera to localize callose, xylan and arbinogalactan in the cell-plate, primary and secondary walls of plant cells. Planta. 1989;178:353–366. doi: 10.1007/BF00391863. [DOI] [PubMed] [Google Scholar]
- 29.Ito M, Kodama H, Komamine A, Watanabe A. Expression of extensin genes is dependent on the stage of the cell cycle and cell proliferation in suspension cultured Catharanthus roseus cells. Plant Mol Biol. 1998;36:343–351. doi: 10.1023/a:1005913818129. [DOI] [PubMed] [Google Scholar]
- 30.Ye Z-H, Varner JE. Tissue-specific expression of cell wall proteins in developing soybean tissues. Plant Cell. 1991;3:23–37. doi: 10.1105/tpc.3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stafstrom JP, Staehelin LA. Cross-linking patterns in salt-extractable extensin from carrot cell walls. Plant Physiol. 1986;81:234–241. doi: 10.1104/pp.81.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapaport H. Ordered peptide assemblies at interfaces. Supramol Chem. 2006;18:445–454. [Google Scholar]
- 33.Smith JJ, Muldoon EP, Willard JJ, Lamport DTA. Tomato extensin precursors P1 and P2 are highly periodic structures. Phytochemistry. 1986;25:1021–1030. [Google Scholar]
- 34.Brady JD, Sadler IH, Fry SC. Di-isodityrosine, a novel tetrameric derivative of tyrosine in plant cell wall proteins: A new potential cross-link. Biochem J. 1996;315:323–327. doi: 10.1042/bj3150323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cejas MA, et al. Collagen-related peptides: Self-assembly of short, single strands into a functional biomaterial of micrometer scale. J Am Chem Soc. 2007;129:2202–2203. doi: 10.1021/ja066986f. [DOI] [PubMed] [Google Scholar]
- 36.Russell JT, et al. Self-assembly and cross-linking of bionanoparticles at liquid-liquid interfaces. Angew Chem Int Ed. 2005;44:2420–2426. doi: 10.1002/anie.200462653. [DOI] [PubMed] [Google Scholar]
- 37.Ohno S. The cardinal principle of like attracting like generates many ubiquitous oligopeptides shared by divergent proteins. Anim Genet. 1994;25:5–11. doi: 10.1111/j.1365-2052.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 38.MacDougall AJ, et al. The effect of peptide-pectin interactions on the gelation behaviour of a plant cell wall pectin. Carbohydr Res. 2001;335:115–126. doi: 10.1016/s0008-6215(01)00221-x. [DOI] [PubMed] [Google Scholar]
- 39.Vincken JP, et al. If homogalacturonan were a side chain of rhamnogalacturonan. I. Implications for cell wall architecture. Plant Physiol. 2003;132:1781–1789. doi: 10.1104/pp.103.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones JDH, et al. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- 41.Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. [Google Scholar]
- 42.Chang SS, et al. Stable genetic transformation of Arabidopsis thaliana by Agrobacterium inoculation in planta. Plant J. 1994;5:551–558. [Google Scholar]
- 43.Chen L, Yu X, Wang D. Cantilever dynamics and quality factor control in AC mode AFM height measurements. Ultramicroscopy. 2007;107:275–280. doi: 10.1016/j.ultramic.2006.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.