Abstract
Because T lymphocytes have the capacity to recognize tumor cells, significant efforts are being devoted towards the development of T cell-based immunotherapy for cancer. Most of this work has centered in the induction of anti-tumor CD8 T cells, which exhibit cytolytic activity towards tumor cells expressing tumor-specific or tumor associated antigens. Unfortunately to this day, T cell-based immunotherapy for cancer remains suboptimal. One of the possible explanations is that these immunotherapies have ignored the role that CD4 T helper lymphocytes play in the generation and persistence of CD8 T cell responses. Thus, we believe that in order to obtain clinical benefits T cell based immunotherapy must stimulate both CD8 and CD4 tumor-reactive T cell responses. During the past seven years our group has focused on the identification of CD4 T cell epitopes from tumor-associated and tumor-specific antigens that could be used to complement the already identified CD8 T cell epitopes to produce effective vaccination strategies against numerous tumor types. We will describe here the strategy we used that resulted in the identification and characterization of numerous CD4 T cell epitopes that are applicable to developing therapies against hematological malignancies and solid tumors.
Introduction
Role of CD4 T lymphocytes in anti-tumor immune responses
Most immune therapy approaches to combat malignant diseases have centered in the induction of CD8 T cell responses against tumor-associated antigens mainly because these cells can be very effective in killing tumor cells. The 2 most commonly thought therapeutic approaches based on the use of anti-tumor CD8 cytotoxic T lymphocytes (CTLs) are the use of various types of vaccines that generate in vivo immune responses (active immunotherapy) or the infusion of in vitro expanded CTLs (adoptive T cell therapy) [1]. Although CTLs have been considered to be the main protagonists in the production of anti-tumor therapeutic effects resulting from vaccines or adoptive T cell therapy, it is clear that CD4 helper T lymphocytes (HTLs) can also participate in the generation of anti-tumor responses through various mechanisms [2, 3]. On one hand, HTLs are critical for the generation and persistence of CTL responses by providing help through multiple interactions with antigen-presenting cells and CD8 T cells during the induction of the immune responses in secondary lymphoid organs [4–6]. Later on, HTLs continue to sustain CTL responses through the maintenance of memory cells in the periphery or by directly providing costimulatory signals to the CTL, which enhance their function and survival at the tumor site [7–9]. In addition, tumor-infiltrating HTLs may function as effector cells either by the local production of cytokines that curtail tumor growth or in some instances by exhibiting anti-tumor cytotoxic activity [10, 11]. Because CD4 HTLs recognize antigen as peptide epitopes presented in the context of MHC class II molecules (MHC-II), the effector activity of these cells will be circumscribed to those tumor cells that express MHC class II molecules or in those instances where conventional antigen-presenting cells (e.g., macrophages, dendritic cells) are able to capture, process and present antigens derived from tumor cells to the HTLs. In view of the above, there has been a considerable interest in identifying MHC-II epitopes for tumor-reactive HTL with the purpose of using this information to develop more effective forms of immunotherapy against malignant diseases.
The predictive approach for defining CD4 T cell epitopes from tumor-associated antigens
The identification of sequence motifs in peptides capable of binding to MHC-II molecules of various alleles has lead to the development of computer-based algorithms to predict the existence of CD4 T cell epitopes within the amino acid sequence of a putative tumor-associated antigen (TAA) [12–14]. In many instances these algorithms have been validated and further refined using quantitative peptide/MHC-II binding assays. We have extensively used one of such algorithms that was designed to identify peptide binders to the products of 3 commonly found human MHC-II alleles: HLA-DR1, DR4 and DR7 [13]. In our studies we tried to select peptide sequences that scored high in the probability scale for their binding capacity for all 3 MHC-II alleles with the goal of identifying promiscuous CD4 T cell epitopes that would result a broad population coverage for a specific clinical indication. The strategy to identify promiscuous CD4 T cell epitopes using the predictive approach would follow these steps:
a) Selection of the protein antigen
There are several characteristics that would make a particular protein interesting and suitable for the analysis. The most obvious one is that the complete amino acid sequence of the target antigen must be known. Second, it is advisable to select large proteins instead of smaller ones since the probability of finding one or more potential MHC class II binding epitopes will increase. Another desirable characteristic of the protein antigen would be to select antigens that are only expressed by tumor cells and not normal cells. The reasons are that immune tolerance mechanisms operating against self-proteins will reduce the effectiveness of immune responses against tumors. In addition, in the event that strong immune responses were to be induced against a self-antigen, there would be a significant risk of autoimmune pathology towards normal tissues expressing that antigen. Unfortunately, these “tumor-specific antigens” (TSA) are not frequently encountered, so in many cases one has to rely in the use of “tumor-associated antigens (TAA) [15]. However, examples of TSA are proteins derived from oncogenic viruses that continue to be expressed by the malignant cells or products of mutated/rearranged genes that are specifically expressed by the tumor cells [16, 17]. Some TAA that are desirable candidates for immunotherapy are those that belong to the “Cancer Testes” (CT) family [18]. These products are expressed in tumor cells and in germinal cells, but not in most normal tissues, limiting the deleterious effects of tolerance and autoimmunity. Other TAA candidates have limited expression in some tissues that are considered not to be vital (prostate, breast, melanocytes). Thus, depending on the tumor type, one may have a single or multiple protein candidates to chose from to submit for prediction analysis for the presence of MHC-II binding peptides.
b) Prediction algorithms for CD4 T cell epitopes
Originally these algorithms were based on the presence of specific residues (or type of residues: hydrophobic, positive/negative charged) present at particular positions within a short peptide sequence (9 to 15 residues long) that were found on T cell epitopes that had been mapped using antigen-specific T cell lines/clones. These motifs were found to be somewhat specific for individual MHC-II alleles but could not be clearly defined. In the case of MHC class I (MHC-I) binding peptides, it became feasible to identify more exact motifs by sequencing peptides eluted from purified MHC molecules because of their restricted size (8–10 residues). On the other hand peptides that bind to MHC-II do not have a constrained size (9–20 residues), making sequencing analysis interpretation much more difficult. A major breakthrough in the design of MHC-II binding algorithms was the development of quantitative MHC-II peptide binding assays. Using this approach, Southwood and collaborators constructed a computer-based algorithm based on a large database of MHC-II binding peptides [13]. This database keeps being refined [19] and the respective algorithms to predict probability of peptides to bind to diverse human (and mouse) MHC-II alleles have been made available to researchers through the Internet (http://www.immuneepitope.org/). We have utilized this algorithm in numerous occasions to predict the existence of peptide sequences from TSA and TAA capable of binding to 3 frequently found MHC-II alleles (HLA-DR1, DR4 and DR7). Results obtained from these algorithms are utilized to limit the number of synthetic peptides that will be tested for their ability to induce T cell responses in labor-intensive cell cultures.
c) In vitro immunization of human lymphocytes using candidate CD4 T cell epitopes
Once a manageable number of peptide sequences have been selected from the algorithm analysis, synthetic peptides are made and tested in tissue culture for their ability to stimulate CD4 T cell responses. The technique to generate antigen-specific CD4 T cell lines and clones used by our group has been described in detail [20]. Briefly, the in vitro immunization method utilizes purified CD4 T cells isolated from peripheral blood of normal volunteers that are stimulated using peptide-pulsed autologous dendritic cells (DCs). The cultures are restimulated periodically with autologous irradiated periheral blood mononuclear cells in the presence of IL-2. T cell lines and clones (isolated by limiting dilution) are then submitted to MHC restriction analysis using HLA-typed antigen-presenting cells (APCs) and mouse fibroblast cells (L-cells) transfected with individual human MHC-II alleles. Peptide-titration curves can be done to estimate the antigen avidity of the T cells. The most important step in this process is to assess whether the peptide-reactive CD4 T cell lines (or clones) have the capacity to recognize the TSA or TAA derived from tumor cells. Only with this validation one can be assured that the peptide in question represents a true tumor-specific CD4 T cell epitope. We believe that this validation is necessary before even considering using the peptide as a vaccine or peptide-reactive T cells for adoptive immunotherapy.
d) Assessing tumor-reactivity of peptide-reactive CD4 T cells
Two types of assays can be done to demonstrate that synthetic peptides capable of eliciting CD4 T cell responses represent real tumor-specific (or tumor-associated) epitopes. The first one is to determine the ability of a CD4 T cell line to recognize antigen presented directly by the tumor cells. For this to take place, one must utilize tumor cell lines that express MHC-II surface molecules, as it occurs with many hematological malignancies (e.g., B and T cell lymphomas). In the case of cell lines derived from solid tumors, expression of MHC-II molecules may be induced by treating the cells for 24–74 h with interferon-gamma (IFNγ), prior to performing the immunological assays. One limiting factor with this type of assay is that the tumor cells must express the same MHC-II allele that restricts the response of the CD4 T cells to peptide. In the second type of assay one estimates the capacity of autologous DCs to capture, process and present tumor antigen proteins. In some instances one may use purified recombinant DNA-produced tumor antigen if it is available. In most cases it is possible to feed the DCs with tumor cell lysates (freeze-thaw) or apoptotic (UV light-induced) tumor cells. Under these circumstances, the tumor cell lines do not require to express MHC-II molecules because the CD4 T cells will recognize the processed epitope in the context of the DC’s MHC-II molecules. Antigen specificity is validated by using control tumor cell lines that do not express the tumor antigen in question and by the demonstration that antibodies to MHC-II molecules can block the reactivity of the CD4 T cells recognizing the antigen presented by the DCs. We strongly argue that only by fulfilling the above-mentioned criteria one can be certain that the peptide epitope originally identified as an MHC-II-binding candidate truly represents a tumor-specific epitope for CD4 T cells. Without this demonstration it would be unwise and perhaps unethical to proceed with clinical development to use a CD4 T cell epitope in cancer patients.
CD4 T cell epitopes for viral-encoded TSA
Our studies have focused on 2 viruses responsible for hematological malignancies in humans, Epstein-Barr virus (EBV) and human T-cell leukemia virus-1 (HTLV-1). For these studies, we selected viral protein antigens that continue to be expressed in the malignant cells and that in most instances are involved in the establishment of the malignant phenotype. EBV is responsible for post-transplant lymphoproliferative disease in immunosuppressed individuals that can progress to B-cell lymphomas, and for other malignancies such as NK/T cell lymphoma and several lymphoepithelioma-like carcinomas including nasopharyngeal carcinoma (NPC) and gastric carcinoma. Our studies have identified CD4 T cell epitopes from 2 viral latent cycle antigens, EBNA2 and LMP1 that could be presented in the context of several MHC-II alleles (Table 1) [21, 22]. The reactivity of the peptide induced CD4 T cell lines and clones was significant against various EBV-transformed cells (examples presented in Figure 1). Other studies from our group helped identify CD4 T cell epitopes from 2 antigens expressed by HTLV-1 (the causative agent of adult T cell leukemia/lymphoma), the Tax and the envelope antigens (Table I) [23, 24]. Again, an important feature of the peptide-induced CD4 T cells was their ability to react with HTLV-1 expressing tumor cells (Figure 1).
Figure 1.
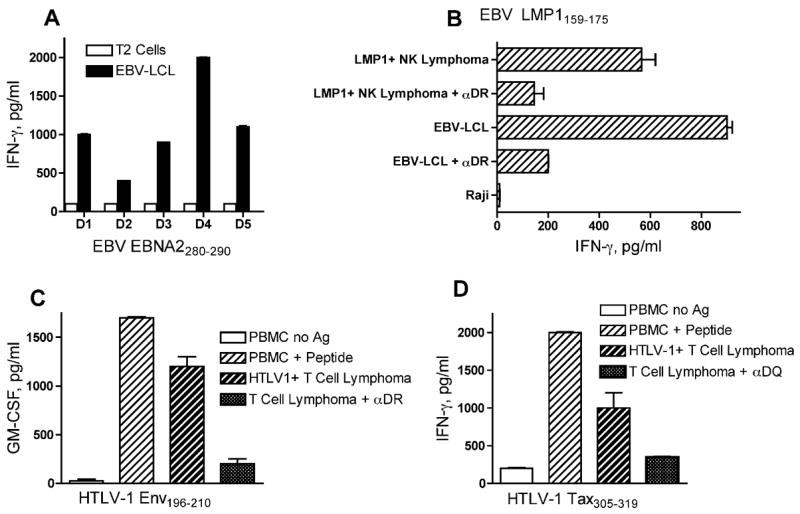
Examples of CD4 T-cell responses to viral-encoded tumor antigens.
A, CD4 HTLs derived from 5 healthy donors (D1-D5) specific for peptide EBNA2280–290 were able to recognize autologous EBV-transformed B lymphoblastoid cell lines (EBV-LCL) but not the MHC-II-negative, EBV-negative Jurkat T cell lymphoma. B, An HLA-DR9-restricted, EBV LMP1159–175-reactive CD4+ HTL clone showing the capacity to directly recognize autologous EBV-LCL and an LMP1+ NK cell lymphoma but not MHC-II allogeneic and unrelated Burkitt’s lymphoma Raji. The reactivity was inhibited by the addition of anti-HLA-DR monoclonal antibodies (αDR) indicating that the peptide-epitope is recognized through MHC-II molecules.
C, An HLA-DR9-restricted, HTLV-1Env196–210-specific CD4 HTL clone recognizes peptide-pulsed autologous peripheral blood mononuclear cells (PBMC) and reacts with a DR9-expressing HTLV-1+ T-cell lymphoma. This reactivity was also blocked with anti-HLA-DR antibodies (αDR). D, An HLA-DQ9-restricted, HTLV-1Tax305–319-specific CD4 HTL clone recognizes peptide-pulsed autologous PBMC and a DQ9-expressing HTLV-1+ T cell lymphoma. Recognition of the HTLV-1 T cell lymphoma by the HTL was blocked by anti-HLA-DQ antibodies (αDQ).
Table I.
| Antigena | Peptide Positionb | Peptide Sequencec | MHC Class II Restrictiond | Effector Functione | Proximal CTL Epitope(MHC-I Restriction)f | Referencesg |
|---|---|---|---|---|---|---|
| EBNA2 | 280 – 290 | TVFYNIPPML | DR1, DR7, DR16, DR52, DQ2 | 1, 2 | [21] | |
| EBV LMP1 | 159 – 175 | YLQQNWWTLLVDLLWLL | DR9, DR15, DR53 | 1, 2 | EBV LMP1159–167 (A2) | [22, 35] |
| HTLV-1 Env | 196 – 210 | LDHILEPSIPWKSKL | DR9 | 1, 2 | [23] | |
| 317 – 331 | AVWLVSALAMGAGVA | DQ6 | 1 | [23] | ||
| 384 – 398 | LLFWEQGGLCKALQE | DR15 | 1 | [23] | ||
| HTLV-1 Tax | 191 – 205 | IEELLYKISLTTGAL | DR1, DR9 | 1 | HTLV-1 Tax181–195 (B14) | [24, 33] |
| 305 – 319 | LHLLFEEYTNIPISL | DR15, DQ9 | 1 | HTLV-1 Tax301–309 (A24) | [24, 43] | |
| HER2/neu | 883 – 899 | KVPIKWMALESILRRRF | DR1, DR4, DR52, DR53 | 1 | [20] | |
| gp100 | 74 – 89 | GPTLIGANASFSIALN | DR7 | 1 | gp10070–78 (C8), gp10087–95 (A3) | [25, 36, 37] |
| 175 – 189 | GRAMLGTHTMEVTVY | DR53, DQ6 | 1, 2 | gp100177–186 (A2) | [25, 34] | |
| 576 – 590 | SLAVVSTQLIMPGQE | DR7 | 1 | gp100570–579 (A2) | [25, 34] | |
| MAGE-A3 | 146 – 160 | FFPVIFSKASSSLQL | DR4, DR7 | 1 | MAGE-A3143–151 (B52), MAGE-A3159–160 (A2) | [26, 39, 41] |
| WT1 | 124 – 138 | QARMFPNAPYLPSCL | DR15, DR53 | 1 | WT1126–134 (A2) | [31, 40, 47] |
| STEAP | 102 – 116 | HQQYFYKIPILVINK | DR1, DR9, DR53 | 1 | STEAP86–94 (A2) | [32, 46] |
| 192 – 206 | LLNWAYQQVQQNKED | DR9, DR53 | 1 | [32] | ||
| TARP | 1 – 14 | MQMFPPSPLFFFLQ | DR53 | 1 | TARP4–13 (A2) | [30, 44] |
| 14 – 27 | QLLKQSSRRLEHTF | DR1, DR9, DR13, DR15 | 1 | TARP27–35 (A2) | [30, 45] | |
| PSMA | 334 – 348 | TGNFSTQKVKMHIHS | DR4 | 1 | [28] | |
| 687 – 701 | YRHVIYAPSSHNKYA | DR9, DR53 | 1 | [28] | ||
| 730 – 744 | RQIYVAAFTVQAAAE | DR53 | 1 | [28] | ||
| CEA | 625 – 639 | YSWRINGIPQQHTQV | DR4, DR53 | 1 | CEA632–640 (B7) | [29, 42] |
| 653 – 667 | YACFVSNLATGRNNS | DR4, DR7 | 1 | CEA652–660 (A2) | [27, 38] |
Protein, used in prediction algorithms to identidy potential MHC-II binding peptides.
Residue positions of peptides used to stimulate T cell responses.
Amino acid sequence of T cell epitopes.
MHC-II restrictions established with either MHC typed human APCs or mouse L cells transfected with individual human MHC-II molecules.
1 = antigen induced cytokine release; 2 = cell-mediated cytotoxicity.
Peptide described to function as a CD8 T cell epitope and the restriction HLA class I allele, in parentheses.
References describing the CD4 and CD8 T cell epitopes.
CD4 T cell epitopes for TAA expressed by solid tumors
Our studies have focused in the identification of CD4 T cell epitopes for tumor types that affect large number of individuals throughout the world, such as breast, prostate, gastrointestinal cancers and malignant melanoma. Proteins that function as TAA because they are either differentiation (tissue-specific) antigens (gp100, PSMA, TARP, STEAP), tumor markers (CEA, HER2/neu, WT1) or belong to the family of cancer-testes antigens (MAGE-A3) were studied as potential sources for CD4 T cell epitopes. Our studies following the strategy described above resulted in the identification of several novel CD4 T cell epitopes for these antigens (Table 1) [20, 25–32]. As mentioned previously, the most critical issue is to demonstrate the reactivity of peptide-induced CD4 T cells against tumor cells that express MHC-II and the TAA or alternatively, towards autologous DCs that are fed with antigens derived from tumor cells (e.g., freeze-thaw lysates). The examples presented in Figure 2 illustrate how effective was the ability of various CD4 T cell lines induced with peptides selected from MHC-II binding predictive algorithms, to recognize antigens derived from tumor cells.
Figure 2.
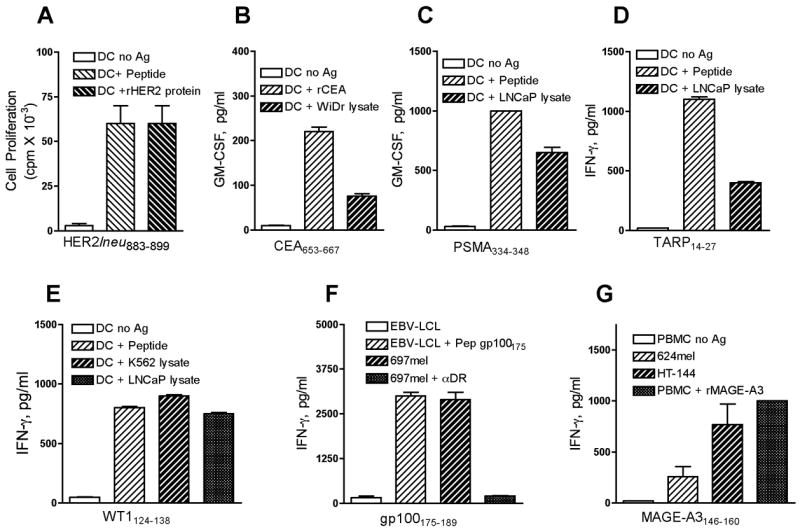
Examples of CD4 T-cell responses to TAA expressed by solid tumors. HTL clones specific for HER2/neu883–899 (A), CEA653–667 (B), PSMA334–348 (C), TARP14–27(D), and WT1124–138-reactive (E) were shown recognize naturally processed antigens derived from recombinant protein or a cell lysate from the prostate tumor LNCaP presented by DCs in the context of MHC-II molecules. A gp100175–189-reactive CD4 HTL clone (F) directly recognizes peptide presented by autologous EBV-LCL and an MHC-II+, gp100+ melanoma tumor mel697 but not the gp100 negative EBV-LCL This recognition is inhibited by anti-HLA-DR antibodies (αDR). MAGE-A3146–160-reactive (G) CD4 HTL clone recognizes directly MHC-II expressing, MAGE-A3+ (HT144 and 624mel) melanomas and react with recombinant MAGE-A3 protein presented by autologous PBMC. STEAP102–116-reactive HTL (H) recognizes peptide or cell lysate from STEAP+ (LNCaP) tumor cells. In addition, this HTL can recognize directly a STEAP+, DR+ prostate tumor cell line (PC3).
CD4 T cell epitopes congregate near CD8 T cell epitopes
Throughout our studies we observed that many of the CD4 T cell epitopes that we identified contain within their sequence, or lie proximal to previously described CD8 T cell epitopes (Table I) [33–47]. Although we do not know whether the close proximity between CD4 and CD8 T cell epitopes is simply a coincidence or holds a biological significance, this observation opens the possibility of using synthetic peptides of relatively small size (15–20 residues) to stimulate CD4 and CD8 T cell responses simultaneously [47–50]. In addition, it has been proposed that large peptides containing MHC-II binding domains may be an effective way to deliver antigens to professional APCs in order to generate more effective immune responses [51].
Closing remarks
The identification of CD4 T cell epitopes for tumor antigens has attracted a high degree of interest because of the belief that immune based therapies against cancer will be more effective if both CD8 and CD4 T lymphocytes participate in curtailing tumor cell growth. CD4 T cells not only are capable of enhancing CD8 T cell responses but may also exhibit anti-tumor effector function. Although many studies in animal models have clearly demonstrated better results when both CD4 and CD8 T cells participate in anti-tumor responses (as compared to results observed with CD8 T cells alone) [52, 53], it remains to be determined whether this strategy will improve the effectiveness of immune therapy in human patients. Nevertheless, there are reports in the area of T cell adoptive therapy that the presence of CD4 T cells in cell products infused into patients, improves the persistence of CD8 T cells and the effectiveness of the therapy. Based on this knowledge we have initiated clinical studies in melanoma patients [54] using peptide gp100175–189 (Table I), which contains an HLA-A2-restricted CD8 T cell epitope imbedded within a promiscuous CD4 T cell epitope [25]. Similarly, our group will soon assess the immunogenicity and safety of peptide vaccines for prostate cancer patients using peptides TARP14–27, which harbors adjoining CD8 and CD4 T cell epitopes and a peptide from PSMA, containing a CD8 T cell epitope, PSMA27–35 [55] fused to a CD4 T cell epitope from the same TAA, PSMA687–701 (Table I). We hope that these studies will provide some evidence of whether simultaneous CD8 and CD4 T cell responses can generate long-lasting immune responses that translate into clinical benefit.
Acknowledgments
Supported by NIH grants P50CA91956, R01CA80782 and R01CA103921 (E. Celis) and Ministry of Education, Sports, and Culture of Japan grant-in-aid 18590360 (H. Kobayashi).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Berzofsky JA, Terabe M, Oh S, Belyakov IM, Ahlers JD, Janik JE, Morris JC. Progress on new vaccine strategies for the immunotherapy and prevention of cancer. J Clin Invest. 2004;113:1515–1525. doi: 10.1172/JCI21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4(+) T cells in the antitumor immune response. J Exp Med. 1998;188:2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha B, Tanchot C. Towards a cellular definition of CD8+ T-cell memory: the role of CD4+ T-cell help in CD8+ T-cell responses. Curr Opin Immunol. 2004;16:259–263. doi: 10.1016/j.coi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Smith CM, Wilson NS, Waithman J, Villadangos JA, Carbone FR, Heath WR, Belz GT. Cognate CD4(+) T cell licensing of dendritic cells in CD8(+) T cell immunity. Nat Immunol. 2004;5:1143–1148. doi: 10.1038/ni1129. [DOI] [PubMed] [Google Scholar]
- 5.Bourgeois C, Rocha B, Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002;297:2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 6.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 7.Giuntoli RL, 2nd, Lu J, Kobayashi H, Kennedy R, Celis E. Direct costimulation of tumor-reactive CTL by helper T cells potentiate their proliferation, survival, and effector function. Clin Cancer Res. 2002;8:922–931. [PubMed] [Google Scholar]
- 8.Kennedy R, Undale AH, Kieper WC, Block MS, Pease LR, Celis E. Direct cross-priming by Th lymphocytes generates memory cytotoxic T cell responses. J Immunol. 2005;174:3967–3977. doi: 10.4049/jimmunol.174.7.3967. [DOI] [PubMed] [Google Scholar]
- •• 9.Kennedy R, Celis E. T helper lymphocytes rescue CTL from activation-induced cell death. J Immunol. 2006;177:2862–2872. doi: 10.4049/jimmunol.177.5.2862. This paper provides direct evidence that CD4 T cells can prevent apoptosis induced by activation in CD8 T cells acting as “true helper lymphocytes” and allowing the survival and persistence of memory T cells. Interestingly, rescue from apoptosis required direct interaction between CD4 and CD8 T cells and appered to be mediated through costimulatory molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn S, Gehri R, Erb P. Mechanism and biological significance of CD4-mediated cytotoxicity. Immunol Rev. 1995;146:57–79. doi: 10.1111/j.1600-065x.1995.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 11.Haigh TA, Lin X, Jia H, Hui EP, Chan AT, Rickinson AB, Taylor GS. EBV Latent Membrane Proteins (LMPs) 1 and 2 as Immunotherapeutic Targets: LMP-Specific CD4+ Cytotoxic T Cell Recognition of EBV-Transformed B Cell Lines. J Immunol. 2008;180:1643–1654. doi: 10.4049/jimmunol.180.3.1643. [DOI] [PubMed] [Google Scholar]
- 12.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 13.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 14.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 15.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science. 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 17.Worley BS, van den Broeke LT, Goletz TJ, Pendleton CD, Daschbach EM, Thomas EK, Marincola FM, Helman LJ, Berzofsky JA. Antigenicity of fusion proteins from sarcoma-associated chromosomal translocations. Cancer Res. 2001;61:6868–6875. [PubMed] [Google Scholar]
- 18.Jager E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothe BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi H, Wood M, Song Y, Appella E, Celis E. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res. 2000;60:5228–5236. [PubMed] [Google Scholar]
- 21.Omiya R, Buteau C, Kobayashi H, Paya CV, Celis E. Inhibition of EBV-induced lymphoproliferation by CD4(+) T cells specific for an MHC class II promiscuous epitope. J Immunol. 2002;169:2172–2179. doi: 10.4049/jimmunol.169.4.2172. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi H, Nagato T, Takahara M, Sato K, Kimura S, Aoki N, Azumi M, Tateno M, Harabuchi Y, Celis E. Induction of EBV-latent membrane protein 1-specific MHC class II-restricted T-cell responses against natural killer lymphoma cells. Cancer Res. 2008;68:901–908. doi: 10.1158/0008-5472.CAN-07-3212. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi H, Nagato T, Yanai M, Oikawa K, Sato K, Kimura S, Tateno M, Omiya R, Celis E. Recognition of adult T-cell leukemia/lymphoma cells by CD4+ helper T lymphocytes specific for human T-cell leukemia virus type I envelope protein. Clin Cancer Res. 2004;10:7053–7062. doi: 10.1158/1078-0432.CCR-04-0897. [DOI] [PubMed] [Google Scholar]
- • 24.Kobayashi H, Ngato T, Sato K, Aoki N, Kimura S, Tanaka Y, Aizawa H, Tateno M, Celis E. In vitro peptide immunization of target tax protein human T-cell leukemia virus type 1-specific CD4+ helper T lymphocytes. Clin Cancer Res. 2006;12:3814–3822. doi: 10.1158/1078-0432.CCR-06-0384. This paper describes 2 novel CD4 T cell epitopes from HTLV-I tax protein that lie proximal to CD8 T cell epitopes Notably, CD4 T cell clones specific for these epitopes could directly recognize HTLV-I tax-expressing T cell lymphoma lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi H, Lu J, Celis E. Identification of helper T-cell epitopes that encompass or lie proximal to cytotoxic T-cell epitopes in the gp100 melanoma tumor antigen. Cancer Res. 2001;61:7577–7584. [PubMed] [Google Scholar]
- 26.Kobayashi H, Song Y, Hoon DS, Appella E, Celis E. Tumor-reactive T helper lymphocytes recognize a promiscuous MAGE-A3 epitope presented by various major histocompatibility complex class II alleles. Cancer Res. 2001;61:4773–4778. [PubMed] [Google Scholar]
- 27.Kobayashi H, Omiya R, Ruiz M, Huarte E, Sarobe P, Lasarte JJ, Herraiz M, Sangro B, Prieto J, Borras-Cuesta F, et al. Identification of an antigenic epitope for helper T lymphocytes from carcinoembryonic antigen. Clin Cancer Res. 2002;8:3219–3225. [PubMed] [Google Scholar]
- 28.Kobayashi H, Omiya R, Sodey B, Yanai M, Oikawa K, Sato K, Kimura S, Senju S, Nishimura Y, Tateno M, et al. Identification of naturally processed helper T-cell epitopes from prostate-specific membrane antigen using peptide-based in vitro stimulation. Clin Cancer Res. 2003;9:5386–5393. [PubMed] [Google Scholar]
- 29.Ruiz M, Kobayashi H, Lasarte JJ, Prieto J, Borras-Cuesta F, Celis E, Sarobe P. Identification and characterization of a T-helper peptide from carcinoembryonic antigen. Clin Cancer Res. 2004;10:2860–2867. doi: 10.1158/1078-0432.ccr-03-0476. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi H, Nagato T, Oikawa K, Sato K, Kimura S, Aoki N, Omiya R, Tateno M, Celis E. Recognition of prostate and breast tumor cells by helper T lymphocytes specific for a prostate and breast tumor-associated antigen, TARP. Clin Cancer Res. 2005;11:3869–3878. doi: 10.1158/1078-0432.CCR-04-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi H, Nagato T, Aoki N, Sato K, Kimura S, Tateno M, Celis E. Defining MHC class II T helper epitopes for WT1 tumor antigen. Cancer Immunol Immunother. 2006;55:850–860. doi: 10.1007/s00262-005-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 32.Kobayashi H, Nagato T, Sato K, Aoki N, Kimura S, Murakami M, Iizuka H, Azumi M, Kakizaki H, Tateno M, et al. Recognition of prostate and melanoma tumor cells by six-transmembrane epithelial antigen of prostate-specific helper T lymphocytes in a human leukocyte antigen class II-restricted manner. Cancer Res. 2007;67:5498–5504. doi: 10.1158/0008-5472.CAN-07-0304. The authors report the existence of a CD4 T cell epitope from TAA STEAP that lies closely to a previously identified CD8 T cell epitope described in reference [46]. [DOI] [PubMed] [Google Scholar]
- 33.Parker CE, Nightingale S, Taylor GP, Weber J, Bangham CR. Circulating anti-Tax cytotoxic T lymphocytes from human T-cell leukemia virus type I-infected people, with and without tropical spastic paraparesis, recognize multiple epitopes simultaneously. J Virol. 1994;68:2860–2868. doi: 10.1128/jvi.68.5.2860-2868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai V, Southwood S, Sidney J, Sakaguchi K, Kawakami Y, Appella E, Sette A, Celis E. Identification of subdominant CTL epitopes of the GP100 melanoma-associated tumor antigen by primary in vitro immunization with peptide-pulsed dendritic cells. J Immunol. 1997;158:1796–1802. [PubMed] [Google Scholar]
- 35.Khanna R, Burrows SR, Nicholls J, Poulsen LM. Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur J Immunol. 1998;28:451–458. doi: 10.1002/(SICI)1521-4141(199802)28:02<451::AID-IMMU451>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Kawashima I, Tsai V, Southwood S, Takesako K, Celis E, Sette A. Identification of gp100-derived, melanoma-specific cytotoxic T-lymphocyte epitopes restricted by HLA-A3 supertype molecules by primary in vitro immunization with peptide-pulsed dendritic cells. Int J Cancer. 1998;78:518–524. doi: 10.1002/(sici)1097-0215(19981109)78:4<518::aid-ijc20>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 37.Castelli C, Tarsini P, Mazzocchi A, Rini F, Rivoltini L, Ravagnani F, Gallino F, Belli F, Parmiani G. Novel HLA-Cw8-restricted T cell epitopes derived from tyrosinase-related protein-2 and gp100 melanoma antigens. J Immunol. 1999;162:1739–1748. [PubMed] [Google Scholar]
- 38.Nukaya I, Yasumoto M, Iwasaki T, Ideno M, Sette A, Celis E, Takesako K, Kato I. Identification of HLA-A24 epitope peptides of carcinoembryonic antigen which induce tumor-reactive cytotoxic T lymphocyte. Int J Cancer. 1999;80:92–97. doi: 10.1002/(sici)1097-0215(19990105)80:1<92::aid-ijc18>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 39.Russo V, Tanzarella S, Dalerba P, Rigatti D, Rovere P, Villa A, Bordignon C, Traversari C. Dendritic cells acquire the MAGE-3 human tumor antigen from apoptotic cells and induce a class I-restricted T cell response. Proc Natl Acad Sci U S A. 2000;97:2185–2190. doi: 10.1073/pnas.040540197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao L, Bellantuono I, Elsasser A, Marley SB, Gordon MY, Goldman JM, Stauss HJ. Selective elimination of leukemic CD34(+) progenitor cells by cytotoxic T lymphocytes specific for WT1. Blood. 2000;95:2198–2203. [PubMed] [Google Scholar]
- 41.Keogh E, Fikes J, Southwood S, Celis E, Chesnut R, Sette A. Identification of new epitopes from four different tumor-associated antigens: recognition of naturally processed epitopes correlates with HLA-A*0201-binding affinity. J Immunol. 2001;167:787–796. doi: 10.4049/jimmunol.167.2.787. [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Celis E. Use of two predictive algorithms of the world wide web for the identification of tumor-reactive T-cell epitopes. Cancer Res. 2000;60:5223–5227. [PubMed] [Google Scholar]
- 43.Harashima N, Kurihara K, Utsunomiya A, Tanosaki R, Hanabuchi S, Masuda M, Ohashi T, Fukui F, Hasegawa A, Masuda T, et al. Graft-versus-Tax response in adult T-cell leukemia patients after hematopoietic stem cell transplantation. Cancer Res. 2004;64:391–399. doi: 10.1158/0008-5472.can-03-1452. [DOI] [PubMed] [Google Scholar]
- 44.Carlsson B, Totterman TH, Essand M. Generation of cytotoxic T lymphocytes specific for the prostate and breast tissue antigen TARP. Prostate. 2004;61:161–170. doi: 10.1002/pros.20091. [DOI] [PubMed] [Google Scholar]
- 45.Oh S, Terabe M, Pendleton CD, Bhattacharyya A, Bera TK, Epel M, Reiter Y, Phillips J, Linehan WM, Kasten-Sportes C, et al. Human CTLs to wild-type and enhanced epitopes of a novel prostate and breast tumor-associated protein, TARP, lyse human breast cancer cells. Cancer Res. 2004;64:2610–2618. doi: 10.1158/0008-5472.can-03-2183. [DOI] [PubMed] [Google Scholar]
- • 46.Alves PM, Faure O, Graff-Dubois S, Cornet S, Bolonakis I, Gross DA, Miconnet I, Chouaib S, Fizazi K, Soria JC, et al. STEAP, a prostate tumor antigen, is a target of human CD8+ T cells. Cancer Immunol Immunother. 2006;55:1515–1523. doi: 10.1007/s00262-006-0165-3. These results confirm that the antigen STEAP can stimulate CD8 T cell responses against tumor cells. One of the epitopes reported lies closely to a CD4 T cell epitope described in reference [32] [DOI] [PMC free article] [PubMed] [Google Scholar]
- • 47.May RJ, Dao T, Pinilla-Ibarz J, Korontsvit T, Zakhaleva V, Zhang RH, Maslak P, Scheinberg DA. Peptide epitopes from the Wilms' tumor 1 oncoprotein stimulate CD4+ and CD8+ T cells that recognize and kill human malignant mesothelioma tumor cells. Clin Cancer Res. 2007;13:4547–4555. doi: 10.1158/1078-0432.CCR-07-0708. Three CD4 T cell epitopes for TAA WT1 were identified where one of them WT1122–140 contains within its sequence a previously defined CD8 T cell epitope. This MHC-II epitope may be the same one described in reference [31]. [DOI] [PubMed] [Google Scholar]
- 48.Knutson KL, Schiffman K, Disis ML. Immunization with a HER-2/neu helper peptide vaccine generates HER-2/neu CD8 T-cell immunity in cancer patients. J Clin Invest. 2001;107:477–484. doi: 10.1172/JCI11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirai M, Pendleton CD, Ahlers J, Takeshita T, Newman M, Berzofsky JA. Helper-cytotoxic T lymphocyte (CTL) determinant linkage required for priming of anti-HIV CD8+ CTL in vivo with peptide vaccine constructs. J Immunol. 1994;152:549–556. [PubMed] [Google Scholar]
- • 50.Odunsi K, Qian F, Matsuzaki J, Mhawech-Fauceglia P, Andrews C, Hoffman EW, Pan L, Ritter G, Villella J, Thomas B, et al. Vaccination with an NY-ESO-1 peptide of HLA class I/II specificities induces integrated humoral and T cell responses in ovarian cancer. Proc Natl Acad Sci U S A. 2007;104:12837–12842. doi: 10.1073/pnas.0703342104. The authors describe a clinical trial in ovarian cancer using a peptide from the TAA NY-ESO-1 containing a CD4 and 2 CD8 T cell epitopes. T cell clones (CD4 and CD8) from vaccinated patients were able to recognize NY-ESO-1 expressing tumor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 51.Bijker MS, van den Eeden SJ, Franken KL, Melief CJ, Offringa R, van der Burg SH. CD8+ CTL priming by exact peptide epitopes in incomplete Freund's adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J Immunol. 2007;179:5033–5040. doi: 10.4049/jimmunol.179.8.5033. This paper demonstrates that vaccines using long synthetic peptides induce far superior immune responses than vaccines composed of the minimal CD8 T cell epitopes. The authors hypothesize that long peptides can only be presented by professional APCs, while the short, minimal epitopes may be presented by non-professional APCs, resulting in T cell anergy. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura T, Iwakabe K, Sekimoto M, Ohmi Y, Yahata T, Nakui M, Sato T, Habu S, Tashiro H, Sato M, et al. Distinct role of antigen-specific T helper type 1 (Th1) and Th2 cells in tumor eradication in vivo. J Exp Med. 1999;190:617–627. doi: 10.1084/jem.190.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 53.Garcia-Hernandez M, de L, Gray A, Hubby B, Kast WM. In vivo effects of vaccination with six-transmembrane epithelial antigen of the prostate: a candidate antigen for treating prostate cancer. Cancer Res. 2007;67:1344–1351. doi: 10.1158/0008-5472.CAN-06-2996. This reports shows remarkable anti-tumor effects of vaccination with the STEAP antigen against prostate tumors using a mouse model system, where both CD4 and CD8 T cells play a critical role. [DOI] [PubMed] [Google Scholar]
- •• 54.Melanoma Study Group of the Mayo Clinic Cancer Center, Celis E. Overlapping human leukocyte antigen class I/II binding peptide vaccine for the treatment of patients with stage IV melanoma: evidence of systemic immune dysfunction. Cancer. 2007;110:203–214. doi: 10.1002/cncr.22744. This paper describes the first clinical study using a synthetic peptide vaccine containing a naturally occurring combination of a CD4 T cell epitope and a CD8 T cell epitope. [DOI] [PubMed] [Google Scholar]
- 55.Lu J, Celis E. Recognition of prostate tumor cells by cytotoxic T lymphocytes specific for prostate-specific membrane antigen. Cancer Res. 2002;62:5807–5812. [PubMed] [Google Scholar]


