Abstract
A method for measuring the microclustering of a class of cell surface receptors called integrins is reported. Integrins are proteins involved in bi-directional signaling across the cell membrane, and are important in cell adhesion, growth and survival. Their activity is regulated by changes in protein conformation and protein clustering. The developed in vivo clustering assay uses fluorescence resonance energy transfer (FRET), and has the benefit of requiring a single cloning step to generate FRET donors and acceptors that can be used to measure the clustering of a series of integrin mutants. The FRET reporters contain extracellular donor or acceptor fluorescent protein attached to native integrin cytoplasmic and transmembrane domains, and these are expressed along with wild-type or mutant integrins. Expression of the FRET reporters has no affect on the ligand binding properties of co-expressed integrins. FRET values are calculated for cell lines spreading on ligand coated surfaces, and these values are independent of fluorescent protein expression. No FRET is observed in cell lines expressing the reporters in the absence of integrins. Integrin dependent FRET values increase approximately 2- to 3-fold when the integrins contain mutations that result in increased ligand binding affinities.
Introduction
The cell membrane contains a complex array of lipids, proteins, carbohydrates, and small molecules, which undergo dynamic organization into microscale and macroscale clusters. This lateral organization varies in the time scale, as well as the length scale, and is important in cell signaling, cell recognition and other basic cellular functions.1 Cell membrane proteins responsible for signal transduction into the cell (outside-in signaling) and into the extracellular environment (inside-out signaling) are termed receptors. The integrin family of cell membrane receptors is important to cell adhesion, mobility, growth, survival, proliferation and differentiation.2 Integrins are composed of non-covalently associated α and β subunits, and are highly conserved from invertebrates to mammals.
With their vital role in cell survival and growth, and their implication in some forms of cancer,3 it is important to understand how integrins' activities are regulated within the cell. Probes for measuring integrin ligand affinity exist.4-6 However, much less is known about the macroscale and microscale dynamics of integrin clusters within the cell membrane of living cells and tissues. Macroscale clusters are defined as those larger than the resolution of an optical microscope, about 200 nm, while microscale clusters cannot be observed using conventional fluorescence microscopy. Studying integrin dynamics requires an in vivo detection technique that minimally disturbs the cells or tissue being studied, which precludes many techniques that could provide microscale resolution, such as atomic force microscopy. Previous experimental designs for elucidating microscale integrin dynamics used fluorescent proteins attached onto the cytoplasmic domains of the integrin α subunit.7, 8 This allowed the use of fluorescence resonance energy transfer (FRET) or bioluminescence resonance energy transfer to study the microclustering properties of the receptors within the cell membrane. A similar experimental setup, with donor and acceptor fluorescent proteins attached to the cytoplasmic domains, has also been used to study the separation distance between the α and β cytoplasmic tails as the integrins undergo conformational changes.9 The attachment of fluorescent proteins onto the amino- or carboxy- terminus of membrane proteins for FRET assays has also been reported for other membrane proteins.10-12 These experiments proved fruitful in measuring the clustering properties of the membrane proteins, however these properties could be altered by direct attachment of the fluorescent reporter proteins. This could affect the interactions of the receptor with cytoplasmic components,13, 14 alter the conformation of the receptor,15, 16 or change its diffusion properties through the additional mass of approximately 26 kD (i.e. one-sixth to one-fourth the weight of an average integrin subunit). Finally, analyzing protein mutants with these experimental setups requires additional, time consuming, cloning steps to generate the FRET donor and acceptor constructs for each mutant. For example, a recent screen to generate mutant integrins with altered function generated over 50 mutants that may have altered clustering properties.17 To study the clustering properties of these 50 mutants with the existing methods would require 100 independent cloning steps to generate FRET donor and acceptor constructs.
We report the development of a FRET assay that requires a single cloning step to study the microclustering of a series of mutant integrin proteins in vivo. The assay is used to study the microclustering of Drosophila αPS2CβPS integrins in S2 cells. The effects of the FRET donor and acceptor on the properties of the integrins are studied, and the application of the developed assay to the study of integrin mutants with enhanced ligand binding affinities is reported. It is proposed that this method for studying protein microclusters can be developed for other membrane proteins and would greatly benefit our understanding of cell membrane protein dynamics.
Experimental
Cell Culture
Drosophila S2 cells were cultured as described previously.18 Cells were co-transfected with plasmids expressing wild-type or mutant αPS2C and βPS integrin subunits and FRET reporters or FRET controls. All constructs were under the regulation of the heat shock 70 inducible promoter, and were co-expressed with bacterial dihydrofolate reductase selectable marker as described previously.19 Cell lines with no integrins were transfected with only the FRET reporters or FRET controls and the selectable marker. Stably transformed cell lines were frozen in several aliquots. Cell lines were used for 2 months and then replaced with a freshly thawed aliquot due to a gradual decrease in expression of the FRET reporters. Removal of endogenous integrins was accomplished using RNA interference against the myospheroid gene (Drosophila βPS integrin) as reported previously.4
Protein Constructs
Wild-type or mutant integrin constructs have been described previously.17, 19, 20 The FRET donor was a monomeric variant of the yellow fluorescent protein, venus (λabs,max = 515 nm; λex,max= 528 nm),21 which was constructed using standard molecular biology techniques to generate a V207L mutation. The FRET acceptor was monomeric dsRED variant cherry22 (λabs,max = 587 nm; λex,max= 610 nm). FRET reporters and FRET controls were constructed by cloning the fluorescent protein into plasmids for the expression of the transmembrane and cytoplasmic domains of βPS (FRET reporters) or mouse mammary CD2 (FRET controls) using standard molecular biology techniques. A serine rich 16 amino acid linker peptide was added between the fluorescent proteins and the transmembrane domain. An incorporated signal sequence insured cell membrane expression of the FRET controls and reporters. Complete protein sequences for the FRET controls and reporters are provided in the supplemental information.
Expression/Affinity Assays
The levels of αPS2C expression were measured in each experiment and for every cell line. An aliquot containing 5 × 105 cells was centrifuged and resuspended in biotinylated αPS2C antibody (CF.2C7) in M3 medium + 12.5% fetal calf serum for 30 minutes on ice, followed by addition of phycoerythrin labeled streptavidin for 30 minutes on ice. Finally the cells were fixed with a 4% v/v formaldehyde solution in phosphate buffered saline, pH 7, and analyzed by flow cytometry for phycoerythrin fluorescence. The levels of YFP expression were measured for each experiment in a similar fashion using green fluorescent protein antibody (Sigma, also binds to venus) and phycoerythrin labeled secondary antibody. Integrin affinity for a soluble binding ligand mimetic, TWOW-1, was measured as previously described4 with the following modifications: (i) the cells were not treated with collagenase/dispase prior to the start of the assay; (ii) the secondary antibody was phycoerythrin labeled goat anti-mouse IgG; (iii) MnCl2 was not used.
Preparation of tiggrin surfaces
Glass microscope slides (Carlson Scientific, Peotone, IL) were sterilized in 70% ethanol for 15 minutes and allowed to air dry in a sterile environment. They were coated with a sterile solution containing 0.5 μg/ml RBB tiggrin23 in phosphate buffered saline, pH 7 for 2 hours. The tiggrin solution was then removed, the slides were rinsed, and the remaining exposed glass surface was blocked using a 10 mg/ml solution of bovine serum albumin in phosphate buffered saline, pH 7. They were stored overnight at 2°C and used within 24 hours.
FRET assay
Cells were given fresh medium 1 to 2 days before each experiment. Cells were heat shocked for 30 minutes at 36°C to induce the expression of integrins and FRET reporters or controls and allowed to recover for 4 hours at 25°C. The cells were counted, pelleted by centrifugation and resuspended in serum free medium at a density of 3 × 105 cells/ml. The cells were placed on a tiggrin coated substrate and allowed to spread for 1 hour. The medium was removed from the surface and replaced with 20 mM BES Tyrodes buffer (137 mM NaCl, 2.9 mM KCl, 1% w/v glucose, 10 μM CaCl2, 1 mM MgCl2) to reduce the amount of background fluorescence generated by the medium. The cells were immediately analyzed using a Ziess Universal microscope with mercury lamp excitation and a Zeiss Axio Cam MRm camera and Zeiss AxioVision 4 software. Three images were captured for each cell that was analyzed: (i) venus filter set (excitation 485 nm band pass, emission 515 – 565 nm), (ii) cherry filter set (excitation 546 nm band pass, emission 590 nm long pass), and (iii) venus excitation filter with cherry emission filter (FRET filters). The exposure time for the cherry image was 2 seconds, and for the FRET and venus images was 10 seconds. The camera gain was set to 2. The S2 cells did not move in the time it took to capture the images. Once the medium was replaced with the BES Tyrodes buffer, all analysis was completed within one hour to ensure cell viability.
Digestion of surface proteins
For experiments involving protease digestion of cell surface proteins, the heat shock step in the FRET assay was replaced by treating 1 × 106 cells in 1 mg/ml dispase/collagenase (Roche Applied Science) in Robb's saline lacking divalent cations (1.2 mM phosphate buffer, pH 6.75, 26 mM NaCl, 20 mM KCl, 5 mM glucose, 50 mM sucrose) at 36°C for 30 minutes. This treatment also served to induce the expression of the integrins and FRET reporters or controls.
Analysis
Data analysis was performed using the program ImageJ 1.34s (NIH). Reported FRET values were calculated as described previously using the following equation24:
| (1) |
Where IDA, IAA, and IDD are the images obtained with the FRET filters, cherry filters, and venus filters, respectively. The constants a, b, c, and d account for bleed through of: cherry into the FRET filters, cherry into the venus filters, venus into the cherry filters, and venus into the FRET filters, respectively. These values were measured in cell lines that contained only the cherry FRET control plus wild type integrins (a and b) or venus FRET control plus wild type integrins (c and d) and were found for the conditions used in this study to be: a = 0.248 ± 0.090; b = 0.084 ± 0.095; c = 0.049 ± 0.056; d = 0.148 ± 0.070. All reported FRET values in the text have been normalized to the values obtained for the cell line expressing wild-type integrins and the FRET reporters. All values represent the average of 3 replicate experiments, and reported errors are ± one standard deviation. For the FRET assays at least 100 cells were analyzed, and for the flow cytometry experiments 30,000 cells were analyzed.
Results and Discussion
A FRET assay was developed to measure the microclustering of wild-type and mutant integrins in vivo. A schematic showing the general assay format is shown in Figure 1. The use of fluorescent proteins as the FRET donor and FRET acceptor satisfy the requirement for in vivo studies. The FRET donor was a monomeric yellow fluorescent protein variant, mvenus. The FRET acceptor was the monomeric version of dsRED, cherry. In order to avoid separate cloning steps for each mutant protein to be studied (i.e. to avoid attaching the FRET donor or FRET acceptor onto each mutant integrin), a general FRET donor and acceptor was developed that can cluster with wild-type or mutant integrins. The developed assay uses donor fluorescent protein and acceptor fluorescent protein cloned onto the transmembrane and cytoplasmic domains of the βPS integrin subunit (TCD-β). Hereon the use of the phrase FRET reporter indicates a population of TCD-β containing the donor fluorescent protein and a separate population of TCD-β containing the acceptor fluorescent protein. It has been found that TCD-β in the absence of fluorescent protein localizes in macroscale clusters with integrins when coexpressed in vivo.25 It has also been shown that the FRET reporters are observed in macroscale integrin clusters in Drosophila S2 cells (see supplemental information). Therefore it was postulated that the FRET reporters would exhibit microscale clustering with the integrins. An additional benefit to the proposed FRET assay is that it does not require any alterations to the integrins, such as attaching the donor or acceptor onto the amino- or carboxy- terminus, which could have an effect on integrin function and integrin-protein interactions. Finally, the FRET reporters do not contain ligand binding domains, which would compete with the integrins for ligand.
Figure 1.
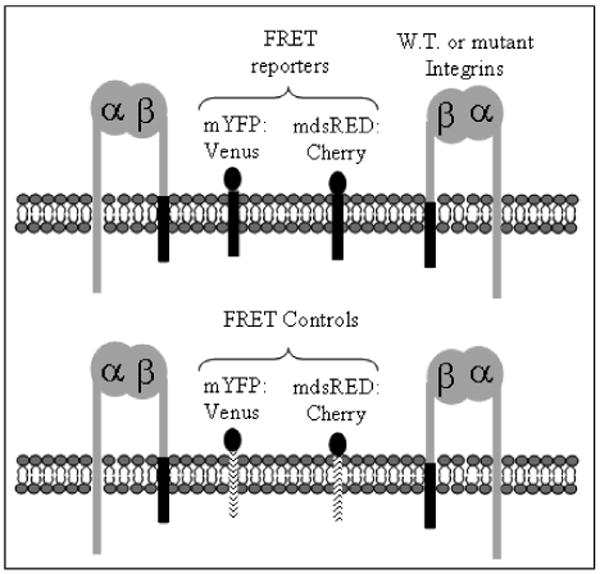
Schematic of the FRET assay used to study integrin microclustering. Integrins were expressed along with FRET reporters containing fluorescent proteins fused to the transmembrane and cytoplasmic domains of the βPS integrin subunit, dark line (top) or FRET controls containing fluorescent proteins fused to the transmembrane and cytoplasmic domains of CD2, dotted line (bottom). A monomeric YFP variant and monomeric dsRED variant were used as the donor and acceptor, respectively.
A set of FRET controls was also developed which were expected to be less effective at microscale clustering with integrins based on macroscale clustering data. The FRET controls contain the transmembrane and cytoplasmic domain of mouse mammary CD2 (TCD-CD2), a cell surface receptor that has no homolog in the Drosophila proteome. It has been shown that TCD-CD2 is not as effective as TCD-β in macroscale clustering with integrins in vivo.25 The FRET reporters or FRET controls were co-expressed with wild-type or mutant αPS2CβPS integrins.
The FRET reporters do not affect the integrins' ligand binding affinity. One ligand for αPS2CβPS integrins is an extracellular matrix protein tiggrin, and TWOW-1 is a monomeric soluble binding ligand mimetic for αPS2CβPS integrins. The results shown in Table 1 were obtained using flow cytometry to measure TWOW-1 binding to seven cell lines expressing wild-type or mutant integrins with or without FRET reporters or controls. All the results are normalized to the cell line expressing wild-type integrins in the absence of FRET reporters or FRET controls (αβ). The cell line expressing wild-type integrins and the FRET reporters (TCD-β; αβ) shows essentially no change in TWOW-1 binding compared to the cell line expressing only the wild-type integrins. The FRET controls also show no affect on the binding of wild-type integrins to TWOW-1 (TCD-CD2; αβ). There is minimal change in TWOW-1 binding for cell lines with and without the FRET reporters when the integrins have an α cytoplasmic mutation (GFFNR>GFANA) with a higher affinity for ligand (αanaβ), or when the integrins have a β mutation (V409D) with a higher affinity for ligand (αβB58). This indicates that the FRET reporters and controls can be used in the FRET assay to study integrin microclustering without affecting the integrins' affinity for ligand, which has been predicted to have an effect on the integrins' clustering properties.26
Table 1.
Normalized binding of the αPS2CβPS integrin soluble binding ligand mimetic (TWOW-1) for the indicated seven cell lines.
| Cell Line | Normalized TWOW-1 Binding |
|---|---|
| αβ | 1.0 ± 0.1 |
| TCD-β; αβ | 1.1 ± 0.1 |
| TCD-CD2; αβ | 1.0 ± 0.1 |
| αanaβ | 2.3 ± 0.3 |
| TCD-β; αanaβ | 2.3 ± 0.1 |
| αβB58 | 2.3 ± 0.1 |
| TCD-β; αβB58 | 2.6 ± 0.3 |
Table 2 shows the calculated FRET results obtained for three cell lines containing the FRET reporters or controls. All results have been normalized to the results obtained for the cell line expressing the FRET reporters and wild-type integrins (TCD-β; αβ), and correspond to an average value obtained from 100 to 250 cells. The calculated FRET value for each cell was obtained using the fluorescence images corresponding to the three filter configurations described in the experimental section and equation 1. A FRET value was obtained for each cell by averaging over every pixel in the image except those pixels corresponding to the nuclear and perinuclear regions, which were rejected in the measurement. Cell membrane proteins are processed in the endoplasmic reticulum, which surrounds the nucleus. A large fluorescence signal was observed in the perinuclear region that corresponds to FRET reporters that are not in the cell membrane, and this interferes with the measurement of integrin clustering in the cell membrane. To remove this interference from the measurement, the fluorescence from the nuclear and perinuclear regions were avoided. It is important to note that a solution based fluorescence measurement, such as flow cytometry, would not be able to reject the internal fluorescence from the cell membrane fluorescence, and would not be useful when measuring the clustering of membrane proteins labeled with fluorescent proteins. FRET was measured on cells that were spread to at least 30 μm in diameter, as the nuclear region could be rejected in these cells. An S2 cell in solution has a diameter of approximately 10 μm.
Table 2.
Normalized FRET results for cells containing FRET reporters (TCD-β) or FRET controls (TCD-CD2) and wild-type integrins (αβ) or no integrins.
| Cell Line | Normalized FRET |
|---|---|
| TCD-β; αβ | 1.00 ± 0.07 |
| TCD-CD2; αβ | 0.52 ± 0.01 |
| TCD-β; none | 0.00 ± 0.00 |
Energy transfer is observed for cells containing wild-type integrins and FRET reporters (TCD-β; αβ), while cells expressing FRET reporters and no integrins (TCD-β; none) showed no measurable energy transfer (Table 2). This result indicates that the clustering of the FRET reporters in cell lines containing integrins is a result of the integrins and not the FRET reporters themselves. The cell line expressing the wild-type integrins with the FRET controls (TCD-CD2; αβ) shows a 50% reduction in the calculated FRET value compared to the cell line containing the wild-type integrins and the FRET reporters (TCD-β; αβ). This confirms the macroscale results that the CD2 transmembrane and cytoplasmic domains are not as effective at clustering with the integrins as the FRET reporters.25 The Förster radius for YFP and dsRED FRET pairs is 3.14.27 If the donor and acceptor are separated by more than 5 nm, there is less than 5% efficiency in energy transfer to the acceptor. Integrin dependent spreading of S2 cells results in the FRET reporters being within 5 nm of each other, using the reasonable assumption that transfer efficiencies below 5% cannot be detected with our camera system. Considering energy transfer is measured only in the presence of integrins and the integrin size,28, 29 it is very likely the FRET being measured using the reporters results from integrin-integrin associations.
To check for macroscale integrin clustering, cells were fixed with formaldehyde and stained with an antibody against the βPS integrin subunit after completion of the FRET analysis on live cells. No significant differences in macroscale integrin clustering were observed for the cells utilized in this study (supplemental information). In the absence of macroscale differences, the most likely explanation for the FRET results is that they are reporting on differences in microscale clustering.
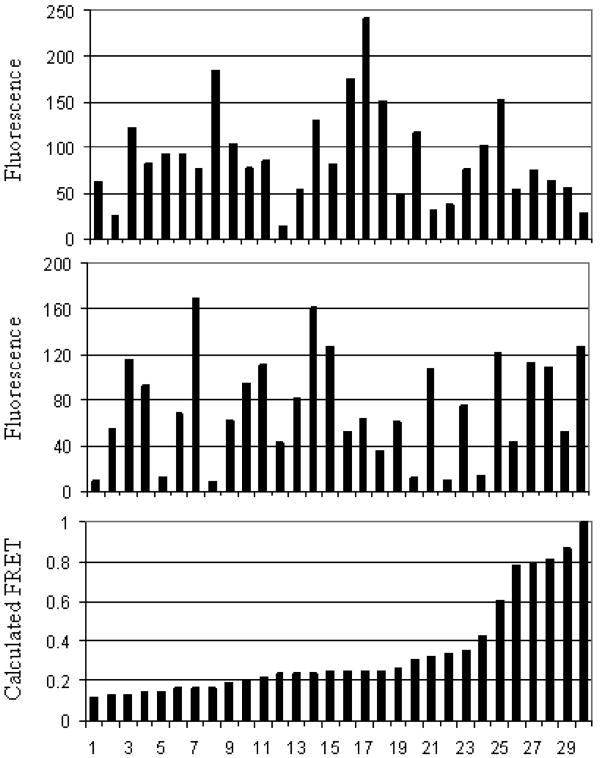
For each experiment and cell line, the average integrin expression and average fluorescent protein expression were measured using αPS2C or venus antibodies, respectively. Flow cytometry was used to obtain an average expression level, and staining was performed in a surface sensitive manner (i.e. protein within the cell was not measured). Shown in Table 3 are the results for three replicate experiments of αPS2C antibody and YFP antibody binding to the cell lines utilized in this study. There are no differences in the expression of αPS2C within the error of the measurement. There is however, considerable variation in the expression of fluorescent protein from one cell line to the next and from cell to cell within the same cell line. This result was confirmed using fluorescence microscopy, where the control cell line (TCD-CD2; αβ) had the highest fluorescence and the cell line with FRET reporters but no integrins (TCD-β; none) had the lowest fluorescence. Shown in Figure 2 is a subset of the data collected from the TCD-β; αβ cell line to determine whether differences in donor and acceptor expression levels affected the calculated FRET values. The cherry fluorescence (top, minus the background signal) and venus fluorescence (middle, minus the background signal) were measured for a number of cells and compared to the calculated FRET value (bottom). The results indicated that there is no correlation between the measured fluorescence and the calculated FRET value, and that differences in fluorescent protein expression do not preclude a comparison of the FRET values obtained for the different cell lines. The results presented in Figure 2 were representative of all the cell lines used in this study (results not shown).
Table 3.
Normalized binding of αPS2c antibody and YFP antibody to the indicated five cell lines.
| Cell Line | Binding of αPS2c antibody | Binding of YFP antibody |
|---|---|---|
| TCD-β; αβ | 1.0 ± 0.3 | 1.0 ± 1.0 |
| TCD-CD2; αβ | 0.7 ± 0.2 | 1.7 ± 0.3 |
| TCD-β; none | 0.1 ± 0.1 | 0.5 ± 0.1 |
| TCD-β; αanaβ | 0.8 ± 0.2 | 1.3 ± 0.8 |
| TCD-β; αβB58 | 1.7 ± 0.7 | 1.0 ± 0.2 |
Figure 2.
(Top) mdsRED fluorescence intensity; (Middle) mYFP fluorescence intensity; (Bottom) calculated FRET value for the cell line TCD-β; αβ. Each data point in a column represents the same cell. Background signals were measured by averaging pixel values where no cells were present, and this value was subtracted from the mdsRED and mYFP data (top and middle). Data was taken from a larger set containing 137 cells.
Mutations that increase the integrins' affinity for monovalent ligand may also result in integrin clustering. The FRET assay was used to measure integrin microclustering for two cell lines containing mutations that have no effect on macroscale integrin clustering compared to wild-type integrins (see supplemental information), but have increased ligand binding affinities. The αanaβ integrins contain an α cytoplasmic mutation that alters the integrins conformation in a manner that is thought to mimic inside-out signaling.4 There is nearly a 2-fold increase in the FRET value for αanaβ integrins compared to wild-type integrins, indicating that there is increased microclustering associated with this mutation (Table 4). The αβB58 integrins contain an extracellular mutation in the β subunit which is thought to drive the integrins' conformation to one that is potentially relevant in outside-in signaling.4 This mutation is associated with an approximately 3-fold higher level of FRET than wild-type integrins, indicating these integrins have increased microclustering compared to both wild-type and αanaβ integrins. This FRET value is a lower limit for αβB58 integrins. S2 cells express endogenous βPS subunits, therefore the reported FRET value was a measure of both αβB58 and αβ microclustering. The results shown in Table 1 indicate that αβB58 integrins are being expressed in these cells since they have a higher affinity for TWOW-1 than cells expressing only wild-type integrins.
Table 4.
Normalized FRET results for cells containing FRET reporters (TCD-β) and the indicated integrins.
| Cell Line | Normalized FRET | Normalized FRET
Treatment† |
|---|---|---|
| TCD-β; αβ | 1.00 ± 0.07 | 1.18 ± 0.8 |
| TCD-β; αanaβ | 1.90 ± 0.50 | 2.31 ± 0.01 |
| TCD-β; αβB58 | 3.19 ± 1.07 | 0.74 ± 0.30 |
Treatment included removal of endogenous integrins through RNA interference against the myospheroid gene and protease treatment of cell surface proteins prior to inducing expression of the indicated integrins and FRET reporters.
The results presented here highlight how the developed FRET assay can be used to study integrin-integrin interactions that are not observed using standard fluorescence microscopy. Coupling this technique with well characterized protein mutants allows elucidation of the molecular mechanism of integrin clustering. Our results indicate that both the outside-in signaling conformation induced by the αβB58 mutation and the inside-out signaling conformation induced by the αanaβ mutation increase integrin-integrin interactions since cells expressing these integrins have increased FRET values compared to cells expressing wild-type integrins. Characterization of more mutants is needed to confirm and extend these preliminary results.
The influence of other membrane proteins in integrin microclustering can also be assessed using the FRET methods described here. Treatment of the cell surface with proteases prior to inducing expression of the FRET reporters and the integrins can be used to test whether other membrane proteins enhance or reduce integrin microclustering. The FRET reporters and integrins are re-expressed quickly and at high levels after surface digestion since they are under the control of an inducible promoter, while expression of other potential interacting proteins is reduced or eliminated during the 5 hour experiment. It has been previously demonstrated that protease treatment does not inhibit ligand binding to wild-type or mutant integrins.4 There is no significant change in the calculated FRET value for cell lines expressing wild-type (1.18 ± 0.80 and 1.00 ± 0.07) and αanaβ (1.90 ± 0.50 and 2.31 ± 0.01) integrins with and without the digestion of other cell surface proteins (Table 4). There is, however, a significant decrease in the FRET value for the cell line expressing αβB58 integrins. The calculated FRET value for cells expressing αβB58 integrins was 3.19 ± 1.07 when other membrane proteins were present, and only 0.74 ± 0.30 after protease treatment of the cell surface. These results indicate that there are other membrane proteins that are required for the increase in integrin microclustering observed with this mutation and not the αanaβ mutation. It could be that other membrane proteins interact directly with the αβB58 conformation to induce clustering, while only cytoplasmic proteins interact with the αanaβ mutation to induce increased integrin microclustering compared to wild-type integrins.
Conclusions
Fluorescent proteins attached to the integrin transmembrane/cytoplasmic domains can be used to measure the microscale clustering of integrins in live cells using FRET. FRET reporters generated with a single cloning step can be used to study many mutant proteins. For example, cloning the FRET reporters can be completed in 1 week and can be used to screen 100s of mutants. If the reporters were cloned directly onto the protein being studied, 200 cloning steps would be required to study 100 mutants. Considerable time and money can be saved using the methods reported here. Future work is aimed at generating FRET reporters that can be used to study the microscale clustering of other classes of cell membrane proteins. This format is expected to be generally applicable, with suitable substitution of the cytoplasmic and transmembrane domains because these domains are important in the clustering of a number of cell membrane proteins.30-33 Such studies could deepen our understanding of cell membrane protein dynamics.
Supplementary Material
Complete protein sequences of FRET reporters and FRET controls; confocal microscope images showing macroscale co-localization of FRET reporters and integrins; wide-field microscope images showing lack of integrin macroclusters for all cell lines used in this study.
Acknowledgments
This work was supported by a National Institutes of Health grant (R01 GM42474). E.A.S. was supported by a National Institutes of Health Training Grant (F32 GM073467) and NIH Cancer Biology Training Grant (T32 CA09213). The authors thank Barb Carolus and Debbie Sakiestewa of the Arizona Research Laboratories Cytometry Service for assistance, Tim Kendall for assistance in preparing plasmids, Dr. Joyce Schroeder and colleagues for providing mouse mammary cDNA, Roger Tsien (Howard Hughes Medical Institute, La Jolla, CA) for dsRED-mCherry plasmid, and Atsushi Miyawaki (Riken, Wako-city, Saitama, Japan) for YFP-Venus plasmid.
References
- 1.Vereb G, Szollosi J, Matko J, Nagy P, Farkas T, Vigh L, Matyus L, Waldmann TA, Damjanovich S. Natl Acad Sci USA. 2003;100:8053–8058. doi: 10.1073/pnas.1332550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 3.Jin H, Varner J. British J of Cancer. 2004;90:561–565. doi: 10.1038/sj.bjc.6601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunch TA, Helsten TL, Kendall TL, Shirahatti N, Mahadevan D, Shattil SJ, Brower DL. J Biol Chem. 2006;281:5050–5057. doi: 10.1074/jbc.M508550200. [DOI] [PubMed] [Google Scholar]
- 5.Shattil SJ, Hoxie JA, Cunningham M, Brass LF. J Biol Chem. 1985;260:11107–11114. [PubMed] [Google Scholar]
- 6.Pampori AL, Hato T, Stupack DGAS, Cheresh DA, Nemerow GR, Shattil SJ. J Biol Chem. 1999;274:21609–21616. doi: 10.1074/jbc.274.31.21609. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Carman CV, TYang W, Salas A, Springer TA. J Cell Biol. 2004;167:1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buensuceso C, de Virgilio M, Shattil SJ. J Biol Chem. 2003;278:15217–15224. doi: 10.1074/jbc.M213234200. [DOI] [PubMed] [Google Scholar]
- 9.Kim M, Carman CV, Springer TA. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 10.Overton MC, Blumer KJ. Methods. 2002;27:324–332. doi: 10.1016/s1046-2023(02)00090-7. [DOI] [PubMed] [Google Scholar]
- 11.Eidne KA, Kroeger KM, Hanyaloglu AC. TRENDS in Endocrinology & Metabolism. 2002;13:415–421. doi: 10.1016/s1043-2760(02)00669-0. [DOI] [PubMed] [Google Scholar]
- 12.Zal T, Zal MA, Gascoigne NR. J Immunity. 2002;16:521–534. doi: 10.1016/s1074-7613(02)00301-1. [DOI] [PubMed] [Google Scholar]
- 13.Alahari SK, Lee JW, Juliano RL. J Cell Biol. 2000;151:1141–1154. doi: 10.1083/jcb.151.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tohyama Y, Katagiri K, Pardi R, Lu C, Springer TA, Kinashi T. Mol Biol Cell. 2003;14:2570–2582. doi: 10.1091/mbc.E02-09-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Femino AM, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 16.Lu C, Takagi J, Springer TA. J Biol Chem. 2001;276:14642–14648. doi: 10.1074/jbc.M100600200. [DOI] [PubMed] [Google Scholar]
- 17.Jannuzi AL, Bunch TA, West RF, Brower DL. Mol Biol Cell. 2004;15:3829–3840. doi: 10.1091/mbc.E04-02-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunch TA, Grinblat Y, Goldstein LSB. Nucleic Acids Research. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jannuzi AL, Bunch TA, Brabant MC, Miller SW, Mukai L, Zavortink M, Brower DL. Mol Biol Cell. 2002;13:1352–1365. doi: 10.1091/mbc.01-08-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunch TA, Brower DL. Development. 1992;116:239–247. doi: 10.1242/dev.116.1.239. [DOI] [PubMed] [Google Scholar]
- 21.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. Nature Biotech. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 22.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Nature Biotech. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 23.Graner MW, Bunch TA, Baumgartner S, Kerschen A, Brower DL. J Biol Chem. 1998;273:18235–18241. doi: 10.1074/jbc.273.29.18235. [DOI] [PubMed] [Google Scholar]
- 24.Xia Z, Liu Y. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Bermudo MD, Brown NH. J Cell Biol. 1996;134:217–226. doi: 10.1083/jcb.134.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carman CV, Springer TA. Curr Opin Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Patterson GH, Piston DW, Barisas BG. Anal Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 28.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker SL, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 30.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, Eisenhaber F. TRENDS in Biochem Sci. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 31.Brand T. Cell Biochem Biophys. 2005;43:95–103. doi: 10.1385/CBB:43:1:095. [DOI] [PubMed] [Google Scholar]
- 32.Sulistijo ES, MacKenzie KR. J Mol Bio. 2006;364:974–990. doi: 10.1016/j.jmb.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 33.Volkmer T, Becker C, Prodohl A, Finger C, Schneider D. Biochem Biophys Acta. 2006;1758:1815–1822. doi: 10.1016/j.bbamem.2006.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete protein sequences of FRET reporters and FRET controls; confocal microscope images showing macroscale co-localization of FRET reporters and integrins; wide-field microscope images showing lack of integrin macroclusters for all cell lines used in this study.