Introduction
Diatoms are unicellular heterokont algae known most notably for their elaborately ornamented cell walls of opaline silica. The scanning electron microscope (SEM) revolutionized diatom systematics, revealing taxonomically important ultrastructural features that were otherwise hidden from the light microscope. The SEM quickly became an important tool for taxon delimitation, and SEM data are now foundational to nearly all levels of the diatom classification system (Round et al. 1990). The introduction of multivariate statistical methods to diatom studies represented a similar advance, revealing complex sets of continuous morphometric characters to taxonomists for the first time. Although less commonly employed than the SEM, morphometric analysis has facilitated the discovery of numerous previously undescribed taxa (e.g., Mann et al. 2004; Theriot and Stoermer 1984). Most recently, the application of molecular biological techniques to systematic studies of diatoms has revealed still more variation, and these methods now play an important role in the discovery and delimitation of diatom species. Each of these tools enhanced our ability to detect and extract subtle differences, and in each case the diatom species category became effectively smaller and more exclusive. This is evident in the increasingly common discovery of "cryptic" species—morphologically similar but genetically distinct species that, at some point, shared the same specific epithet. In some cases, DNA sequence data have indeed provided compelling evidence for the existence of cryptic species, in which the rates of morphological and molecular evolution have apparently been decoupled. In other cases, DNA sequence data retrained the eye to reveal subtle morphological differences, so there was corroborating evidence from morphology and molecules to differentiate multiple species from what previously had been considered one. These species are sometimes referred to as "semi-" or "pseudo-cryptic," but the term "previously undetected" seems to apply equally well.
The power of the molecular systematic approach to species research, though fully established (Avise 2000), is just being realized for diatoms. This article focuses on the burgeoning use of molecular data to discover and delimit diatom species. The reader is referred to Mann (1999) for valuable historical context and a more thorough account of the traditional approaches to diatom taxonomy. This review instead will focus on studies that use phylogenetic analysis of DNA sequences to identify and delimit species of diatoms. The approach has a proven history of success, particularly in animal systems. A simple glance at the animal literature on species-level molecular systematics, and the closely related field of phylogeography, demonstrates the great potential of this approach to transform species research in diatoms. The molecular systematic approach is not without its challenges, however, as is clear from the general literature (e.g., Funk and Omland 2003) and the handful of diatom studies undertaken so far. This review will draw from those examples to highlight both the promises and problems of the molecular systematic approach to species delimitation. First, the extent to which common diatom species concepts influence or constrain species delimitation, and the advantages of adopting a lineage-based species concept, will be discussed. Second, different genetic markers can provide conflicting evidence about species boundaries, highlighting the importance of marker choice, particularly for single-locus studies. Several advantageous properties of mitochondrial DNA (mtDNA) for species-level studies are reviewed, which together recommend mtDNA as a better "first pass" molecular marker over the traditional nuclear ribosomal DNA (rDNA). Finally, the importance of species monophyly is discussed. To date, most species-level molecular systematic studies of diatoms have focused on the detection of cryptic species, which are, with rare exceptions (Slapeta et al. 2006), very likely to be recently diverged and therefore unlikely to exhibit reciprocal monophyly in their gene trees. This underscores the need to develop and evaluate numerous low-copy nuclear markers for systematics research in diatoms.
Species Concepts in Diatom Systematics
A brief overview of species concepts is necessary to the extent that they have impacted molecular systematic studies of diatoms. As phylogenetic analysis of DNA sequences becomes commonplace in the delimitation of diatom species, the traditional species concept can be expanded to more fully incorporate phylogeny, which in turn should lead to a more natural species-level classification of diatoms. Diatom species traditionally are diagnosed and classified based on morphological features of the siliceous cell wall, whereby names are assigned to more-or-less discrete, morphologically similar phenotypes. As a result, diatom taxonomy often straddles the boundary between phylogenetics and phenetics, depending on whether the emphasis is placed on a species' unique features or, more vaguely, its overall dissimilarity to previously recognized species. Despite any conceptual shortcomings, the approach has been successful, resulting in descriptions of ≥25,000 species to date (according to the computerized database of verified diatom names at the California Academy of Sciences, E. Fourtanier and J.P. Kociolek, pers. comm., 23 January 2008). It is important to note that phylogenetic analysis applies equally well to both molecular and morphological characters, including the traditional qualitative and quantitative features of the diatom cell wall (e.g., Cox and Williams 2006; Edgar and Theriot 2004; Theriot 1992). Hopefully the recent infusion of phylogenetic analysis into molecular diatom studies will spill over into morphological studies as well.
The Biological Species Concept
Although most species boundaries are drawn from multiple lines of evidence, reproductive isolation is often considered the hallmark of diatom speciation, consistent with the biological species concept (BSC) (Coyne and Orr 2004; Mayr 2000). Although not universally embraced (Mishler and Theriot 2000; Williams and Reid 2006), the BSC is central to many species-level molecular systematic studies of diatoms (Amato et al. 2007; Behnke et al. 2004, 2005b, 2007; Evans et al. 2008; Sarno et al. 2005; Vanormelingen et al. 2007, 2008). Primacy of the BSC was evident in the suggestion that consistent character differences, both morphological and molecular, might lack "biological meaning" in the absence of corroborating evidence for reproductive isolation from in vitro (laboratory) breeding experiments (Amato et al. 2007). Since virtually every diatom species is inextricably linked to a morphological, character-based definition, candidates for mating experiments are chosen on the basis of their characters, which suggests some level of confidence in the character-based classification. Note also that classic heritability studies still provide an excellent way to explore the potential evolutionary importance of morphological characters in diatoms (Edgar and Theriot 2003; Wood et al. 1987). A positive trend in BSC-based diatom research is the increasing number of studies that cite direct evidence from mating experiments (Amato et al. 2007; Behnke et al. 2004; Mann et al. 2004; Vanormelingen et al. 2007, 2008), rather than indirect character-based evidence, to support species boundaries under the BSC. Still, data from mating experiments will always be wanting because it remains unclear how the realization of breeding potential under controlled, laboratory conditions extrapolates to what occurs in vivo (in nature). Genetic data can help bridge this gap by providing valuable clues about the reproductive history of natural populations (Beszteri et al. 2007; Evans et al. 2004; Rynearson and Armbrust 2005; Rynearson et al. 2006).
Several other criticisms of the BSC underscore its limitations for diatom taxonomy and systematics. For example, the BSC is relevant only for species that co-occur in nature and therefore have the real potential to interbreed naturally (Mayr 2000; Wiens 2004). Accordingly, in vitro breeding experiments involving sympatric populations are valuable (Amato et al. 2007; Behnke et al. 2004; Vanormelingen et al. 2008), whereas under the BSC, breeding experiments involving allopatric populations (e.g., Vanormelingen et al. 2007) are less valuable. There is, of course, ongoing disagreement about whether any diatom species are truly allopatric (Finlay et al. 2002; Vyverman et al. 2007). The resilient siliceous cell wall of diatoms affords the luxury of integrating fossil information into evolutionary hypotheses (Sims et al. 2006). Since the reproductive characteristics of fossil diatoms cannot be observed directly, the BSC is directly relevant for extant species only, which excludes a large proportion of the total diatom species diversity. Likewise, it is safe to assume that mating experiments will not be performed for the overwhelming majority of the ≥200,000 species thought to exist (Mann and Droop 1996). A sound species concept that could be applied more universally across diatoms would be preferable. Wheeler and Meier (2000) and Wiens (2004) provide further general critiques of the BSC, and Reid and Williams (2007) provide commentary on the BSC as it relates to the existence of cryptic diatom species.
To be sure, breeding experiments can provide valuable information about species boundaries in diatoms. Reproductive data should not, however, overturn morphological, molecular, and other evidence that might be indicative of speciation, nor is it necessary to hedge well supported, character-based species boundaries on pending evidence for reproductive isolation. If we shift towards considering species as evolutionary lineages, reproductive isolation becomes one of several lines of evidence for lineage separation, or in this case, speciation (de Queiroz 2007).
Towards a Lineage-Based Species Concept for Diatoms
Efforts to reconcile the multitude of competing species concepts consider the species as an evolutionary lineage—a single line of direct ancestor–descendant relationship, or a single branch on a phylogenetic tree (de Queiroz 2007; Wiens 2004). Under this paradigm, the species concept is differentiated from the evidence (e.g., reproductive isolation, autapomorphic features, reciprocal monophyly, etc.) used to diagnose the species itself (de Queiroz 2007). Species boundaries are drawn in light of compelling evidence for lineage separation (de Queiroz 2007), and the systematist must decide which and how much evidence is sufficient to conclude that two lineages have diverged to the point that they should be considered separate species. Regardless of the species concept, species delimitation will always require an element of subjectivity (Silva 2008). Multiple, congruent lines of evidence provide stronger support for lineage separation and will lead to the establishment of more robust species boundaries (de Queiroz 2007); that is, the species hypothesis is less likely to be rejected as new data become available. Note that in this context, congruence requires a phylogenetic approach to character interpretation (Patterson 1988). Under the lineage-based concept, no single piece of evidence is the sine qua non of speciation, which effectively dethrones reproductive isolation as the decisive criterion for species delimitation of diatoms (Amato et al. 2007; Mann 1999).
In practice, this shift would have minimal impact on the already common, multifaceted approach to species-level systematic studies of diatoms. Adoption of a lineage-based species concept will simply provide a clearer context for interpretations about the different kinds of commonly gathered evidence (e.g., morphological and molecular characters, the degree of reproductive isolation, physiological data, etc.). Current practice promotes the misconception that different kinds of evidence support competing species concepts, when in fact the distinction often has less to do with the nature of the organisms and their properties and more to do with how those properties (i.e., characters) are treated or interpreted. For example, the distinction between morphological and phylogenetic species concepts (e.g., Amato et al. 2007; Lundholm et al. 2006) is an artificial one, since both concepts are fundamentally based on character evidence. In practice, however, phylogenetic analysis of DNA sequences typically underlies species delimitation under a phylogenetic species concept (Amato et al. 2007; Lundholm et al. 2006), whereas morphological characters rarely are subject to phylogenetic analysis. So treated equally, any character—morphological or molecular— could support species delimitation under a phylogenetic species concept. Given their mutual reliance on character data, species delimitation under a morphological species concept is not conceptually different from species delimitation under a hypothetical "second-codon-position" species concept, which most would dismiss outright. Adoption of a lineage-based concept will help alleviate this confusion by providing a clearer conceptual framework for the integration of multiple lines of evidence into species delimitation, which is often one of the stated goals in molecular systematic studies of diatoms (Mann 1999; Sarno et al. 2005; Vanormelingen et al. 2007).
Molecular Data
Non-repetitive DNA sequences are the primary source of data for molecular systematic studies of diatoms at all taxonomic levels. Other sequences, particularly microsatellites, have proven extremely powerful for overall assessment of intraspecific variation (e.g., Evans et al. 2005) and fine-scale population-level differentiation (e.g., Rynearson and Armbrust 2004). Microsatellites can reveal variation that is undetectable by rDNA (Rynearson and Armbrust 2004) and in many cases might provide a more powerful means of diagnosing cryptic diversity and biogeographic patterns. Nevertheless, non-repetitive DNA studies dominate, and this trend is likely to continue well into the foreseeable future. One great advantage of non-repetitive DNA sequences is their amenability to phylogenetic analysis (Hillis et al. 1996). Theoretical advances and a growing number of analytical tools for phylogenetic analysis allow the practicing systematist to take a rigorous, hypothesis-driven approach to DNA-based species discovery. A welcome benefit of this approach is that phylogenetic criteria can be incorporated into the establishment of species boundaries, which should lead to a more natural, maximally informative species-level classification of diatoms.
Ribosomal DNA and its Limits
For various reasons, portions of the nuclear rDNA cistron remain the most widely sequenced markers for many organisms, including diatoms. First, a high copy number in the genome and availability of universal primers make PCR amplification and sequencing of rDNA relatively easy. Second, the presence of highly conserved regions and a growing number of rRNA secondary structure predictions can help guide multiple sequence alignment (e.g., Alverson et al. 2006; Amato et al. 2007; Behnke et al. 2004), whereas hypervariable regions provide a large number of phylogenetically informative characters. Third, the growing database of diatom rDNA sequences facilitates broad comparative analyses, at least for the more conserved regions. Finally, different portions of the rDNA cistron can resolve vastly different levels of phylogenetic relationships. For example, small subunit (SSU or 18S) rDNA is useful for reconstructing higher level relationships across the entire phylogeny of diatoms (e.g., Alverson et al. 2006; Sorhannus 2004), whereas large subunit (LSU or 28S) D1–D3 and internal transcribed spacer (ITS) regions can resolve species-and sometimes population-level relationships (e.g., Behnke et al. 2004; Beszteri et al. 2005b; Godhe et al. 2006; Vanormelingen et al. 2007; Vanormelingen et al. 2008). All in all, rDNA has clearly been instrumental in jump-starting the field of molecular systematics of diatoms, but it also has several important drawbacks that warrant re-examination of its future role in the field.
A single rDNA cistron is iterated hundreds to thousands of times in tandem within the genome, and rDNA loci are sometimes distributed sporadically among multiple chromosomes (Alvarez and Wendel 2003). Given sufficient time, concerted evolution should homogenize nucleotide polymorphisms that differentiate paralogous cistrons (Alvarez and Wendel 2003), but empirical studies have demonstrated that most diatoms harbor a substantial amount of intragenomic polymorphism along virtually the entire length of the rDNA cistron, including the SSU (Alverson and Kolnick 2005), LSU d1–d3 (Beszteri et al. 2005b, 2007; Kooistra et al. 2008), and ITS regions (Behnke et al. 2004; Beszteri et al. 2005b; Vanormelingen et al. 2007). Intragenomic rDNA polymorphism levels vary from less than one to greater than seven percent in diatoms (Alverson and Kolnick 2005; Behnke et al. 2004). High copy number combined with sometimes large intragenomic polymorphism levels make orthology virtually impossible to determine at the outset, the result being that rDNA has the strong potential to obscure species boundaries (Alvarez and Wendel 2003) and biodiversity estimates (Thornhill et al. 2007). Although conflicts between organismal trees and rDNA gene trees are commonly reported for vascular plants (Alvarez and Wendel 2003), no examples of such conflict have been reported for diatoms, which likely reflects the overall greater sequencing effort in plants to date. Another consequence of the high copy number of rDNA is the widespread occurrence of pseudogenes (Alvarez and Wendel 2003; Thornhill et al. 2007), which can complicate both alignment and phylogenetic inference. The presence of several highly divergent, phylogenetically anomalous ITS rDNA suggested that some Cyclotella meneghiniana strains probably contain ITS rDNA pseudogenes (Beszteri et al. 2005b).
It is clear that many of the advantages offered by rDNA are balanced by disadvantages that can render rDNA data difficult to analyze and interpret, particularly at the species-level. Although some regions evolve very rapidly and can therefore be useful for discriminating species-level relationships, in practice these regions are notoriously difficult to align and often must be excluded from phylogenetic analyses (Behnke et al. 2004; Lundholm et al. 2006). As an extreme example, just a fraction of the total ITS rDNA sites could be aligned for several closely related demes within Sellaphora pupula (Behnke et al. 2004), which later were recognized as distinct species (Mann et al. 2004). Consequently, although the database of ITS and LSU rDNA sequences for diatoms has grown quite large, the overwhelming majority of these sequences cannot be aligned outside of a very narrow phylogenetic range. The result is an expansive set of sequences of very limited use. Although no single gene and no one genetic compartment can guarantee accurate identification of species boundaries, other markers might prove to be more broadly useful, easier to analyze, and even more informative than rDNA. In the meantime, however, a number of laboratory safeguards and analytical considerations have been outlined to help ensure that rDNA data are used and interpreted as carefully as possible (Feliner and Rossello 2007).
The Case for Mitochondrial Markers
Many of the properties that make mitochondrial DNA the data of choice for discerning cryptic species and teasing apart fine-scale phylogeographic patterns in animals (Funk and Omland 2003) should also apply to diatoms. A brief summary of these properties, and some promising early data, recommend replacement of the traditional rDNA with mtDNA for single-gene and "first pass" species-level systematic studies of diatoms. Virtually all animal mitochondrial genomes share the same complement of protein-coding genes, and importantly, all of these genes are single-copy, which circumvents the myriad of problems that burden phylogenetic studies based on the large rDNA gene family. The fully sequenced mitochondrial genomes of Thalassiosira pseudonana (Armbrust et al. 2004) and Nitzschia leucosigma (E. Ruck, unpublished data) show that, as for animals, gene complement and gene copy-number is similarly conserved across diatoms, so in the absence of any compelling evidence for widespread mitochondrial heteroplasmy in diatoms, assumptions of orthology will be valid. Among the >30 protein-coding genes in diatom mitochondrial genomes, only cytochrome oxidase subunit I (cox1) and cytochrome b (cob) have been investigated for their phylogenetic utility, and results show that mtDNA can resolve multiple levels of phylogenetic relationship within diatoms. Ehara et al. (2000a) obtained cox1 sequences from a broad sample of diatoms and found overall congruence between their phylogenetic results and those based on the more traditional SSU rDNA sequences (Alverson and Theriot 2005). At the opposite extreme, Evans et al. (2007) found that cox1 sequences were more divergent than chloroplast rbcL sequences and easier to align than ITS rDNA sequences, demonstrating the utility of cox1 for resolving low-level, intra-and inter-specific relationships within Sellaphora. The cob gene shows similar promise for resolving species-level relationships. A study of two dinoflagellate species with identical rDNA (ITS, SSU, and partial LSU) showed 1.9% divergence in their mitochondrial cob sequences (Logares et al. 2007). Within diatoms, two Surirella strains with near-identical nuclear SSU and LSU d1–d2 rDNA (7/2172 sites = 0.4%) and chloroplast psbC and rbcL (3/2629 sites = 0.1%) show striking divergence in their mitochondrial cob (49/701 sites = 6.9%) and cox1 (35/632 sites = 5.5%) sequences. (E. Ruck, unpublished data). In addition to the strong resolving capacity of mtDNA, these studies also highlight one of the inherent, though understated, advantages of using protein-coding sequences for phylogenetic inference: alignment of protein-coding sequences is trivial, in most cases along the entire length of the gene. This saves time and allows most or all of the nucleotide positions to be considered, even for broadly inclusive analyses (Ehara et al. 2000a). In stark contrast, alignment of ITS rDNA sequences among closely related Sellaphora species was so difficult that phylogenetic analyses could not be expanded to include outgroups (Behnke et al. 2004).
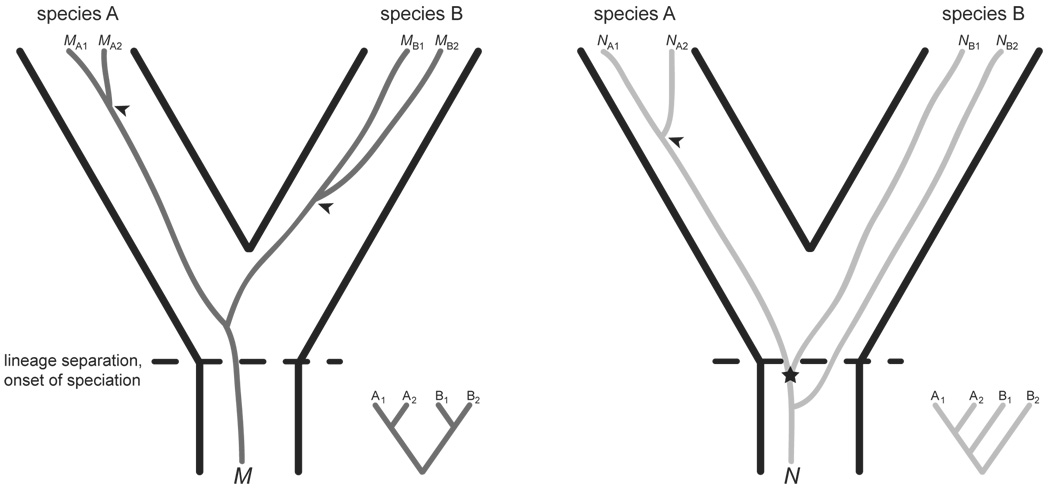
Beyond the practical advantages of mtDNA, its longstanding use in animal systems has motivated an extensive literature detailing the successes, challenges, expectations, and theoretical considerations for species delimitation based on mtDNA (Avila et al. 2006; Avise 2000, 2004; Funk and Omland 2003; Hudson and Coyne 2002; McGuire et al. 2007). In most cases, mtDNA is a uniparentally inherited, haploid genetic marker, which translates into a much lower overall effective population size (Ne) than that of nuclear markers (Funk and Omland 2003). As a result, coalescent times of organelle loci should generally be shorter, making mtDNA-based species delineation less vulnerable to the effects of incomplete lineage sorting, which often manifests as species-level paraphyly or polyphyly (Fig. 1; Funk and Omland 2003; Hudson and Coyne 2002; Hudson and Turelli 2003). The assumption of a reduced Ne for organelle loci should be validated empirically (Lynch et al. 2006), which for most diatoms will require the traditional approach of comparative sequencing to estimate the standing level of neutral variation in the genome as well as determination of the mode of mitochondrial inheritance, about which very little is currently known. The surprisingly frequent biparental inheritance of chloroplasts in Pseudo-nitzschia delicatissima suggests that cpDNA might not confer all of the advantages usually associated with organelle markers (Ghiron et al. 2008). Even uniparentally inherited organelle markers are not immune to the effects of incomplete lineage sorting, particularly for cases of very rapid or recent speciation, which are the same ones that often require sequence data to detect (Funk and Omland 2003). Furthermore, incomplete lineage sorting and introgression—the process by which different species hybridize and produce viable offspring, which subsequently backcross with the parental species—will have the same phylogenetic signature, making the two scenarios difficult (Funk and Omland 2003; Wendel and Doyle 1998) but not impossible (McGuire et al. 2007; Morando et al. 2004) to discern empirically. Finally, the comparatively shorter coalescent times for organelle loci can arguably lead to false confidence in species boundaries, since organelle loci can show reciprocal monophyly in the absence of any such evidence from the nuclear genome (Fig. 1; Hudson and Coyne 2002). These issues are discussed in more detail in the "Species Monophyly" section below.
Figure 1.
Conflicting evidence from two loci (M and N) about the monophyly of two species (A and B). In this example, each species has two different alleles (1 and 2) per locus. M and N might represent two unlinked autosomal loci, or one autosomal (N) and one organelle locus (M). In most cases, mitochondrial and chloroplast loci (e.g., M) will coalesce sooner than nuclear loci (e.g., N), which has important consequences for species delimitation from gene phylogenies. In this example, species A retains a single ancestral allelic lineage for both M and N, with all of the polymorphisms coalescing to a point that postdates the speciation event (marked by arrowheads). As a result, species A is monophyletic in both gene trees. The same scenario applies to the M locus for species B (also marked by arrowhead). Species B, however, retains two ancestral allelic lineages for N, one of which (NB1) shares a more recent common ancestor with alleles from species A (marked by a star). As a result, species B is paraphyletic in the N gene tree. Note that if allele NB1 was not sampled, species B would appear genetically distinct in both M and N gene trees. Figure concept adapted from Wendel and Doyle (1998) and Felsenstein (2004).
Although no universal primers exist for diatom mitochondrial genes, the available cox1 sequences span a broad enough phylogenetic range (Ehara et al. 2000a) that specific primers can be designed for any group of diatoms (Evans et al. 2007). The large and variably present group II intron in the cox1 gene requires a PCR strategy that either accommodates or avoids the intron altogether (Ehara et al. 2000a, b), but introns fortunately appear to be rare in other mitochondrial genes (Armbrust et al. 2004). Additional markers can be developed from the fully sequenced diatom (Armbrust et al. 2004) and brown algal (Oudot-Le Secq et al. 2006) mitochondrial genomes. This underscores the need for more diatom organelle genome sequences, which greatly facilitate the development of new phylogenetic markers (Timme et al. 2007).
In short, although not a panacea, mitochondrial DNA offers a large and virtually untapped reservoir of phylogenetic characters that are informative at multiple levels, including the species level and below. Mitochondrial genes are not encumbered by the same tenuous assumptions of orthology as rDNA, and unlike rDNA, multiple sequence alignment of protein-coding mtDNA is trivial, allowing for broadly inclusive comparative analyses. In addition, the mtDNA-based studies of phylogeography and speciation in animals provide a useful blueprint for parallel studies in diatoms. So all in all, mtDNA lends itself as an excellent molecular marker for species discovery in diatoms. Chloroplast DNA might offer similar advantages, but more work should be done to determine its mode inheritance across a broader sample of diatoms. Also, limited data suggest that cpDNA offers fewer informative characters than mtDNA (E. Ruck, unpublished data; Evans et al. 2007).
The Hunt for "Hidden" Species: Empirical Examples
The large amount of intraspecific variation revealed by allozyme electrophoresis studies showed early on that the tools of molecular biology offered much promise to species-level systematic studies of diatoms (Brand et al. 1981; Gallagher 1980, 1982; Murphy 1978; Murphy and Guillard 1976). Above all, the discovery of strong genetic differentiation between morphologically indistinguishable, seasonally isolated populations of Skeletonema costatum in Narragansett Bay (Rhode Island, USA) set the direction for much of the work that would follow (Gallagher 1980, 1982). Once considered an abundant, morphologically variable and widely distributed species (Hasle 1973), S. costatum has since been subdivided among seven additional species (Medlin et al. 1991; Sarno et al. 2005, 2007; Zingone et al. 2005), owing largely to the introduction of DNA sequence data to diatom systematics (Medlin et al. 1991). For Skeletonema, the DNA sequence data served a primary role in the species discovery process, revealing surprisingly high levels of structured genetic variation within S. costatum, which undoubtedly helped streamline the successful search for morphological features that differentiate the new species (Sarno et al. 2005, 2007). Some of the challenges of the molecular systematic approach were evident from the beginning, however. For example, although rDNA data have supported monophyly of S. pseudocostatum since its description, the initial split left S. costatum—still a taxonomic potpourri at that point— paraphyletic in rDNA gene trees (Medlin 1997; Medlin et al. 1991). In all cases, there was some initial evidence to suggest that each of the new species was morphologically distinct and monophyletic in rDNA phylogenies (Medlin et al. 1991; Sarno et al. 2005, 2007), but as intraspecific sampling efforts intensified, some of the species boundaries were fortified while others dissolved. Among the newly described species, S. ardens, S. grevillei, and (arguably) S. pseudocostatum continue to satisfy the very strict criterion of reciprocal monophyly in the most densely sampled rDNA phylogeny to date (Kooistra et al. 2008). In all other cases, either the focal species itself or some part of its sister taxon became paraphyletic with increased sampling, which greatly obscures species boundaries and makes it unclear which evidence, if any, supports the conclusion that paraphyletic species should nevertheless be considered "genetically distinct" (Kooistra et al. 2008). In the context of a phylogenetic study, it is unclear what "genetically distinct" might otherwise equate to, if not monophyly.
Pseudo-nitzschia has also been the focus of several species-level molecular systematic studies (Amato et al. 2007; Lundholm et al. 2002b, 2003, 2006; Lundholm and Moestrup 2002), motivated in part by the pressing need to understand which and how many species produce domoic acid, the cause of amnesic shellfish poisoning in humans (Bates et al. 1989). Pseudo-nitzschia poses a different set of challenges than Skeletonema, and accordingly, DNA sequence data have sometimes served a different purpose in these studies. Nitzschioid diatoms are notoriously difficult to identify and classify by traditional means alone, which is evident in their unstable genus-and species-level classifications (Lundholm et al. 2002a; Mann 1999). In several cases, therefore, species descriptions based primarily on morphology were augmented by phylogenetic analysis of rDNA sequences to help place the new species within the imperfect genus-level classification (Lundholm et al. 2002b; Lundholm and Moestrup 2002). In another study, rDNA sequences were considered alongside detailed morphological observations to differentiate two new species, P. decipiens and P. dolorosa, from the widespread and morphologically variable species, P. delicatissima (Lundholm et al. 2006). Further, more intensive population-level sampling of P. delicatissima from the Gulf of Naples showed as many as five distinct ITS rDNA lineages (Orsini et al. 2004). An expanded follow-up study based on rbcL and ITS rDNA sequences showed that populations of P. delicatissima and P. pseudodelicatissima in the Gulf of Naples consist of up to three and five phylogenetically distinct lineages, respectively (Amato et al. 2007). In one of the strongest tests of the BSC so far, in vitro mating experiments confirmed that these intraspecific lineages were reproductively isolated as well (Amato et al. 2007). Microsatellite and ITS rDNA studies have shown that two other Pseudo-nitzschia species, P. pungens and P. multiseries, harbor substantial amounts of genetic variation as well (Casteleyn et al. 2008; Evans et al. 2004, 2005).
Intraspecific genetic variation has been documented for several other common and geographically widespread diatom species. Molecular data have played a supporting role in the delineation of species boundaries within the Sellaphora pupula species complex, which consists of several well-characterized morphological and reproductive demes (now species—Mann et al. 2004) and is the model for BSC-based species research in diatoms (Behnke et al. 2004; Mann 1999). Behnke et al. (2004) used Sellaphora to test the general prediction that rDNA sequence divergence is correlated with reproductive isolation, and further, that compensatory nucleotide substitutions in rDNA helices correlate with reproductive isolation and speciation in algae (Coleman 2000; Medlin 1997). An overall correlation was found, which provided strong corroborating evidence for the existence of several distinct species lineages within S. pupula (Behnke et al. 2004). Species boundaries were further substantiated by phylogenetic analysis of nuclear SSU rDNA, mitochondrial cox1 and chloroplast rbcL sequences (Evans et al. 2007, 2008).
Another set of studies examined Cyclotella meneghiniana, an exemplar for the ubiquitous dispersal model of freshwater diatom diversity (Finlay et al. 2002). Beszteri et al. (2005b) followed up a detailed morphometric analysis of 20 sympatric strains by sequencing several portions of the rDNA cistron, which revealed evidence for as many as four distinct rDNA lineages, more than was evident from morphometric analysis alone (Beszteri et al 2005a, b). Expanded geographic sampling showed as many as eight genetic lineages within C. meneghiniana (Beszteri et al. 2007). These studies used evidence from DNA sequences and AFLP comparisons to support species separation under the BSC, but to date, none of the putative species have been formally split away from C. meneghiniana (Beszteri et al. 2005a, b).
Taken together, these studies highlight both the promises and challenges of the molecular systematic approach to species delimitation. Phylogenetic analysis of DNA sequence data is a powerful discriminatory tool that has led, either directly or indirectly, to the formal recognition of at least a dozen diatom species to date, mostly in cases where traditional taxonomy failed to provide similar resolution. Species boundaries that relied heavily on DNA sequence data have in some cases, however, shown a sobering level of phylogenetic instability as new data became available. It is constructive, therefore, to identify potential sources of instability, and to the extent possible, account for them in future studies. For example, given the potential problems outlined above, do the rDNA gene trees provide an accurate reflection of the underlying organismal phylogeny? Cloning of intragenomic ITS rDNA variants revealed that one of 20 C. meneghiniana strains examined contained divergent ITS rDNA types that spanned putative species boundaries suggested by LSU rDNA sequences (Beszteri et al. 2005b). In this case, incomplete, differential assortment of ancestral ITS rDNA polymorphism in the descendant lineages obscured species boundaries based on any single rDNA region alone. The confounding effects of rDNA will always be difficult to rule out completely, which by itself recommends its complete replacement with other, less ambiguous markers. Do chloroplast, mitochondrial, and other unlinked nuclear loci support the same species boundaries, or do different markers support different interpretations of species boundaries? Kooistra et al. (2008) noted that unpublished cpDNA phylogenies support monophyly for two of the three Skeletonema species that are paraphyletic in rDNA phylogenies. This result might, in fact, be expected given what is known about the very different population-genetic environments of nuclear and organelle loci. As discussed in more detail below, nuclear and organelle gene genealogies can provide very different perspectives on the underlying organismal phylogeny, especially for recently diverged species (Funk and Omland 2003; Hudson and Coyne 2002). Aside from the underlying history of the surrogate gene genealogies, the current disconnect between Skeletonema taxonomy and phylogeny might also be symptomatic of over-or under-classification at the species level (Kooistra et al. 2008). This, more than anything, reflects the propensity of different systematists to either retain inclusive, genetically diverse species or to subdivide the variation among smaller, genetically exclusive species lineages.
Species Delimitation—Where to Draw the Line?
Sequence Divergence, Ribosomal DNA, and the Diatom Species
Since the introduction of DNA sequences to diatom systematics, a diatom's SSU rDNA sequence has been tightly coupled to its species identity (Medlin 1997; Medlin et al. 1991; Sarno et al. 2005), so much so that identical SSU rDNA sequences are often considered de facto evidence that otherwise genetically distinct populations belong to the same species (Rynearson and Armbrust 2004, 2005). Although divergence in any gene provides positive evidence for lineage separation (see "Towards a Lineage-Based Species Concept for Diatoms" above), sequence divergence is a phenetic discriminator that by itself will not yield robust, phylogenetically informative species boundaries. It is also conceivable, if not likely, that incipient species lineages could separately evolve for a considerable period of time before showing any such evidence for separation in their SSU rDNA sequences. Rynearson and Armbrust (2004) proposed just such a scenario for three populations of Ditylum brightwellii that were highly differentiated by microsatellites but identical in their SSU rDNA. Sequence divergence is a particularly problematic measure for the SSU rDNA gene because of the high levels of intragenomic polymorphism known in diatoms. In some cases, the level of sequence divergence between paralogous SSU rDNA types within an individual can exceed the level of divergence between species (Alverson and Kolnick 2005).
Tied to the perception of a special ability of SSU rDNA to reflect species boundaries is the notion that compensatory base changes in rDNA helices also hold special importance for species delimitation of algae, including diatoms (Coleman 2000; Muller et al. 2007). Briefly, nucleotide pairings in rRNA helices are maintained despite evolutionary changes in the nucleotide sequences. Substitutions in one of the two base-paired nucleotides are often coordinated with a substitution at the second paired nucleotide (i.e., a compensatory base change) to maintain canonical pairing, which preserves the integrity of the helix. Some data suggest a general correlation between rDNA divergence and the degree of reproductive isolation between related species (Behnke et al. 2004; Coleman 2000; Coleman and Mai 1997; Muller et al. 2007), which has led to the bold claim that compensatory base changes in rDNA helices can predict reproductive compatibility and therefore define species boundaries under the BSC (Medlin 1997). As species lineages diverge, the expectation is that a multitude of traits will diverge at a rate proportional to the length of time the two lineages have been separately evolving. Any apparent correlation between the presence of compensatory base changes and the degree of reproductive isolation owes solely to the length of time that has elapsed since the two species diverged, not to any direct link between the two measures (Coleman 2000). Both provide independent measures of divergence and strengthen the case for recognition of the two lineages as distinct species under the lineage-based concept outlined above. de Queiroz (2007) provides a useful illustration of this principle in the context of species concepts.
Balancing Intraspecific Homogeneity and Heterogeneity
A fundamental question underlies species-level systematics, whether based on morphological or molecular characters: Given that species will always harbor some variation, how much intraspecific variation can exist before it should be formally partitioned between one or more new species? A recurring theme in diatom systematics is that a classification system should maximize the internal morphological and molecular homogeneity of its constituent taxa. For example, Kooistra et al. (2003) noted that the removal of Toxarium from Pennales had the advantage of increasing the overall morphological homogeneity of Pennales. Amato et al. (2007) concluded that rbcL was not a "perfect discriminator" of species because reproductively compatible strains of P. delicatissima did not share identical rbcL sequences (sequences differed at 4/1452 sites). Kooistra et al. (2008, p. 188) went further, suggesting that two species, S. costatum and S. grethae, that are paraphyletic in rDNA trees nevertheless can be considered genetically distinct species "because their intraspecific LSU rDNA sequence variation is small." This example illustrates that, applied as a criterion for classification, intraspecific homogeneity (or intrafamilial, intraordinal, etc.) will inevitably lead to a non-natural classification of diatoms. Ultimately, despite use of an explicit species concept or adoption of formal criteria (e.g., reciprocal monophyly) for separating species, all decisions to formally recognize new species will bear some degree of subjectivity (Silva 2008).
Species Monophyly
In most molecular systematic studies, DNA sequence data and phylogenetic analysis go hand-in-hand, with resulting gene phylogenies providing a surrogate measure of the organismal phylogeny. Extrapolating up from gene monophyly to species monophyly is safe provided that, within each species, all of the alleles at the study locus are monophyletic (Funk and Omland 2003). This can be a tenuous assumption when intraspecific taxon sampling is sparse, as evidenced from the Skeletonema species that initially appeared monophyletic but which increased taxon sampling revealed to be paraphyletic (Chen et al. 2007; Kooistra et al. 2008; Sarno et al. 2005, 2007). The problem is not unique to diatoms, of course. A literature survey found that fully 23% of >2000 animal species investigated showed species-level paraphyly or polyphyly (Funk and Omland 2003).
The consequences of inaccurate species delimitation and non-natural classification system are important, however, because the species unit underlies most applied diatom sciences (Kociolek 2005; Stoermer and Smol 1999) and general hypotheses about the distribution and diversity of diatoms worldwide (Finlay et al. 2002; Mann and Droop 1996; Pither 2007; Pither and Aarssen 2005a, b; Telford et al. 2006a, b). A paraphyletic species is not characterized by a discrete set of biological traits; that is, the traits of some individuals will more closely match those of another species than they do their own conspecifics, which share a more recent common ancestor. In short, paraphyletic species have no predictive value with respect to their biological attributes, including those of greatest interest to diatom biologists. So while we are encouraged to reinterpret the extensive body of taxonomic, experimental, and physiological literature surrounding S. costatum in light of the new classification (Kooistra et al. 2008; Sarno et al. 2005), it remains unclear how to do so given its current state. The mixture of paraphyletic and monophyletic Skeletonema species predicts that the phylogenetic distribution of physiological, morphological, and other biological traits will be equally mixed, as illustrated for the girdle band characters originally thought to distinguish S. marinoi and S. dohrnii, the latter of which grades into S. marinoi in densely sampled rDNA phylogenies (Chen et al. 2007; Ellegaard et al. 2008; Kooistra et al. 2008). Geographic distributions of paraphyletic species are equally difficult to interpret. Species paraphyly might, for example, account for some of the apparent disjunctions in the geographic distributions and temperature preferences of Skeletonema species (Kooistra et al. 2008). An alternative explanation is that Skeletonema, despite all of the taxonomic work to date, is still under-classified at the species level (Kooistra et al. 2008).
The consequences of species-level paraphyly are clear, but the transcendent challenge is to identify and understand the factors that cause species-level paraphyly so they can be accounted for in the design of future studies. Clearly multiple individuals (i.e., clonal culture strains) must be sampled from each species to provide a sufficiently strong test of monophyly for the study locus, and hence, the species. This is straightforward enough in the context of known or existing species boundaries but presents a greater challenge for studies of cryptic speciation, in which species boundaries are unknown or ambiguous. Numerous variables factor in the sampling design of species-level molecular systematic studies, so unfortunately there is no magic number of individuals beyond which one can safely assume species monophyly. This is particularly true for single-locus studies.
As species diverge over time, lineage sorting will transition their allelic lineages from an initial state of polyphyly, through paraphyly, to eventual reciprocal monophyly (Funk and Omland 2003; Wendel and Doyle 1998). At this point, shared ancestral polymorphisms have been "fully sorted" in the descendant species, and all standing polymorphisms coalesce to a point that postdates the speciation event (Fig. 1; Funk and Omland 2003; Wendel and Doyle 1998). The expectation, therefore, is that very recently diverged species will exhibit paraphyletic or polyphyletic gene trees (Funk and Omland 2003; Wendel and Doyle 1998). Incomplete lineage sorting as the cause of species-level paraphyly is difficult to demonstrate empirically, particular for phylogenies based on a single locus (Funk and Omland 2003; Wendel and Doyle 1998). Data from multiple, unlinked nuclear loci are necessary to determine the extent to which incomplete lineage sorting may or may not be obfuscating species boundaries (Felsenstein 2006; Hudson and Coyne 2002; Knowles and Carstens 2007). One potential advantage of uniparentally inherited mitochondrial (and chloroplast) loci is that the progression from polyphyly to reciprocal monophyly is considerably faster, so mtDNA has the potential to fully resolve recently diverged species lineages much sooner than autosomal loci (Hudson and Coyne 2002, but see "The Case for Mitochondrial Markers" above). This begs the question of whether two taxa that exhibit reciprocal monophyly at organelle loci but non-monophyly at some or all autosomal loci should be considered separate species lineages (Fig. 1; Hudson and Coyne 2002). A recent simulation study demonstrated the power of applying probabilistic models to this kind of problem, namely, how to resolve species boundaries in the face of widespread incomplete lineage sorting (Knowles and Carstens 2007). Using this approach, hypothetical species boundaries were accurately inferred even for the most recently diverged lineages, where the probability of observing reciprocal monophyly for any single locus, let alone multiple loci, was essentially zero (Knowles and Carstens 2007).
These issues highlight several future directions for species research in diatoms. For example, the progression from polyphyly to reciprocal monophyly is longer for species with larger Ne (Hudson and Coyne 2002), which might very well be the case for many diatoms. To my knowledge, Ne has not been measured for any diatom species, despite its potential importance for species delimitation. A single population bloom of D. brightwelii consisted of ≥2400 distinct clonal lineages, which suggests that Ne might be quite high for some species (Rynearson and Armbrust 2005). Effective population size can easily be estimated from the level of standing neutral variation (e.g., from substitutions at synonymous sites in protein-coding genes) in a population. The already large collections of Pseudo-nitzschia DNA samples and Skeletonema culture strains present good opportunities to estimate Ne, so we can begin to understand its impact on species-level systematic studies of diatoms. Finally, a large and growing body of evidence underscores that data from multiple, unlinked loci are necessary for accurate delineation of recently diverged species lineages (Felsenstein 2006; Hudson and Coyne 2002; Knowles and Carstens 2007). Development of these markers poses a critically important, though formidable, challenge for species research in diatoms. Several low-copy nuclear markers have been developed and successfully applied to a range of diatoms (Harper et al. 2005), but these might not be informative for species-level comparisons. Near-universal primers are available for diatom silicon transporter genes (Thamatrakoln et al. 2006), which do resolve closely related species (Alverson 2007). The increasing number of diatom EST and whole-genome projects should help facilitate the development of more autosomal markers, as has been shown for plants (Xu et al. 2004).
Conclusions
The proven efficiency and discriminatory power of the molecular systematic approach ensure an increasing, if not central, future role in the discovery and delimitation of diatom species. It is also clear, however, that DNA sequence data are not a panacea, and in anticipation of their continued use in species-level systematic studies, it is worth exploring more fully some of the theoretical considerations outlined above. To this end, at least three good candidate systems have emerged: Sellaphora, Pseudo-nitzschia, and Skeletonema, each of which offers its own advantages and possibilities for studying diatom speciation. Sellaphorais a relatively small freshwater genus for which both the phylogeny and fossil record are well characterized (Evans et al. 2008); the reproductive biology of Sellaphora is also extremely well known (Behnke et al. 2004; Mann et al. 2004), so these issues could be explored in the context of the BSC. The marine planktonic genus Pseudo-nitzschia is another strong candidate for more in-depth speciation research in diatoms. Pseudo-nitzschia has well-characterized reproductive biology (Amato et al. 2007), known plastid inheritance (Ghiron et al. 2008), several intensively sampled population-level studies already completed (Casteleyn et al. 2008; Evans and Hayes 2004; Evans et al. 2005), and a forthcoming nuclear genome sequence, which will provide an invaluable resources for the development of new phylogenetic markers. Skeletonema is another planktonic, predominantly marine genus with a large number diverse culture strains available (Kooistra et al. 2008), a full genome sequence and EST library of a close relative, T. pseudonana (Armbrust et al. 2004), and an exemplary set of morphological observations tied to each species (Sarno et al. 2005, 2007). Importantly, the phylogenetic instability of several Skeletonema species provides the ideal context in which to explore many of these issues. Do mitochondrial loci support monophyly for species that appear paraphyletic at rDNA loci? Do any nuclear loci support monophyly of these species? What are the estimates of Ne for these different populations, and how do they bear on species boundaries inferred from autosomal versus organelle loci? With a better sense of the species boundaries, do reinterpretations of older data provide new insights into the morphology, physiology and biogeography of Skeletonema? The groundbreaking work in Skeletonema raised many of these important questions, and Skeletonema stands as a strong candidate to help address them.
The molecular systematic approach has opened up exciting new possibilities for species research in diatoms. Among other things, DNA sequence data have shown that some diatom species harbor extensive intraspecific variation that can be partitioned among several new, phylogenetically distinct species. These species boundaries have, in some cases, proven sensitive to intraspecific sampling, choice of genetic marker, and strength of independent corroborating evidence for their initial separation. These findings underscore that species are testable hypotheses. Increased attention to study design, including the number of sampled individuals and number and choice of genetic loci for study, can lead to more robust species hypotheses that are less likely to be falsified by new data.
Acknowledgements
I thank Elizabeth Ruck for sharing unpublished results. Comments by David Mann, Elizabeth Ruck, Virginia Sanchez-Puerta, Ed Theriot, and one anonymous reviewer greatly improved an earlier version of this manuscript. Financial support during preparation of this manuscript came from an NIH Ruth L. Kirschstein NRSA Postdoctoral Fellowship (1F32GM080079-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Alverson AJ. Strong purifying selection in the silicon transporters of marine and freshwater diatoms. Limnol Oceanogr. 2007;52:1420–1429. [Google Scholar]
- Alverson AJ, Kolnick L. Intragenomic nucleotide polymorphism among small subunit (18S) rDNA paralogs in the diatom genus Skeletonema (Bacillariophyta) J Phycol. 2005;41:1248–1257. [Google Scholar]
- Alverson AJ, Theriot EC. Comments on recent progress toward reconstructing the diatom phylogeny. J Nanosci Nanotechnol. 2005;5:57–62. doi: 10.1166/jnn.2005.007. [DOI] [PubMed] [Google Scholar]
- Alverson AJ, Cannone JJ, Gutell RR, Theriot EC. The evolution of elongate shape in diatoms. J Phycol. 2006;42:655–668. [Google Scholar]
- Amato A, Kooistra WHCF, Ghiron JHL, Mann DG, Pröschold T, Montresor M. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WWY, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Avila LJ, Morando M, Sites JW. Congeneric phylogeography: hypothesizing species limits and evolutionary processes in Patagonian lizards of the Liolaemus boulengeri group (Squamata : Liolaemini) Biol J Linn Soc. 2006;89:241–275. [Google Scholar]
- Avise JC. Phylogeography: the History and Formation of Species. Cambridge: Harvard University Press; 2000. [Google Scholar]
- Avise JC. Molecular Markers, Natural History and Evolution. 2nd ed. Sunderland: Sinauer Associates, Inc; 2004. [Google Scholar]
- Bates SS, Bird CJ, Defreitas ASW, Foxall R, Gilgan M, Hanic LA, Johnson GR, McCulloch AW, Odense P, Pocklington R, Quilliam MA, Sim PG, Smith JC, Rao DVS, Todd ECD, Walter JA, Wright JLC. Pennate diatom Nitzschia pungens as the primary source of domoic acid, a toxin in shellfish from eastern Prince Edward Island, Canada. Can J Fish Aquat Sci. 1989;46:1203–1215. [Google Scholar]
- Behnke A, Friedl T, Chepurnov VA, Mann DG. Reproductive compatibility and rDNA sequence analyses in the Sellaphora pupula species complex (Bacillariophyta) J Phycol. 2004;40:193–208. [Google Scholar]
- Beszteri B, Acs E, Medlin L. Conventional and geometric morphometric studies of valve ultrastructural variation in two closely related Cyclotella species (Bacillariophyta) Eur J Phycol. 2005a;40:89–103. [Google Scholar]
- Beszteri B, Acs E, Medlin LK. Ribosomal DNA sequence variation among sympatric strains of the Cyclotella meneghiniana complex (Bacillariophyceae) reveals cryptic diversity. Protist. 2005b;156:317–333. doi: 10.1016/j.protis.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Beszteri B, John U, Medlin LK. An assessment of cryptic genetic diversity within the Cyclotella meneghiniana species complex (Bacillariophyta) based on nuclear and plastid genes, and amplified fragment length polymorphisms. An. Eur J Phycol. 2007;42:47–60. [Google Scholar]
- Brand LE, Murphy LS, Guillard RRL, Lee HT. Genetic variability and differentiation in the temperature niche component of the diatom Thalassiosira pseudonana. Mar Biol. 1981;62:103–110. [Google Scholar]
- Casteleyn G, Chepurnov VA, Leliaert F, Mann DG, Bates SS, Lundholm N, Rhodes L, Sabbe K, Vyverman W. Pseudo-nitzschia pungens (Bacillariophyceae): a cosmopolitan diatom species? Harmful Algae. 2008;7:241–257. [Google Scholar]
- Chen GF, Wang GC, Zhang BY, Fan XL. Morphological and phylogenetic analysis of Skeletonema costatum-like diatoms (Bacillariophyta) from the China Sea. Eur J Phycol. 2007;42:163–175. [Google Scholar]
- Coleman AW. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist. 2000;151:1–9. doi: 10.1078/1434-4610-00002. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Mai JC. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J Mol Evol. 1997;45:168–177. doi: 10.1007/pl00006217. [DOI] [PubMed] [Google Scholar]
- Cox EJ, Williams DM. Systematics of naviculoid diatoms (Bacillariophyta): a preliminary analysis of protoplast and frustule characters for family and order level classification. Syst Biodivers. 2006;4:385–399. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland: Sinauer Associates, Inc; 2004. [Google Scholar]
- de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Edgar SM, Theriot EC. Heritability of mantle areolar characters in Aulacoseira subarctica (Bacillariophyta) J Phycol. 2003;39:1057–1066. [Google Scholar]
- Edgar SM, Theriot EC. Phylogeny of Aulacoseira (Bacillariophyta) based on molecules and morphology. J Phycol. 2004;40:772–788. [Google Scholar]
- Ehara M, Watanabe KI, Ohama T. Distribution of cognates of group II introns detected in mitochondrial cox1 genes of a diatom and a haptophyte. Gene. 2000b;256:157–167. doi: 10.1016/s0378-1119(00)00359-0. [DOI] [PubMed] [Google Scholar]
- Ehara M, Inagaki Y, Watanabe KI, Ohama T. Phylogenetic analysis of diatom coxI genes and implications of a fluctuating GC content on mitochondrial genetic code evolution. Curr Genet. 2000a;37:29–33. doi: 10.1007/s002940050004. [DOI] [PubMed] [Google Scholar]
- Ellegaard M, Godhe A, Harnstrom K, McQuoid M. The species concept in a marine diatom: LSU rDNA-based phylogenetic differentiation in Skeletonema marinoi/dohrnii (Bacillariophyceae) is not reflected in morphology. Phycologia. 2008;47:156–167. [Google Scholar]
- Evans KM, Hayes PK. Microsatellite markers for the cosmopolitan marine diatom Pseudo-nitzschia pungens. Mol Ecol Notes. 2004;4:125–126. [Google Scholar]
- Evans KM, Kuhn SF, Hayes PK. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzschia pungens (Bacillariophyceae) populations. J Phycol. 2005;41:506–514. [Google Scholar]
- Evans KM, Wortley AH, Mann DG. An assessment of potential diatom "barcode" genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora(Bacillariophyta) Protist. 2007;158:349–364. doi: 10.1016/j.protis.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Evans KM, Bates SS, Medlin LK, Hayes PK. Microsatellite marker development and genetic variation in the toxic marine diatom Pseudo-nitzschia multiseries (Bacillariophyceae) J Phycol. 2004;40:911–920. [Google Scholar]
- Evans KM, Wortley AH, Simpson GE, Chepurnov VA, Mann DG. A molecular systematic approach to explore diversity within the Sellaphora pupula species complex (Bacillariophyta) J Phycol. 2008;44:215–231. doi: 10.1111/j.1529-8817.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- Feliner GN, Rossello JA. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol Phylogenet Evol. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring Phylogenies. Sunderland: Sinauer Associates, Inc; 2004. [Google Scholar]
- Felsenstein J. Accuracy of coalescent likelihood estimates: Do we need more sites, more sequences, or more loci? Mol Biol Evol. 2006;23:691–700. doi: 10.1093/molbev/msj079. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Monaghan EB, Maberly SC. Hypothesis: The rate and scale of dispersal of freshwater diatom species is a function of their global abundance. Protist. 2002;153:261–273. doi: 10.1078/1434-4610-00103. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2003;34:397–423. [Google Scholar]
- Gallagher JC. Population genetics of Skeletonema costatum (Bacillariophyceae) in Narragansett Bay. J Phycol. 1980;16:464–474. [Google Scholar]
- Gallagher JC. Physiological variation and electrophoretic banding patterns of genetically different seasonal populations of Skeletonema costatum (Bacillariophyceae) J Phycol. 1982;18:148–162. [Google Scholar]
- Ghiron JHL, Amato A, Montresor M, Kooistra W. Plastid inheritance in the planktonic raphid pennate diatom Pseudo-nitzschia delicatissima (Bacillariophyceae) Protist. 2008;159:91–98. doi: 10.1016/j.protis.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Godhe A, McQuoid MR, Karunasagar I, Karunasagar I, Rehnstam-Holm AS. Comparison of three common molecular tools for distinguishing among geographically separated clones of the diatom Skeletonema marinoi Sarno et Zingone (Bacillariophyceae) J Phycol. 2006;42:280–291. [Google Scholar]
- Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- Hasle GR. Morphology and taxonomy of Skeletonema costatum (Bacillariophyceae) Norw J Bot. 1973;20:109–137. [Google Scholar]
- Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. 2nd ed. Sunderland: Sinauer Associates, Inc; 1996. [Google Scholar]
- Hudson RR, Coyne JA. Mathematical consequences of the genealogical species concept. Evolution. 2002;56:1557–1565. doi: 10.1111/j.0014-3820.2002.tb01467.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Turelli M. Stochasticity overrules the "three-times rule": Genetic drift, genetic draft, and coalescence times for nuclear loci versus mitochondrial DNA. Evolution. 2003;57:182–190. doi: 10.1111/j.0014-3820.2003.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Knowles LL, Carstens BC. Delimiting species without monophyletic gene trees. Syst Biol. 2007;56:887–895. doi: 10.1080/10635150701701091. [DOI] [PubMed] [Google Scholar]
- Kociolek JP. Taxonomy and ecology: further considerations. Proc Calif Acad Sci. 2005;56:99–106. [Google Scholar]
- Kooistra W, De Stefano M, Mann DG, Salma N, Medlin LK. Phylogenetic position of Toxarium, a pennate-like lineage within centric diatoms (Bacillariophyceae) J Phycol. 2003;39:185–197. [Google Scholar]
- Kooistra WHCF, Sarno D, Balzano S, Gu H, Andersen RA, Zingone A. Global diversity and biogeography of Skeletonema species (Bacillariophyta) Protist. 2008;159:177–193. doi: 10.1016/j.protis.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Logares R, Rengefors K, Kremp A, Shalchian-Tabrizi K, Boltovskoy A, Tengs T, Shurtleff A, Klaveness D. Phenotypically different microalgal morphospecies with identical ribosomal DNA: a case of rapid adaptive evolution? Microb Ecol. 2007;53:549–561. doi: 10.1007/s00248-006-9088-y. [DOI] [PubMed] [Google Scholar]
- Lundholm N, Moestrup Ø. The marine diatom Pseudo-nitzschia galaxiae sp. nov. (Bacillariophyceae): morphology and phylogenetic relationships. Phycologia. 2002;41:594–605. [Google Scholar]
- Lundholm N, Daugbjerg N, Moestrup Ø. Phylogeny of the Bacillariaceae with emphasis on the genus Pseudo-nitzschia (Bacillariophyceae) based on partial LSU rDNA. Eur J Phycol. 2002a;37:115–134. [Google Scholar]
- Lundholm N, Hasle GR, Fryxell GA, Hargraves PE. Morphology, phylogeny and taxonomy of species within the Pseudo-nitzschia americana complex (Bacillariophyceae) with descriptions of two new species, Pseudo-nitzschia brasiliana and Pseudo-nitzschia linea. Phycologia. 2002b;41:480–497. [Google Scholar]
- Lundholm N, Moestrup Ø, Hasle GR, Hoef-Emden K. A study of the Pseudo-nitzschia pseudodelicatissima/cuspidata complex (Bacillariophyceae): What is P. pseudodelicatissima? J Phycol. 2003;39:797–813. [Google Scholar]
- Lundholm N, Moestrup Ø, Kotaki Y, Hoef-Emden K, Scholin C, Miller P. Inter- and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses. J Phycol. 2006;42:464–481. [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Mann DG. The species concept in diatoms. Phycologia. 1999;38:437–495. [Google Scholar]
- Mann DG, Droop SJM. Biodiversity, biogeography and conservation of diatoms. Hydrobiologia. 1996;336:19–32. [Google Scholar]
- Mann DG, McDonald SM, Bayer MM, Droop SJM, Chepurnov VA, Loke RE, Ciobanu A, Du Buf JMH. The Sellaphora pupula species complex (Bacillariophyceae): morphometric analysis, ultrastructure and mating data provide evidence for five new species. Phycologia. 2004;43:459–482. [Google Scholar]
- Mayr E. The Biological Species Concept. In: Wheeler QD, Meier R, editors. Species Concepts and Phylogenetic Theory: A Debate. New York: Columbia University Press; 2000. pp. 17–29. [Google Scholar]
- McGuire JA, Linkem CW, Koo MS, Hutchison DW, Lappin AK, Orange DI, Lemos-Espinal J, Riddle BR, Jaeger JR. Mitochondrial introgression and incomplete lineage sorting through space and time: Phylogenetics of crotaphytid lizards. Evolution. 2007;61:2879–2897. doi: 10.1111/j.1558-5646.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- Medlin LK. Can molecular techniques help define species limits? Diatom Res. 1997;13:19–23. [Google Scholar]
- Medlin LK, Elwood HJ, Stickel S, Sogin ML. Morphological and genetic variation within the diatom Skeletonema costatum (Bacillariophyta): evidence for a new species, Skeletonema pseudocostatum. J Phycol. 1991;27:514–524. [Google Scholar]
- Mishler BD, Theriot EC. The Phylogenetic Species Concept (sensu Mishler and Theriot): Monophyly, Apomorphy, and Phylogenetic Species Concepts. In: Wheeler QD, Meier R, editors. Species Concepts and Phylogenetic Theory: A Debate. New York: Columbia University Press; 2000. pp. 44–54. [Google Scholar]
- Morando M, Avila LJ, Baker J, Sites JW. Phylogeny and phylogeography of the Liolaemus darwinii complex (Squamata : Liolaemidae): evidence for introgression and incomplete lineage sorting. Evolution. 2004;58:842–861. doi: 10.1111/j.0014-3820.2004.tb00416.x. [DOI] [PubMed] [Google Scholar]
- Müller T, Philippi N, Dandekar T, Schultz J, Wolf M. Distinguishing species. RNA. 2007;13:1469–1472. doi: 10.1261/rna.617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LS. Biochemical taxonomy of marine phytoplankton by electrophoresis of enzymes. II. Loss of heterozygosity in clonal cultures of centric diatoms Skeletonema costatum and Thalassiosira pseudonana. J Phycol. 1978;14:247–250. [Google Scholar]
- Murphy LS, Guillard RRL. Biochemical taxonomy of marine phytoplankton by electrophoresis of enzymes .I. Centric diatoms Thalassiosira pseudonana and Thalassiosira fluviatilis. J Phycol. 1976;12:9–13. [Google Scholar]
- Orsini L, Procaccini G, Sarno D, Montresor M. Multiple rDNA ITS-types within the diatom Pseudo-nitzschia delicatissima (Bacillariophyceae) and their relative abundances across a spring bloom in the Gulf of Naples. Mar Ecol Prog Ser. 2004;271:87–98. [Google Scholar]
- Oudot-Le Secq MP, Goer SL, Stam WT, Olsen JL. Complete mitochondrial genomes of the three brown algae (Heterokonta : Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr Genet. 2006;49:47–58. doi: 10.1007/s00294-005-0031-4. [DOI] [PubMed] [Google Scholar]
- Patterson C. Homology in classical and molecular biology. Mol Biol Evol. 1988;5:603–625. doi: 10.1093/oxfordjournals.molbev.a040523. [DOI] [PubMed] [Google Scholar]
- Pither J. Comment on "Dispersal limitations matter for microbial morphospecies". Science. 2007;316 doi: 10.1126/science.316.5828.1124a. [DOI] [PubMed] [Google Scholar]
- Pither J, Aarssen LW. Environmental specialists: their prevalence and their influence on community-similarity analyses. Ecol Lett. 2005a;8:261–271. [Google Scholar]
- Pither J, Aarssen LW. The evolutionary species pool hypothesis and patterns of freshwater diatom diversity along a pH gradient. J Biogeogr. 2005b;32:503–513. [Google Scholar]
- Reid G, Williams DM. Some Commentary on Molecules and Morphology, Species and Higher Taxa in Diatoms, with a Note on the Relationships of the Genus Cistula Cleve. In: Kusber W-H, Jahn R, editors. Proceedings of the 1st Central European Diatom Meeting 2007. Berlin: Botanic Garden and Botanical Museum Berlin-Dahlem; 2007. pp. 135–138. [Google Scholar]
- Round FE, Crawford RM, Mann DG. The Diatoms: Biology & Morphology of the Genera. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- Rynearson TA, Armbrust EV. Genetic differentiation among populations of the planktonic marine diatom Ditylum brightwellii (Bacillariophyceae) J Phycol. 2004;40:34–43. [Google Scholar]
- Rynearson TA, Armbrust EV. Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Mol Ecol. 2005;14:1631–1640. doi: 10.1111/j.1365-294X.2005.02526.x. [DOI] [PubMed] [Google Scholar]
- Rynearson TA, Newton JA, Armbrust EV. Spring bloom development, genetic variation, and population succession in the planktonic diatom Ditylum brightwellii. Limnol Oceanogr. 2006;51:1249–1261. [Google Scholar]
- Sarno D, Kooistra W, Balzano S, Hargraves PE, Zingone A. Diversity in the genus Skeletonema (Bacillariophyceae): III. Phylogenetic position and morphological variability of Skeletonema costatum and Skeletonema grevillei, with the description of Skeletonema ardens sp. nov. J Phycol. 2007;43:156–170. [Google Scholar]
- Sarno D, Kooistra W, Medlin LK, Percopo I, Zingone A. Diversity in the genus Skeletonema (Bacillariophyceae). II. An assessment of the taxonomy of S. costatum-like species with the description of four new species. J Phycol. 2005;41:151–176. [Google Scholar]
- Silva PC. Historical review of attempts to decrease subjectivity in species identification, with particular regard to algae. Protist. 2008;159:153–161. doi: 10.1016/j.protis.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Sims PA, Mann DG, Medlin LK. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia. 2006;45:361–402. [Google Scholar]
- Slapeta J, Lopez-Garcia P, Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol Biol Evol. 2006;23:23–29. doi: 10.1093/molbev/msj001. [DOI] [PubMed] [Google Scholar]
- Sorhannus U. Diatom phylogenetics inferred based on direct optimization of nuclear-encoded SSU rRNA sequences. Cladistics. 2004;20:487–497. doi: 10.1111/j.1096-0031.2004.00034.x. [DOI] [PubMed] [Google Scholar]
- Stoermer EF, Smol JP, editors. The Diatoms: Applications for the Environmental and Earth Sciences. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Telford RJ, Vandvik V, Birks HJB. Dispersal limitations matter for microbial morphospecies. Science. 2006a;312:1015–1015. doi: 10.1126/science.1125669. [DOI] [PubMed] [Google Scholar]
- Telford RJ, Vandvik V, Birks HJB. How many freshwater diatoms are pH specialists? A response to Pither & Aarssen (2005) Ecol Lett. 2006b;9:E1–E5. doi: 10.1111/j.1461-0248.2005.00875.x. [DOI] [PubMed] [Google Scholar]
- Thamatrakoln K, Alverson AJ, Hildebrand M. Comparative sequence analysis of diatom silicon transporters: Toward a mechanistic model of silicon transport. J Phycol. 2006;42:822–834. [Google Scholar]
- Theriot E, Stoermer EF. Principal component analysis of Stephanodiscus: observations on two new species from the Stephanodiscus niagarae complex. Bacillaria. 1984;7:37–58. [Google Scholar]
- Theriot EC. Clusters, species concepts, and morphological evolution of diatoms. Syst Biol. 1992;41:141–157. [Google Scholar]
- Thornhill DJ, Lajeunesse TC, Santos SR. Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol Ecol. 2007;16:5326–5340. doi: 10.1111/j.1365-294X.2007.03576.x. [DOI] [PubMed] [Google Scholar]
- Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- Vanormelingen P, Chepurnov VA, Mann DG, Cousin S, Vyverman W. Congruence of morphological, reproductive and ITS rDNA sequence data in some Australasian Eunotia bilunaris (Bacillariophyta) Eur J Phycol. 2007;42:61–79. [Google Scholar]
- Vanormelingen P, Chepurnov VA, Mann DG, Sabbe K, Vyverman W. Genetic divergence and reproductive barriers among morphologically heterogeneous sympatric clones of Eunotia bilunaris sensu lato (Bacillariophyta) Protist. 2008;159:73–90. doi: 10.1016/j.protis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Vyverman W, Verleyen E, Sabbe K, Vanhoutte K, Sterken M, Hodgson DA, Mann DG, Juggins S, De Vijver BV, Jones V, Flower R, Roberts D, Chepurnov VA, Kilroy C, Vanormelingen P, De Wever A. Historical processes constrain patterns in global diatom diversity. Ecology. 2007;88:1924–1931. doi: 10.1890/06-1564.1. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Doyle JJ. Phylogenetic Incongruence: Window into Genome History and Molecular Evolution. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular Systematics of Plants II: DNA Sequencing. Boston: Kluwer Academic Publishers; 1998. pp. 265–296. [Google Scholar]
- Wheeler QD, Meier R, editors. Species Concepts and Phylogenetic Theory: A Debate. Ed 230. New York: Columbia University Press; 2000. [Google Scholar]
- Wiens JJ. What is speciation and how should we study it? Am Nat. 2004;163:914–923. doi: 10.1086/386552. [DOI] [PubMed] [Google Scholar]
- Williams DM, Reid G. Amphorotia nov. gen., a New Genus in the Family Eunotiaceae (Bacillariophyceae), Based on Eunotia clevei Grunow in Cleve et Grunow. Vaduz, FL: Gantner Verlag; 2006. [Google Scholar]
- Wood AM, Lande R, Fryxell GA. Quantitative genetic analysis of morphological variation in an Antarctic diatom grown at two light intensities. J Phycol. 1987;23:42–54. [Google Scholar]
- Xu W, Briggs WJ, Padolina J, Timme RE, Liu W, Linder CR, Miranker DP. Using MoBIoS' scalable genome join to find conserved primer pair candidates between two genomes. Bioinformatics. 2004;20:i355–i362. doi: 10.1093/bioinformatics/bth929. [DOI] [PubMed] [Google Scholar]
- Zingone A, Percopo I, Sims PA, Sarno D. Diversity in the genus Skeletonema (Bacillariophyceae). I. A reexamination of the type material of S. costatum with the description of S. grevillei sp nov. J Phycol. 2005;41:140–150. [Google Scholar]