Abstract
Shotgun proteomics typically uses multidimensional LC/MS/MS analysis of enzymatically digested proteins, where strong cation-exchange (SCX) and reversed-phase (RP) separations are coupled to increase the separation power and dynamic range of analysis. Here we report an on-line multidimensional LC method using an anion- and cation-exchange (ACE) mixed-bed for the first separation dimension. The mixed-bed ion exchange resin improved peptide recovery over SCX resins alone and showed better orthogonality to RP separations in two-dimensional separations. The Donnan effect, which was enhanced by the introduction of fixed opposite charges in one column, is proposed as the mechanism responsible for improved peptide recovery by producing higher fluxes of salt cations and a lower populations of salt anions proximal to the SCX phase. An increase in orthogonality was achieved by a combination of increased retention for acidic peptides and moderately reduced retention of neutral to basic peptides by the added anion-exchange resin. The combination of these effects led to ∼100% increase in the number of identified peptides from an analysis of a tryptic digest of a yeast whole cell lysate. The application of the method to phosphopeptide-enriched samples increased by 94% phosphopeptide identifications over SCX alone. The lower pKa of phosphopeptides led to specific enrichment in a single salt step resolving acidic phosphopeptides from other phospho- and nonphospho-peptides. Unlike previous methods that use anion-exchange to alter selectivity or enrich phosphopeptides, the proposed format is unique in that it works with typical acidic buffer systems used in electrospray ionization making it feasible for online multidimensional LC/MS/MS applications.
Introduction
Shotgun or bottom-up proteomics requires the analysis of thousands of peptides generated by an enzymatic digestion of a complex protein mixture. At present, multidimensional LC methods coupled with tandem mass spectrometry such as MudPIT1, 2 (Multidimensional Protein Identification Technology, the improved successor of DALPC3) are one of the most widely accepted techniques for obtaining amino acid sequence information from complex peptide mixtures. In contrast to conventional gel-based proteomics methods where separation is performed at the protein level, MudPIT's ability to identify a protein based on one or more well-behaved peptides makes it capable of detecting proteins of low abundance and extreme hydrophobicity, pI, and molecular weight as long as the complex peptide mixture can be effectively fractionated.1-3 To meet the technical challenge of resolving thousands of generated peptides, MudPIT takes advantages of the greater separation power of multidimensional LC proposed initially by Giddings in 1984.4 In theory, the peak capacity of the system is a product of the peak capacities of each separation dimension provided the separation systems that are coupled are truly orthogonal. A recent study showed the total peak capacity of a 2-dimensional (2D) LC separation (SCX and RP) could exceed ∼1,500 by using an ultrahigh-pressure LC system and sub-micron separation media.5 It has been further shown that the addition of more separation dimensions6 or the coupling of different modes of separations7 were also an effective strategy to increase total separation power. The Human proteome, however, is extremely complex consisting of more than 25,000 proteins with a variety of splicing variants and post translational modifications.8 To meet the challenges of complexity, dynamic range, and sensitivity, further improvements in separation performance are required.
Due to its compatibility and orthogonality to reversed-phase (RP) LC separation, strong cation-exchange (SCX) has often been the first choice for the upstream LC dimension of multidimensional LC methods.1-3, 9 SCX separations of peptides are typically operated at low pH to block the dissociation of peptide carboxylic groups so that interactions between the protonated basic amino acid residues and the sulfonate groups of SCX resins are promoted.3, 10 Importantly, these eluent conditions are ideal to couple with RP separations and subsequent mass spectrometry analysis. In RP separations, a low pH prevents the deprotonation of residual silanol groups in silica resins and reduces undesired interactions between the analytes and the resin surface. Additionally, a low pH increases protonation of the analytes resulting in enhanced sensitivity in electrospray ionization.11 This situation is in clear contrast to anion-exchange media that are operated at less desirable neutral to basic pHs hindering their use in online 2D LC/MS/MS applications. Furthermore peptide elution in SCX can be accomplished using volatile organic salts such as ammonium acetate,2, 11 which is more compatible with on-line elution directly into a mass spectrometer.
The orthogonality between SCX and RP separations is based on SCX's use of electrostatic interactions to retain peptides.12, 13 In practice, retention in SCX peptide separations is a combination of electrostatic (main) and hydrophobic (sub) interactions,14, 15 the latter of which results from the hydrophobic nature of the sulfonyl polymer backbone. This “mixed-mode” property has been recognized as one of the reasons why SCX can separate structurally similar peptides possessing the same net charge.15 Consequently SCX requires the addition of an organic modifier in the mobile phase to improve peptide resolution and recovery.16 The addition of an organic modifier not only precludes the use of high concentration salt buffers, but also limits SCX's compatibility in online formats that are less tolerant to high organic modifiers (e.g. RP). The compromise is to add a small amount (5-10%) of organic modifier to moderate hydrophobic interactions with the SCX phase but not so much as to impede retention on the RP. Indeed, this limitation has been one of the criticisms of online SCX/RP multidimensional LC approaches where each separation dimension can not be independently optimized.17 Due to these inherent difficulties, the optimization of SCX separation in online formats has been overlooked in contrast to efforts to improve the RP separation.9, 18-23
The idea of using “mixed-bed” resins, typically a blend of anion- and cation-exchanges, for protein or peptide separation was first described by El Rassi et al. in the mid 80s as a way to separate acidic and basic proteins in a single chromatographic run.24 The working principle was identical to that of water purification methods reported in the 1950's.25 Anion- and cation-exchangers are both charged at the neutral pH of the elution buffer so that the mixed-bed can interact simultaneously with both cation and anion species in the sample. In their later work,26 the retention behavior of proteins in mixed-bed systems was investigated by using a few standard proteins. Despite their careful experimental design using the same silica base materials for both ion exchange resins, the principles governing the retention of proteins in these mixed-bed systems could not be elucidated. Since then, mixed-bed formats appear to have been almost completely ignored, likely due to the complex and unpredictable nature of the retention mechanism. More recently, a similar idea of using a pair of opposite charges for the separation of ionic species has been explored and has been termed electrostatic or zwitterionic ion chromatography.27 In these methods, ion-exchange sites of two opposite charges are typically bound to the same ligand, so that the system contains a chemically homogeneous resin (i.e., not a mixed-bed). It has been demonstrated that both anions and cations or zwitterionic analytes could interact with the zwitterionic stationary phase simultaneously.28 To our knowledge, however, no application using these zwitterionic stationary phases for large-scale peptide separations has been reported.
Several concepts have been proposed to explain the ion-exchange chromatography of ionic species with the Donnan equilibrium theory being the most widely accepted. According to the theory, the mechanism of ion-exchange phenomena can be considered as a Donnan exclusion process, i.e., ions move from one phase to the other in a heterogeneous system in order to balance electrostatic potential. The IUPAC terminology defines the Donnan effect as “the reduction in concentration of mobile ions within an ion exchange membrane due to the presence of fixed ions of the same sign as the mobile ions”. The advanced MudPIT format reported in this paper took advantage of this principle by blending anion- and cation-exchanger resins in one column. By creating oppositely charged fixed fields inside the column, it was expected that the separation of salt cations/anions during the salt pulse would be promoted by the Donnan effect. The anticipated outcome was improved recovery of peptides from SCX resins by (1) increased activity of salt cations proximal to the SCX phase and (2) fewer salt anions in the SCX phase resulting in lower peptide-anion ion-pair populations. The other potential advantage of mixed-bed ion exchange was better orthogonality with the RP separation resulting in improved resolution in a 2D separation. In this report, we detail the use and performance of mixed-bed ion exchange separations for complex peptide and phosphopeptide mixtures.
Experimental Procedures
Materials
Sequence-grade modified trypsin was purchased from Promega Corporation (Madison, WI). Endoproteinase Lys-C was obtained from Roche Diagnostics (Indianapolis, IN). A protease-deficient S. cerevisiae strain BJ546029 was purchased from American Type Culture Collection (Manassas, VA). HeLa cell nuclear extract was purchased from Bio Vision Inc (Mountain View, CA).
Tryptic Digests of S. cerevisiae
S. cerevisiae strain BJ5460 was grown, lysed, and digested as reported previously.1, 9
Phosphopeptides enriched from HeLa cell nuclear extract
The enrichment of phosphopeptides from HeLa cell nuclear extract was performed as will be described elsewhere.30 Briefly, proteins were extracted from 1 mg of HeLa cell nuclear extract by chloroform-methanol precipitation and digested by trypsin. Phosphopeptides were then enriched from the digest by precipitation at three different pHs. For this study, two pH fractions (3.5 and 4.6) were selected for further analysis by splitting each fraction in half and loading them individually on SCX and ACE mixed-bed ion-exchange columns for comparison (∼50 μg / analysis).
Preparation of Packed Capillary Columns
Fused-silica capillary analytical columns (100 μm i.d.) and biphasic trapping columns (250 μm i.d., RP/SCX or RP/ACE mixed-bed) were prepared by slurry packing using an in-house pressure vessel driven by He gas. Capillaries for analytical columns were pulled beforehand by a laser puller (Sutter Instrument Co., Novato, CA) to form a fritless emitter capillary with a ∼5 μm opening.31 Analytical capillary columns were packed with 3 μm octadesyl silica (ODS) materials (Aqua C18, Phenomenex, Torrance, CA) to approximately 10 cm. The biphasic column was composed of 2.5 cm of 5 μm Aqua C18 (Phenomenex, Torrance, CA) (as upstream) and 2.5 cm of 5 μm Partisphere SCX resins (Whatman, Clifton, NJ) or a mixture of Partisphere SCX and PolyWAX LP (PolyLC Inc., Columbia, MD). A modified Kasil® frit preparation method was used to prepare a fritted filter at the end of biphasic capillary columns to retain resins in the capillary. The Kasil® solutions were a generous gift from PQ Corporation (Berwyn, PA).
Multidimensional Protein Identification Technology (MudPIT) Analysis
Digested proteins were analyzed using a modified 13-step MudPIT-type separation as described previously.1, 2 Briefly, 0.5 μg of the protein digest was pressure-loaded onto a biphasic trapping column, and the column was washed with buffer A (see below) for more than 10 sample volumes. After desalting, the biphasic column was attached to an analytical capillary column with a through-hole union (Upchurch Scientific, Oak Harbor, WA), and the entire split-column (biphasic column–union–analytical column) was placed inline with an Agilent 1100 quaternary HPLC pump (Palo Alto, CA). The buffer solutions used were as follows: water/acetonitrile/formic acid (95:5:0.1, v/v/v) as buffer A (pH ∼2.6), water/acetonitrile/formic acid (20:80:0.1, v/v/v) as buffer B, and buffer A including 500 mM ammonium acetate as buffer C (pH ∼6.8), and water/acetonitrile/trifluoroacetic acid (95:5:0.05, v/v/v) as buffer D (pH ∼2.1) unless otherwise specified.
Gradient Conditions
The gradient profile of Step 1 was as follows: 5 min of 100% buffer A, a 5 min gradient from 0-15% buffer B, a 60 min gradient from 15-45% buffer B, a 10 gradient from 45-75% buffer B, and 5 min of 75% buffer B. Steps 2-12 had the following profile: 1 min of 100% buffer A, 4 min of X% buffer C followed by the same gradient as Step 1. The 4 min buffer C percentages (X) were 5, 10, 15, 20, 25, 30, 40, 50, 75, and 100%, respectively, for the Step 2-11 (25, 50, 75, 100, 125, 150, 200, 250, 375, 500 mM as ammonium acetate concentrations). The salt pulse for the Step 12 was 90% buffer C + 10% buffer B (450 mM ammonium acetate in 12.5% acetonitrile). For the final step (Step 13), the gradient contained: 1 min of 100% buffer A, 4 min of 100% buffer D, 4 min of 90% buffer C + 10% buffer B followed by the gradient same as Step 1.
Mass Spectrometry Conditions
An LCQ-Deca mass spectrometer (Thermo Fisher Scientific, Waltham, MA) was used for the majority of this study while an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific) was used for the phosphopeptide analysis. In both cases, peptides eluted from the microcapillary fritless column were directly electrosprayed into the mass spectrometer with the application of a distal 2.4 kV spray voltage.31 In the experiments with an LCQ-Deca, a cycle of one full-scan mass spectrum (m/z 400-1400) followed by 3 data-dependent tandem mass spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. The number of microscans was 3 for both MS and MS/MS scans. The dynamic exclusion settings used were as follows: repeat count: 1, repeat duration: 0.50 min, exclusion list size: 25, and exclusion duration: 10.00 min. For the phosphopeptide analysis, an LTQ mass spectrometer was programmed to conduct neutral-loss triggered data-dependent MS/MS for the peptides that produced a 49 or 98 amu neutral loss from their precursor ions. Similar to the experiments with LCQ-Deca, one full-scan mass spectrum (m/z 400-1800) was followed by 7 data-dependent MS/MS and up to 7 data dependent MS3 spectra at a 35% normalized collision energy. The number of microscans was 1 for MS and MSn scans. The dynamic exclusion settings used were: repeat count: 1, repeat duration: 0.50 min, exclusion list size: 50, exclusion duration: 1.00 min. Application of mass spectrometer scan functions and gradient generation was controlled by the Xcalibur datasystem (Thermo Fisher Scientific).
Data Analysis
Collected MS/MS spectra were processed using RawExtract32 and then filtered with the PARC algorithm.32 The filtered tandem mass spectra were searched with the SEQUEST algorithm (ver 27) against a yeast database [SGD, 12-16-2005] that was concatenated with reversed protein sequences to estimate a false positive rate.17 SEQUEST results were then filtered with DTASelect 2.0 using XCorr and ΔCn values that correspond to user-defined false positive rates.33 The false positive rates were estimated by the program from the number and quality of spectral matches to the decoy database. The minimum number of peptides to identify proteins was set at 1 or 2 depending on the purpose of the experiment. Only half- or fully tryptic peptides were accepted to identify peptides/proteins unless otherwise specified. Additional database search criteria are as follows; (1) mass tolerance of 6 mass units for the precursor peptide was used in the SEQUEST database search, (2) the [M+H]+ value used in the search was calculated using average mass from the precursor ion, (3) monoisotopic masses were used for the predicted fragment ions, (4) and the database search was performed without enzyme specificity. Cysteine residues were considered to have a static modification of + 57 mass units. Oxidized methionine was searched as a dynamic modification of + 16 mass units.
Recovery Estimation
The detailed experimental procedure is provided as Supporting Information (Procedure S-1)
Results and Discussion
Concept and postulated mechanism of the anion- and cation-exchange (ACE) mixed-bed system for improved peptide recovery and 2D orthogonality
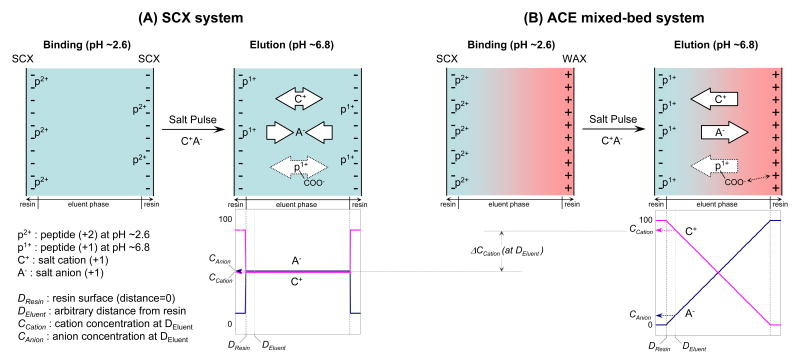
Illustrated in Figure 1 is the concept and postulated mechanism of the anion- and cation-exchange (ACE) mixed-bed system designed to improve peptide recovery and orthogonality in 2-D LC separation. We postulate a Donnan effect may be occurring because of the close proximity of the SCX and anion-exchanges (AEX) resins in the mixed-bed. The Donnan effect, also known as the Donnan exclusion effect, may be creating anionic and cationic micro gradients during the salt wash resulting in improved selectivity and elution of peptides. In theory, the “fixed” opposite charges of the resins in the mixed-bed creates a potential gradient (=Donnan membranes) that promotes unequal distribution of cationic and anionic species within the ion exchange bed. Thus, when a salt pulse is applied to the ACE mixed-bed, salt cations will be forced to the SCX surface by repulsion from the positive charges on the AEX surface resulting in the predicted concentration profile shown in Figure 1 (B, right bottom). At the same time, salt anions will be repulsed by negative charges on the surface of the SCX by the same principle (the Donnan exclusion plus the attraction by AEX resins). A lower concentration of anions near the SCX surface should lead to a lower population of peptide-anion ion-pairs during the peptide displacement processes. Thus, in the ACE mixed-bed format, the displacement of peptides from the SCX resin was expected to be facilitated by a greater flux of salt cations to the SCX surface (=higher activity) at a lower overall salt concentration. In principle, AEX could be either weak anion-exchange (WAX) or strong anion-exchange (SAX), but WAX was chosen for this study as preliminary analyses showed better performance for peptides (data not shown).
Figure 1. Proposed mechanism of the ACE mixed-bed system (B) compared to the conventional SCX format (A); a simplified schematic to illustrate the concept based on the Donnan effect for improved peptide recovery from SCX resins and an additional anion-exchange interaction for unique selectivity in the ion-exchange separation.
During the peptide binding phase at pH∼2.6 (by the mobile phase), the majority of tryptic peptides are doubly charged and bound to the SCX resin in both systems. When a salt pulse is applied to elute the peptides, salt cations displace peptides by competing with the peptides for SCX's negative charge sites. Salt anions have a mixed effect; the elution of peptides is promoted by neutralization of the peptides' positive charge(s) while the resultant ion-pairs increases peptides' hydrophobicity and discourage elution due to the hydrophobic property of SCX resins. In the ACE mixed-bed format, salt cations and anions are separated inside of the column by the Donnan (exclusion) effect that is enhanced by the fixed opposite charges of both resins. Through repulsion with anion-exchange resins, salt cations are consistently pushed toward the SCX phase resulting in the schematic concentration profile shown in (B) bottom. Conversely, salt anions are pulled away from the SCX surface by the same principle. The net consequence is a greater flux of salt cations and fewer salt anions proximate to the SCX surface compared to the SCX-only format. The displacement of peptides in the ACE mixed-bed format is expected to be facilitated by the sum of those effects. The additional interaction between acidic group(s) of peptides and anion-exchanger resins is unique to the ACE mixed-bed format at the elution phase at pH∼6.8. The ACE mixed-bed format can provide unique selectivity and improved orthogonality by this interaction. The white arrows in the figure refer solely to the direction of the attractive interaction and are not quantitative.
Another feature of the ACE mixed-bed was the additional electrostatic interaction between acidic groups of peptides and anion-exchange resins. During the salt pulse in online multidimensional LC, the pH of the mobile phase is almost neutral through the buffering effect of ammonium acetate ions. As a result, the carboxylic acid groups of peptides will be ionized in this particular environment and peptides lose one net positive charge. Since every peptide has at least one carboxylic acid group at its C-terminus, the ACE mixed-bed format should, in principle, produce additional electrostatic interactions between the resin and all peptides. This interaction should appear during the salt pulse and disappear once the salt solution has cleared the ion-exchange column and the acidic reversed-phase buffer lowers the pH and protonates the acidic groups of peptides. This means that except for very acidic peptides with a pKa below the buffer pH (∼2.6 in this study by 0.1% formic acid), the ACE mixed-bed should elute the majority of peptides during salt pulses. Therefore, the ACE mixed-bed format should be an attractive alternative to SCX in online multidimensional LC by providing an anion-exchange mode of separation with no change to the buffer systems. Another potential advantage to a mixed-bed would be the enrichment of phosphopeptides as they are acidic by nature. Theoretically, phosphopeptides and any acidic peptides that are ionized in the buffer (pH ∼2.6) should produce more interaction with ACE mixed-beds than would be normally observed with SCX media.
Peptide recovery from SCX resins
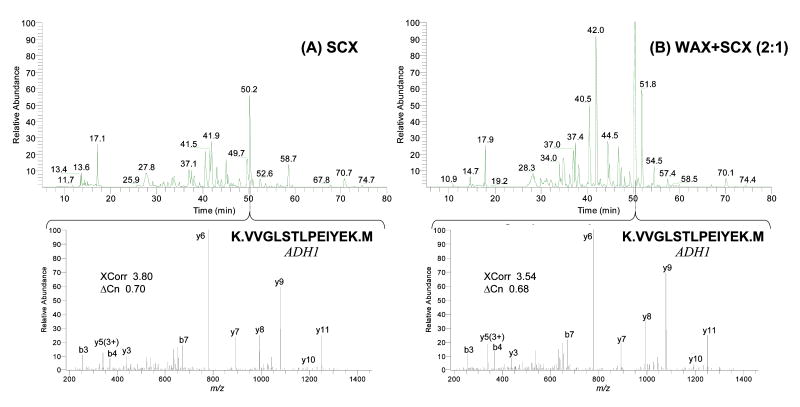
To evaluate the recovery of peptides, both ACE and SCX ion-exchange resins were challenged with a mixture of peptides (Figure 2, see Supporting Information (Procedure S-1) for details). Briefly, tryptic digests of five standard proteins were each eluted from an ion-exchange column by a 1-step high-concentration salt pulse and subsequently analyzed by an online RP gradient LC/MS/MS run proteins (0.1 fmol – 1 pmol on column, see Table S-1 for a list of proteins). The recoveries were estimated from the total peak areas of base peak chromatograms by normalizing them with those derived from single-phase RP runs. A 4-min salt pulse of 450 mM ammonium acetate including 12.5% acetonitrile (90% buffer C + 10% buffer B) was chosen to elute “all” peptides from the ion-exchange resins since these conditions were previously determined to be the most effective for SCX. The ACE mixed-bed used in this evaluation was a blend of WAX and SCX at 2:1 weight ratio, which we also determined performed the best under these conditions as will be discussed in the next sections (denoted as WAX+SCX(2:1) in the figure). To ensure that the two formats were compared objectively, the total amount of SCX resin in each column was adjusted so that the total volume of SCX between the two formats would be identical; i.e., the length of SCX bed was 1/3 of that of the WAX+SCX(2:1) bed.
Figure 2. Improved peptide recovery from ion-exchanger resins – chromatographic comparison between SCX and ACE mixed-bed formats.
Typical base peak chromatograms and MS/MS spectra from triplicate recovery experiments are shown. Tryptic digests of a five protein standard mixture were eluted from each ion-exchanger column using a 1-step high-concentration salt pulse and subsequently analyzed by online reversed-phase gradient LC/MS/MS. The majority of peptides observed in the ACE mixed-bed platform (B) had greater intensities than those of the SCX format (A). The amounts of packed SCX resin were adjusted to be identical for both cases to make the comparison valid (i.e., the SCX bed was 1/3 length of the WAX+SCX(2:1) bed).
As evidenced by the increased peak heights for the majority of observed peptides shown in Figure 2 (B, top panel), the ability of the ACE mixed-bed to facilitate the elution of peptides from ion-exchange resins using typical high-concentration salt elution was confirmed. Peptide recoveries were 82.1+/-8.4% and 47.2+/-13% for ACE mixed-bed and SCX, respectively, when the results from single-phase RP runs were used as the baseline for full recovery (all triplicates). It should be mentioned that the salt elution condition applied was slightly advantageous for the SCX format since very acidic peptides, if any, would not have been eluted from the ACE mixed-bed under the conditions used.
Table S-2 summarizes the recovery experiment and includes a comparison of sensitivity, dynamic range, and sequence coverage. The results clearly show that the ACE mixed-bed outperformed SCX alone in all aspects which is almost certainly a consequence of improved recovery. To address the question of whether the salt concentration was high enough to elute peptides from SCX resins, 2D LC/MS/MS experiments using salt pulses of twice the salt concentration (1 M ammonium acetate) were also conducted and the higher salt concentration gave no noticeable improvements in peptide recovery for either format (data not shown).
To investigate possible differences in the properties of the peptides recovered from both formats, the acquired data was statistically analyzed in terms of the distribution of charge states and pI of the identified peptides (Figure S-1). The results indicated that there were no statistically significant differences in the distributions of charge state and pI of peptides identified in either format (t-Test, P<0.05). Hence, we concluded from this experiment that the ACE mixed-bed format produces better peptide recovery under typical salt elution conditions without a significant change in the distribution of pI or charge state.
Effect of ACE mixed-bed format on peptide/protein identifications by MudPIT
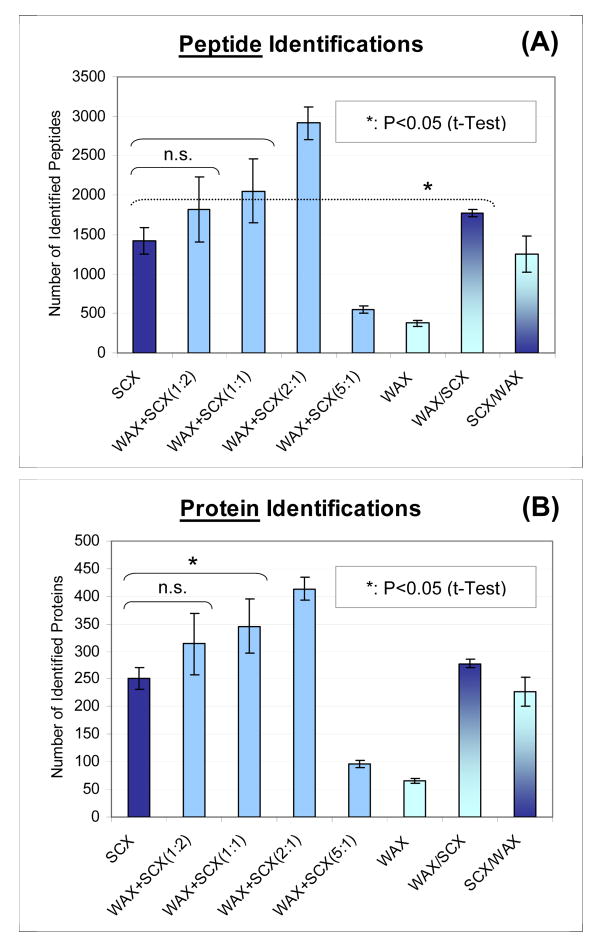
The improvement in peptide identifications using the ACE mixed-bed format in MudPIT analysis is clearly shown in Figure 3. The graphs were generated from quadruplicate 13-step MudPIT runs (=online 2D LC/MS/MS) for each resin combination (See Experimental Procedures for details. Protein lists are provided as Table S-3∼10). Tryptic digests of a soluble yeast cell lysate were used as a test sample having moderate complexity (0.5 μg/run). We find that the ACE mixed-bed, namely, the blends of WAX and SCX resins, resulted in a dramatic increase in peptide/protein identification compared to the conventional format of SCX alone. In this particular example, the number of identified peptides nearly doubled from that of the SCX benchmark when compared to the best ACE mixed-bed combination WAX+SCX (2:1). The WAX only format gave rise to the lowest number of identifications, presumably because the resin did not bind peptides in the acidic buffer system used. It should be mentioned that the marked improvement in the results from ACE mixed-beds were not only the result of improved recovery. This interpretation is based on the observation that the recovery improvements in WAX+SCX(1:2) and (1:1) combinations, as estimated in the same manner as in the previous section, were not as significant as those expected from the identification increases shown in Figure 3 (data not shown). As will be discussed in subsequent sections, the improved orthogonality in 2D separations was also likely to be a significant contributing factor to the increased number of protein identifications by allowing more efficient precursor sampling in tandem mass spectrometry.
Figure 3. Improved peptide (A) and protein (B) identifications in 13-step MudPIT – the effect of ACE mixed-beds on two-dimensional (2D) LC/MS/MS strategy.
0.5-μg of yeast tryptic digests were analyzed by 13-step MudPIT runs for each ion-exchange resin combination (N=4). Peptide/protein identification criteria were: (1) at least 2 peptides identification per protein, (2) peptides must be half- or fully-tryptic, and (3) filtering criteria must result in false positive rates at less than 2% at the protein level as estimated by a decoy reversed-database approach. The ACE mixed-beds are denoted as WAX+SCX(A:B), where WAX and SCX resins were mixed with A:B weight ratios and used as columns for the first-dimension separation. WAX/SCX and SCX/WAX are tandem bed configurations by the packing order of “upstream/downstream” (2.5 cm each in length).
The tandem column combinations denoted as WAX/SCX and SCX/WAX, in which two columns packed with each resin were connected in series, were tested to verify and probe the working principle of the ACE mixed-bed system. It is worth mentioning that the WAX/SCX combination exhibited a slight but statistically significant increase in peptide identification compared to SCX (t-Test, P<0.05). On the contrary, SCX/WAX led to a slight decrease in the number of identifications. These results can be explained by the separation of salt cations and anions during the salt pulse for WAX/SCX and an additional interaction between peptide acidic groups and anion-exchange for SCX/WAX. The details of these interpretations are discussed in the following sections.
Ion-exchange separation profile estimated by the number of identified peptides in each salt step
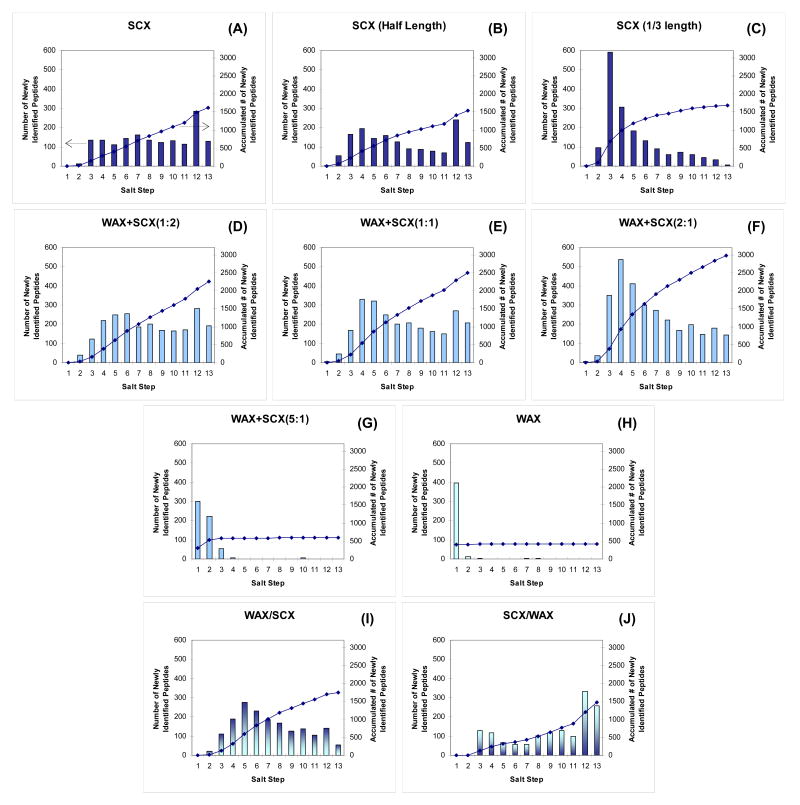
To understand the influence of ion-exchange separation efficiency on the improved peptide identifications in the ACE mixed-bed format, ion-exchange separation profiles were evaluated. Figure 4 (A–J) were generated by plotting the number of newly identified peptides in each step of 13-step MudPIT runs performed with different ion-exchange combinations. Since the method employed step salt gradients to elute peptides from ion-exchange resins, this was an effective way to overview peptide elution profiles in ion-exchange separations. It should be clarified, however, that the intensities of the bars do not directly represent the actual numbers of peptides eluted in the corresponding salt step. This is because these numbers are based on successful identification of peptides from collected tandem mass spectra and not the number of peptides eluting at that point in the chromatogram. In other words, the height of the bar could be significantly affected by the orthogonality of two separation dimensions and the complexity of the final RP separation. For example, if the ion-exchange column eluted peptides of more diverse hydrophobicity in one particular salt step, the number of identifications for that step would increase because the subsequent RP separation should improve the sampling of peptide precursor ions in the tandem mass spectrometer. To minimize the influence of such factors, the sample amount was carefully chosen as 0.5 μg per analysis to provide a margin for the final RP separation and mass spectrometric sampling. Although the extent of undersampling, if any, cannot be clearly distinguished in this profile analysis, the graphs in Figure 4 are a good tool to observe trends in the ion-exchange salt step gradients and also to understand the working principle of the ACE mixed-bed system.
Figure 4. Ion-exchange elution profile comparison of 13-step MudPIT runs with different ion-exchanger combinations.
The graphs were generated from the datasets that provided the best peptide/protein identifications in Figure 3 for each resin combination (where applicable). The runs denoted as SCX(half length) (B) and SCX(1/3 length) (C) used columns of half or 1/3 length of the others so that they contained the same amount of SCX resin and could be compared directly to the WAX+SCX(1:1) or (2:1), respectively. Peptide identifications criteria were modified to 1 peptide per protein with estimated false positive rates < 1% at peptide level. The y-axis indicates the number of peptides newly identified in each step so that peptides already identified in previous salt steps were excluded from the profile. Peptides identified in multiple charge states were counted only once.
For a better profile comparison, the graphs in Figure 4 were generated from the data presented in Figure 3 from which the best protein identifications among the quadruplicates were achieved. Peptide identification criteria were modified to 1 peptide per protein with a more stringent false positive rate cutoff (<1.0% at peptide level) since we filtered the dataset at the peptide level rather than the protein level. To avoid duplicate counts on the same peptides identified at different charge states, only the charge state at which the peptide was first identified was considered. The salt pulse conditions (concentrations and durations) for the step 1 through 11 were the standard ones used in our laboratory (0–500 mM ammonium acetate in 5% acetonitrile, 4-minute rectangle pulses, see Experimental Procedures for detail). The salt pulse for Step 12 was a combination of a high concentration of salt (450 mM ammonium acetate) and high organic modifier (12.5% acetonitrile), which was added to probe the hydrophobic interaction between peptides and SCX resins. The salt pulse for Step 13 was designed to elute the acidic peptides interacting with anion-exchangers and relied on a buffer containing trifluoroacetic acid (pH ∼2.1).
In the case of SCX (Figure 4 (A)), the number of newly identified peptides was nearly evenly distributed across the salt steps (except for the first two and 12th steps) which was in good agreement with the results reported by Wolters et al.2 As expected, the WAX showed a single-phase RP-like profile (Figure 4 (H)) supporting the previous assumption that the resin does not bind peptides under the acidic buffer conditions used since most of the peptides were positively charged in the acidic buffer as were the functional groups of anion-exchange resins. Thus, peptides were presumably repelled from the anion-exchange resin by charge repulsion. The contrast between SCX (A) and WAX (H) clearly highlighted the power of multidimensional LC approaches, i.e., the addition of the SCX separation dimension reduced the complexity of the RP separation by distributing peptides over several salt fractions and thereby allowing more efficient precursor ion sampling and MS/MS identification.
From the comparison of ACE mixed-beds of different blend ratios (Figure 4 (D-G)), two trends in the peptide elution profile appeared. In line with the results from Figure 3, greater the amounts of WAX in the ACE mixed-bed led to more peptides being identified (right y-axis) until it exceeded the optimum ratio (WAX+SCX(2:1)). Beyond a ratio of 2:1 the profile was similar to that of WAX alone and resulted in fewer peptide identifications. The second trend was the shift of the “peak” in the profiles to early salt steps as the amount of WAX resin was increased. This shift can be explained by the total amount of SCX resin in the ACE mixed-bed column since the length of the ion-exchange bed was fixed in this comparison. To confirm the influence of the SCX bed volume on the ion-exchange elution profile, half- and 1/3-length SCX beds were also examined in the same manner (as Figure 4 (B) and (C) to compare with the elution profiles of WAX+SCX(1:1) and (2:1), respectively. The results showed that ion-exchange columns containing identical SCX bed volumes had similar peptide identification profiles, although the information content (i.e., the properties of identified peptides in each salt step) was different due to the improved orthogonality of the WAX (related discussion is in the next section). The other important observation was that the amount (or length) of the SCX resin itself had no effect on the total number of identified peptides in the case of the SCX only formats (see the right y-axis for Figure 4 (A)–(C)), which was in clear contrast to the ACE mixed-bed formats that exhibited greater numbers of peptide identifications using different WAX and SCX ratios (Figure 4 (D)–(F)).
Additional evidence supporting a Donnan effect on peptide elution was seen in the result from the WAX/SCX tandem column (Figure 4 (I)). In this case, peptides were trapped at the top of the SCX bed after the loading step (with no salt pulse) because the peptides did not bind to the WAX resin at the pH of the acidic buffer system as shown in Figure 4 (H). Thus, this configuration could be considered a conventional SCX resin equipped with an “ion separation filter” that is provided by the WAX phase. This “filter” would be expected to separate salt cations and anions during every salt pulse. If filtering was occurring, a higher concentration of salt cations would be expected to displace SCX-bound peptides more effectively than in a normal salt pulse where the salt anions coexist at higher concentrations. This effect was indeed observed in the elution profiles apparently until the salt concentration exceeded the capacity of the WAX resin. To further confirm this effect, the eluent from the column was measured using a conductivity detector (Figure S-2). Ammonium ions (NH4+) were observed to elute earlier than acetate ions (CH3COO-) when a salt pulse of 100 mM ammonium acetate was applied to a WAX column having the identical dimensions to that found in Figure 4 (H) (Figure S-2 (A), with the separation factor ∼1.3). An opposite elution order was observed from an SCX column (i.e., CH3COO- eluted earlier under the same condition) (Figure S-2 (B)), although the separation was not as great as that of the WAX column.
The profile of the SCX/WAX tandem column configuration (Figure 4 (J)) provided evidence for a peptide-WAX interaction based on the properties of the WAX resin that should also govern its behavior in the context the ACE mixed-bed system. If there was no interaction between the WAX resin and the peptides released from the upstream SCX column, Figure 4 (J) would have shown a profile similar to that of the SCX resin alone (Figure 4 (A)). Instead, Figure S-2 (B) suggests that peptides released from the SCX resin in the SCX/WAX tandem configuration co-eluted with salt buffer ions. Thus, the pH inside the WAX column was neutral until the salt ions were cleared from the column and as a result, the acidic groups of the peptides, mainly carboxylic acids, ionized and interacted with the basic residues of WAX resin under these conditions. The complexity of the elution profile would suggest that the strength of this interaction is determined by many factors including ionic strength, pI of the peptides, pKa of acidic groups, etc. Although the detailed ion-exchange retention mechanism for individual peptides could not be determined solely from this analysis, the profile showed clear evidence for the existence of an additional interaction between peptides and WAX resin during the salt pulse.
Orthogonality improvements in 2D LC separation
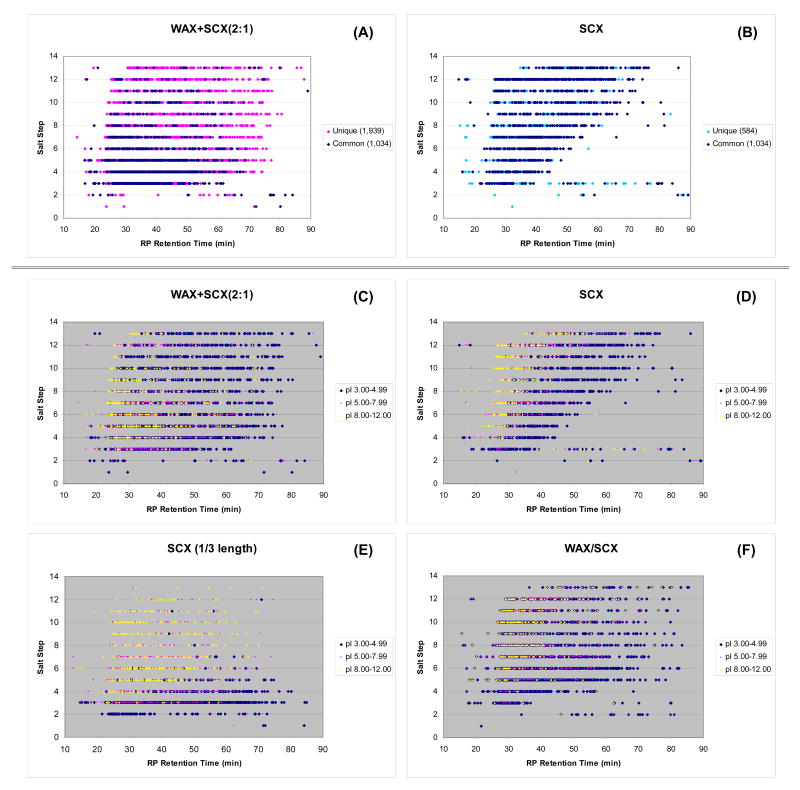
To determine if the improved separation of peptides is a result of better orthogonality, the datasets used in the previous section (those used in Figure 4) were re-analyzed by 2D view plots (shown in Figure 5 (A), (B) for WAX+SCX(2:1) and SCX, respectively). In the figures, peptides in common to both formats were represented as dark blue dots (1,034 data points), while those uniquely identified in only one format were shown as pink and blue dots for the ACE mixed-bed (1,939 data points) and SCX (584 data points), respectively. An obvious feature in the 2D plots was the improved separation of peptides suggesting the ACE mixed-bed is more orthogonal to RP than SCX alone. Unlike the SCX separation which shows a blank triangular space at the right-hand bottom corner seemingly due to the hydrophobic nature of the SCX resin (Figure 5 (B)), the ACE mixed-bed was able to use the 2D separation plane more efficiently (Figure 5 (A)). The difference in the distribution of peptides identified in both ACE and SCX (dark blue dots) can be explained by the aforementioned elution shift due to the reduced amount of SCX resin in the column. The uniquely identified peptides found in the later salt steps of the ACE mixed-bed format (shown as pink dots) are likely to be from improved recovery and increased retention provided by the ACE mixed-bed.
Figure 5. 2D view plots of 13-step MudPIT runs with different ion-exchanger combinations.
The same datasets used in Figure 4 were plotted in two-dimensional planes that consisted of salt step in which the peptide eluted and its RP retention time. Plots (A) and (B) are a comparison of the ACE mixed-bed (WAX+SCX(2:1)) with SCX with an emphasis on the distribution of common and uniquely identified peptides in the two formats. The plots (C)–(F) were generated in the same way except that the identified peptides were categorized according to their theoretical pI.
To understand the nature of the additional interaction in the ACE mixed-bed, the same data was further analyzed by grouping the identified peptides according to their pI (Figure 5 (C) and (D)). The three pI groups used were 3.00-4.99, 5.00-7.99, and 8.00-12.00 to represent acidic, neutral, and basic peptides, respectively. Comparable 2D plots for 1/3 length SCX and WAX/SCX tandem columns are shown as Figure 5 (E) and (F), respectively.
By comparing WAX+SCX (2:1) and SCX shown in Figure 5 (C) and (D), respectively, it is clear that the ion-exchange retention of neutral to basic peptides was moderately decreased in the ACE mixed-bed format. In contrast, acidic peptides were observed throughout the 2D plane suggesting retention on the ion-exchange resin was greatly affected by the ACE mixed-bed. Peptide properties shown in the 2D plot for the SCX (1/3 length) resin (Figure 5 (E)) is in clear contrast to the ACE mixed-bed. Since the amount of SCX resin is the same in the two formats (WAX+SCX (2:1) and SCX (1/3 length)), the nature of the additional WAX-peptide interaction in the ACE mixed-bed system was expected to be better highlighted in this comparison. In fact, Figures 5 (C) and (E) demonstrate the ACE mixed-bed increases retention of acidic peptides while maintaining the retention profiles of neutral to basic peptides. From this observation, it is strongly suggested that the nature of the additional interaction provided by the ACE mixed-bed is mostly between acidic functional groups of peptides and the WAX resin, and that the contribution of C-terminal carboxylic groups was relatively insignificant. Improving ion-exchange separation of acidic peptides is beneficial for MudPIT applications since acidic peptides are frequently observed in typical tryptic digests7, 34 and often elute as clusters7, 17 as readily seen in Figure 5 (E).
The above observations were also verified at the individual peptide level by analyzing the elution “shift” of each peptide in the ion-exchange step separations (see Figure S-3 for details). An LTQ-Orbitrap was used solely for this purpose to attain high-confidence 1-peptide hit assignments by taking advantage of its high-accuracy mass measurement capability35, 36. As shown in the histograms from Figure S-3 (A) and (B), the increased retention for acidic to neutral peptides in the ACE mixed-bed format was confirmed at the individual peptide level. In the comparisons between SCX formats of different column lengths (half and 1/3 length) (Figure S-3 (D)-(F)), the reversed but reasonable trend was observed in accordance with the retention mechanism of SCX, i.e., basic peptides were retained longer in the longer SCX column, while the retention of acidic peptides was minimally affected by the column length.
The apparent inconsistency observed in the relationships between the SCX column length and the retention of acidic peptides is worth mentioning. In contrast to the large difference observed in the comparison between SCX (full length) and SCX (half length) in Figure 5 (D) and (E), respectively, negligible differences were seen between SCX (half length) and SCX (1/3 length) shown as Figure S-3 (D). This difference can be readily explained by the difficulty in optimizing peptide separation and retention in the conventional “SCX-only” online 2D LC formats. The ion-exchange peptide retention in 2D LC methods can be manipulated by the concentration of the salt pulse, the length of column (=resin volume), and the duration of salt pulse. However, since the retention principle of SCX depends on basic residues, the resolution of acidic peptides cannot be easily improved compared to that of neutral to basic peptides. Indeed, the data indicate that the retention of acidic peptides did not increase when the salt pulse duration was relatively long with respect to the length of the column used (Figure S-3 (D)). Once the balance was shifted toward a shorter salt pulse and exceeded a critical point, the separation of acidic peptides appeared to have improved but this was mostly the result of the secondary hydrophobic interactions that becomes dominant under these conditions (as seen in the elution profile of Figure 5 (D)). As discussed earlier, the 2D separation plane for the SCX-only separation will not be used as effectively as that of the ACE mixed-bed format in this case. In contrast, all of the tested ACE mixed-beds provided superior 2D separation orthogonality through the combination of moderately reduced SCX retention for neutral to basic peptides and the increased retention for acidic peptides (data not shown).
The 2D view plot of a WAX/SCX tandem column (Figure 5 (F)) is presented as further evidence that the working principle of the ACE mixed-bed format is based on the effect of salt cation/anion separation on peptide displacement from SCX resin. As discussed in the previous section, a WAX column placed upstream of a SCX column could serve as an ion filter that supplied a cation(NH4+)-rich salt pulse to the adjacent SCX phase. The effect of such a unique salt pulse was clearly visualized by comparing the 2D plots with and without the “filter” (Figure 5 (F) and (D), respectively). Under these conditions which were identical to the conventional SCX format except for the presence of an upstream WAX column, the elution of the peptides was considerably facilitated when using the “filter” format (Figure 5 (F)). The comparison of the 2D plots of commonly identified peptides is shown in Figure S-4 to highlight the differences more distinctly. We believe this is strong evidence supporting our hypothesis that the increased concentration of salt cations produced by their separation from their original counter ions greatly affects the elution of peptides from SCX resins. At present, the contribution of ion-paired peptides on the hydrophobic elution behavior is unclear since it could not be separated from the hydrophobic property of backbone peptides. This question as well as a better understanding of the relationships between improved 2D orthogonality and peptide recovery as a function of blend ratios will be explored in future work.
Application to phosphopeptides analysis
The ACE mixed-bed system was also applied to the analysis of phosphopeptides from a HeLa nuclear extract enriched using a precipitation method that will be described elsewhere.30 Considering the acidic nature of phosphorylated peptides, it was logical to think that the ACE mixed-bed system would work well for enriching phosphopeptides. In fact, anion-exchange resins have been used for the offline pre-fractionation of phosphopeptides prior to their enrichment by IMAC37 (not as a mixed-bed). To the best of our knowledge, however, online usage of anion-exchange for the enrichment of phosphopeptides has not yet been reported. The pKa of phosphate groups is lower than that of the buffer system used (pH ∼2.6 with 0.1% formic acid). Thus, we modified the last salt step using a low pH (∼1.73) buffer containing a high concentration of salt (500 mM ammonium acetate) to recover phosphopeptides from the ACE mixed-bed resin.
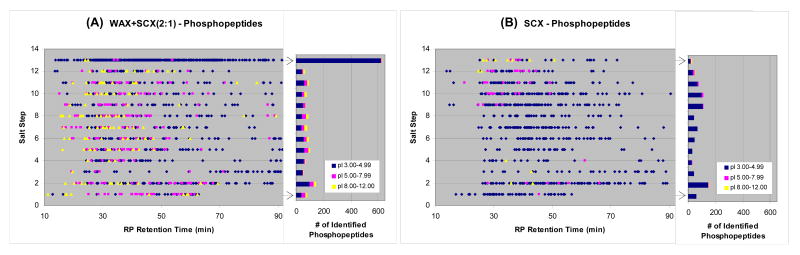
The advantages of the ACE mixed-bed for phosphopeptides analysis over SCX are highlighted in Figure 6, which shows the 2D profiles of phosphopeptides identified by the two formats. The distinct contrasts between the two methods were the enrichment of acidic phosphopeptides (pI 3.00-4.99 as backbone peptide) and the number of total phosphopeptides identified. Interestingly, the identification of neutral to basic phosphopeptides was also distinctly improved by the ACE mixed-bed format, presumably due to the improved recovery of the method. As expected, the ACE mixed-bed exhibited an ability to enrich phosphopeptides, especially those with a low pI, which was evidenced by the dense dots (left panel) and the highest bar (right panel) in Step 13 where the combination of low-pH and high-concentration salt pulse was applied. At this time, very acidic phosphopeptides were eluted in one salt step and not further fractionated due to a concern about the durability of silica-based packing resins for repeated injections of very acidic buffers. The use of acid-resistant packing materials will solve this problem and allow further fractionation of those acidic phosphopeptides. Since the total peak capacity can be improved by exploring the additional separation dimension, the optimization of elution buffer and packing materials (SCX, WAX and RP) should be considered in future research.
Figure 6. 2D distribution of phosphopeptides identified in (A) WAX+SCX(2:1) and (B) SCX formats using modified 13-step MudPIT.
Phosphopeptides enriched from HeLa cell nuclear extract30 were analyzed in the both ion-exchanger formats (∼50 μg/run). The phosphopeptides were identified from MS/MS spectra of either the precursor peptide or its neutral loss species, and the plots were generated by combining those identifications. The salt pulse for the Step 13 in this analysis was modified as follows: a 2 min salt pulse of 95% buffer D' + 5% buffer B, where buffer D' was water/acetonitrile (95:5, v/v) containing 500 mM ammonium acetate (pH 1.73 by trifluoroacetic acid). Peptide/protein identification criteria were: (1) one or more peptide identification per protein, (2) peptides must be fully tryptic, and (3) estimated false positive rates less than 0.5% at protein level as determined by a decoy reversed-database approach.
Table 1 is a summary of the comparative phosphopeptide analysis and includes results from the pH 4.6 fraction. Data regarding non-phosphopeptide-based identifications and the combination of phospho- and non-phosphopeptide-based identifications are also provided for further discussion. An obvious contrast was that the number of spectra that had passed the pre-filtering criteria and the number of identified peptides, and proteins were greater in the ACE mixed-bed format in all cases. From the fact that the phospho- and non-phosphorylated peptides exhibited similar fold increases, it was suggested that the improved peptide recovery was largely responsible for the improved identification of phosphopeptides. Due to the relatively high amount of sample analyzed (∼50 μg/analysis), the 2D separation planes were nearly fully occupied by both phospho- and non-phosphopeptides (Supporting Information Figure S-5). Thus, it was inferred that the orthogonality improvement might have had little contribution to the identification improvements of phosphopeptides in this specific example. However, it should be emphasized that its contribution was not subtle as seen in the phosphopeptide identifications from the pH 3.5 fraction. In this case, the fold increase of phosphopeptide identification (1.94) was greater than that of non-phosphorylated peptides (1.81), suggesting that the phosphopeptide analysis was indeed enhanced by the chromatographic phosphopeptide enrichment. The reason why this did not hold true for the pH 4.6 fraction was presumably because the fraction included less acidic peptides that could be enriched by the format (data not shown). From the principle of the enrichment method30 used in this evaluation, the pH 4.6 fraction undoubtedly included fewer acidic phosphopeptides than the pH 3.5 counterpart. Thus, the 2D chromatographic enrichment in this case would not be expected to have been as effective as that of the pH 3.5 fraction. In other words, the ACE mixed-bed format would perform best when combined with pH fraction-based phosphopeptide enrichment methods because it provides better separation for abundant acidic phosphopeptides.
Table 1. Summary of phosphopeptide analysis by the ACE mixed-bed systema).
| (1) Phosphopeptide-based Identification | ||||||
| pH 3.5 fractionb) | pH 4.6 fractionb) | |||||
|
|
|
|
||||
| Number of: |
WAX+SCX
(2:1) |
SCX | fold increasec) |
WAX+SCX
(2:1) |
SCX | fold increasec) |
|
|
|
|
||||
| Spectrad) | 2,566 | 1,777 | 1.44 | 1,870 | 1,383 | 1.35 |
| Peptides | 1,655 | 852 | 1.94 | 1,063 | 682 | 1.56 |
| Proteins | 569 | 349 | 1.63 | 435 | 288 | 1.51 |
| (2) Nonphosphopeptide-based Identification | ||||||
| pH 3.5 fractionb) | pH 4.6 fractionb) | |||||
|
|
|
|
||||
| Number of: |
WAX+SCX
(2:1) |
SCX | fold increasec) |
WAX+SCX
(2:1) |
SCX | fold increasec) |
|
|
|
|
||||
| Spectrad) | 24,939 | 16,172 | 1.54 | 24,271 | 14,929 | 1.63 |
| Peptides | 15,985 | 8,830 | 1.81 | 14,038 | 6,776 | 2.07 |
| Proteins | 2,804 | 2,037 | 1.38 | 2,682 | 1,792 | 1.50 |
| (3) Phospho- and Nonphosphopeptide combined Identifacation | ||||||
| pH 3.5 fractionb) | pH 4.6 fractionb) | |||||
|
|
|
|
||||
| Number of: |
WAX+SCX
(2:1) |
SCX | fold increasec) |
WAX+SCX
(2:1) |
SCX | fold increasec) |
|
|
|
|
||||
| Spectrad) | 26,792 | 17,952 | 1.49 | 25,392 | 16,228 | 1.56 |
| Peptides | 17,262 | 9,682 | 1.78 | 14,701 | 7,424 | 1.98 |
| Proteins | 2,891 | 2,115 | 1.37 | 2,731 | 1,863 | 1.47 |
Results of four independent 13-step MudPIT runs for two pH fractions by two ion-exchanger formats (∼50μg/run). Number of spectra, identified peptides and proteins are based on the sum of MS/MS and neutral loss MS/MS data. Database: EBI-IPI_human_3.04_03-07-2005, false positive rate: <0.5% at protein level (based on decoy database approach), minimum number of peptide to identify protein: 1, tryptic status requirements: complete tryptic peptides only.
Prepared following ref. (30) by Ruse CI et al.
As ratios of WAX+SCX(2:1) / SCX
Those passed the pre-filtering criteria
This application also serves to demonstrate that the ACE mixed-bed format holds advantages over SCX even when a faster, more sensitive mass spectrometer (LTQ vs. LCQ-Deca) is used for the analysis of relatively large amounts of peptide mixtures (∼50 μg/run). Although the fold increases of peptide identifications appeared slightly diminished compared to those obtained by a slower instrument (Figure 3 by LCQ-Deca), the results still support the suitability of the ACE mixed-bed format for broad proteomic applications using “real” samples. We did, however, notice that a further increase in sample amounts did not always improve protein identifications, an observation we attribute to peak broadening in the final RP separation. We believe that the improved peptide recovery by the ACE mixed-bed format can lead to an overloading of RP resins at the given conditions (resin chemistry, column bore size, ion-pair reagent, etc). To investigate such potential limitations of the method for future improvements, a follow-up study including loading capacity evaluation is currently underway. It should be emphasized, however, that the ACE mixed-bed format will be particularly useful when complex protein mixtures are to be analyzed but their amounts are limited which is often the case in many biological samples.
Conclusions
An advanced MudPIT format based on a novel concept is reported as a new approach to improve peptide identification and phosphopeptide analysis for online multidimensional LC/MS/MS analysis. The recovery of peptides from SCX resin and orthogonality in two-dimensional separations was significantly improved by replacing the SCX resin with a mixed-bed of anion-exchange and SCX (termed ACE mixed-bed for Anion- and Cation-Exchange). Experimental data suggested that the Donnan effect, which was generated inside the column by a pair of opposite fixed charges, was responsible for the improved peptide recovery as a result of higher fluxes of salt cations and a lower population of salt anions proximate to the SCX resin. Improved orthogonality was achieved by the combination of the increased retention of acidic peptides and the moderately reduced retention of neutral to basic peptides by the added anion-exchange resin. The proposed ACE mixed-bed system is effective, simple to implement and useful for a variety of analyses including online chromatographic enrichment of acidic phosphopeptides. Overall, the ACE mixed-bed format is an excellent alternative to the conventional SCX-based 2D LC/MS/MS analytical formats.
Supplementary Material
Additional information as noted in the text is available free of charge at http://pubs.acs.org/ac.
Acknowledgments
The authors thank Dr. Daniel Cociorva for his great and continuous supports on computational data processing and analysis, GE Healthcare (Piscataway, NJ) for providing the MDLC system equipped with a conductivity detector, Dr. Edwin P. Romijn for introducing the Kasil® frit generation protocol and Dr. Greg T. Cantin for his critical reading of the manuscript. AM is supported by Shiseido Co., Ltd. for his sabbatical research at The Scripps Research Institute. CIR is supported by NIH 5R01MH067880-02. JRY is supported by NIH 5R01MH067880-02 and P41RR11823-10.
References
- 1.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 2.Wolters DA, Washburn MP, Yates JR., 3rd Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 3.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 4.Giddings JC. Anal Chem. 1984;56:1258A–1260A. 1262A, 1264A. doi: 10.1021/ac00276a003. [DOI] [PubMed] [Google Scholar]
- 5.Gilar M, Daly AE, Kele M, Neue UD, Gebler JC. J Chromatogr A. 2004;1061:183–192. doi: 10.1016/j.chroma.2004.10.092. [DOI] [PubMed] [Google Scholar]
- 6.Issaq HJ, Chan KC, Janini GM, Conrads TP, Veenstra TD. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;817:35–47. doi: 10.1016/j.jchromb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 7.Gilar M, Olivova P, Daly AE, Gebler JC. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 8.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Motoyama A, Venable JD, Ruse CI, Yates JR., 3rd Anal Chem. 2006;78:5109–5118. doi: 10.1021/ac060354u. [DOI] [PubMed] [Google Scholar]
- 10.Garcia MC. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;825:111–123. doi: 10.1016/j.jchromb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 11.Cole RB, editor. Electrospray Ionization Mass Spectrometry: Fundamentals, Instrumentation, and Applications. John Wiley and Sons, Inc.; New York: 1997. [Google Scholar]
- 12.Simpson RJ. Purifying Proteins for Proteomics: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Woodbury, NY: Jan, 2004. [Google Scholar]
- 13.Cunico RL, Gooding KM, Wehr T. Basic HPLC and CE of Biomolecules. Bay Bioanalytical Laboratory, Inc.; Hercules, CA: 1998. [Google Scholar]
- 14.Burke TW, Mant CT, Black JA, Hodges RS. J Chromatogr. 1989;476:377–389. doi: 10.1016/s0021-9673(01)93883-x. [DOI] [PubMed] [Google Scholar]
- 15.Alpert AJ, Andrews PC. J Chromatogr. 1988;443:85–96. doi: 10.1016/s0021-9673(00)94785-x. [DOI] [PubMed] [Google Scholar]
- 16.Wagner Y, Sickmann A, Meyer HE, Daum G. J Am Soc Mass Spectrom. 2003;14:1003–1011. doi: 10.1016/S1044-0305(03)00399-4. [DOI] [PubMed] [Google Scholar]
- 17.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 18.MacNair JE, Lewis KC, Jorgenson JW. Anal Chem. 1997;69:983–989. doi: 10.1021/ac961094r. [DOI] [PubMed] [Google Scholar]
- 19.MacNair JE, Patel KD, Jorgenson JW. Anal Chem. 1999;71:700–708. doi: 10.1021/ac9807013. [DOI] [PubMed] [Google Scholar]
- 20.Tolley L, Jorgenson JW, Moseley MA. Anal Chem. 2001;73:2985–2991. doi: 10.1021/ac0010835. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Anal Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang K, Pasa-Tolic L, Veenstra TD, Lipton MS, Udseth HR, Smith RD. Anal Chem. 2001;73:1766–1775. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 23.Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. Anal Chem. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 24.el Rassi Z, Horvath C. J Chromatogr. 1986;359:255–264. doi: 10.1016/0021-9673(86)80079-6. [DOI] [PubMed] [Google Scholar]
- 25.Kunin R, McGarvey FX. Industrial & Engineering Chemistry. 1951;43 [Google Scholar]
- 26.Maa YF, Antia FD, el Rassi Z, Horvath C. J Chromatogr. 1988;452:331–345. doi: 10.1016/s0021-9673(01)81458-8. [DOI] [PubMed] [Google Scholar]
- 27.Fritz JS. J Chromatogr A. 2005;1085:8–17. doi: 10.1016/j.chroma.2004.12.087. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Irgum K. Anal Chem. 2002;74:4682–4687. doi: 10.1021/ac020293+. [DOI] [PubMed] [Google Scholar]
- 29.Jones EW. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 30.Ruse, C. I.; McLuchy, D. B.; Park, S. K.; Cociorva, D.; Lu, B.; Motoyama, A.; Wohlschlegel, J.A.; Yates, J. R., 3rd (manuscript in preparation).
- 31.Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., 3rd Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 32.Bern M, Goldberg D, McDonald WH, Yates JR., 3rd Bioinformatics. 2004;20 1:I49–I54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 33.Cociorva, D.; Xu, T.; Yates, J. R., 3rd (manuscript in preparation).
- 34.Dai J, Shieh CH, Sheng QH, Zhou H, Zeng R. Anal Chem. 2005;77:5793–5799. doi: 10.1021/ac050251w. [DOI] [PubMed] [Google Scholar]
- 35.Scigelova M, Makarov A. Proteomics. 2006;6:16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]
- 36.Yates JR, Cociorva D, Liao L, Zabrouskov V. Anal Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 37.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Mol Cell Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information as noted in the text is available free of charge at http://pubs.acs.org/ac.