Abstract
Site-specific conjugation of small molecules and enzymes to monoclonal antibodies has broad utility in the formation of conjugates for therapeutic, diagnostic, or structural applications. Precise control over the location of conjugation would yield highly homogenous materials that could have improved biological properties. We describe for the first time chemical reduction and oxidation methods that lead to preferential cleavage of particular monoclonal antibody interchain disulfides using the anti-CD30 IgG1 monoclonal antibody cAC10. Alkylation of the resulting cAC10 cysteine thiols with the potent antimitotic agent monomethyl auristatin E (MMAE) enabled the assignment of drug conjugation location by purification with hydrophobic interaction chromatography followed by analysis using reversed-phase HPLC and capillary electrophoresis. These analytical methods demonstrated that treating cAC10 with reducing agents such as DTT caused preferential reduction of heavy-light chain disulfides, while reoxidation of fully reduced cAC10 interchain disulfides caused preferential reformation of heavy-light chain disulfides. Following MMAE conjugation, the resulting conjugates had isomeric homogeneity as high as 60−90%, allowing for control of the distribution of molecular species. The resulting conjugates are highly active both in vitro and in vivo, and are well tolerated at efficacious doses.
Monoclonal antibodies (mAbs) have been used extensively as carriers of fluorophores, radionuclides, cytotoxic agents, and enzymes, yielding conjugates that find utility in therapeutic (1-3) and imaging applications (4, 5), ELISA-based assays (6), as well as for the investigation of protein structure and dynamics (7). The methods employed for making mAb-based conjugates can be classified in two general categories: those that involve the random modification of mAb amino acid residues, and those that are highly regioselective. Examples of random modification procedures include the acylation of lysine ε-amino groups (8), alkylation of tyrosines (9), and amidation of carboxylates (10). The biological and functional properties of these conjugates are often acceptable, however random modification of mAbs may impair antigen binding and leads to conjugate heterogeneity.
In the past several years, a number of selective methods have been described to introduce molecules of interest onto mAbs. The ability to control the location and stoichiometry of conjugation can significantly improve the properties of mAb conjugates in some applications. The greatest selectivities are obtained using recombinant technologies for the production of fusion proteins (11-14). Selective modification has also been reported for such chemically based methods as reductive amination of oxidized mAb carbohydrates (15), photoaffinity labeling of unconventional mAb binding sites (16), and reduction-alkylation of antibody interchain disulfides (17, 18).
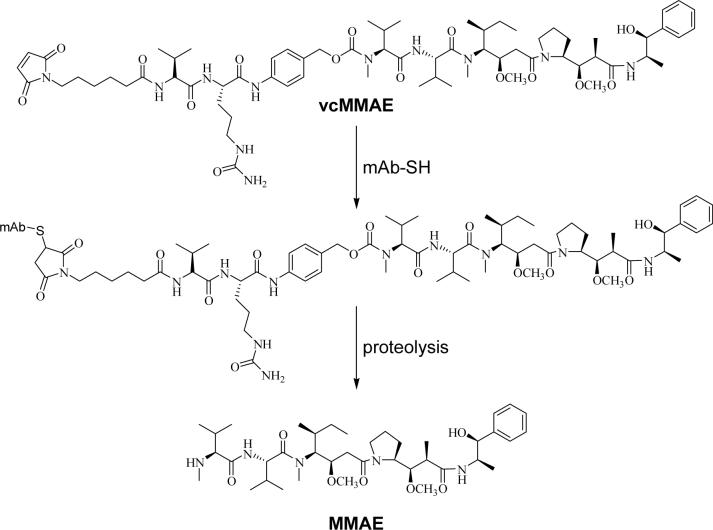
We have previously described the preparation of mAb-drug conjugates for use as antitumor agents (17, 19). The potent antimitotic agent monomethyl auristatin E (MMAE) was conjugated to the chimeric anti-CD30 mAb cAC10, an IgG1 mAb with 4 interchain disulfides (Figure 1). Conjugates were formed through full reduction of all interchain disulfides, followed by alkylation with the drug-linker complex. The resulting mAb-drug conjugates were homogeneous in composition, with about 8 drugs/mAb. Since mAb interchain disulfides are distant from the antigen binding site and are generally not required to maintain mAb integrity (20), this site-specific conjugation strategy yielded conjugates that were potent and selective for CD30-positive hematologic malignancies (17, 19).
1.
Conjugation strategy. The drug-linker vcMMAE reacts with a mAb cysteine to form the ADC. The potent antimitotic agent MMAE is released from the ADC following proteolysis. As many as 8 molecules of vcMMAE can react with each mAb following reduction of the 4 interchain disulfides present in cAC10.
We recently demonstrated that drug-load stoichiometry significantly influenced conjugate pharmacokinetics, and that conjugates with fewer drugs/mAb had larger therapeutic windows (21). Specifically, conjugates with 4 drugs/mAb were highly active and significantly less toxic than their counterparts with 8 drugs/mAb. However, such partially-loaded conjugates are not homogeneous, and the number of drugs on each mAb vary from 0−8, with several isomers at each drug substitution level. In order to minimize the heterogeneity of these partially loaded conjugates with 4 drugs/mAb, we explored various reduction/alkylation strategies and evaluated the distribution of species formed. We recognized that the absolute drug loading and the isomeric distribution could play a role in efficacy and toxicity. However, the literature does not describe how to chemically control the isomeric distribution of drug loading, nor does it illustrate how to determine which of the various mAb thiols are drug substituted. To address these issues, analytical technologies were established to determine the sites of drug substitution, and conjugation methods were developed that allowed for isomeric homogeneities as high as 60−90%. The in vitro and in vivo properties of these conjugates are also described.
Materials and Methods
Materials
cAC10, vcMMAE, and cAC10 with 8 MMAE/mAb (E8) were prepared as previously described (17, 22). DTT, DTPA, and 4,4’-dithiodipyridine were obtained from Sigma-Aldrich (St. Louis, MO). EDTA and sodium chloride were obtained from Cambrex (Rockland, ME). Sodium borate, sodium phosphate, and citric acid were obtained from Mallinckrodt (Phillipsburg, NJ). DTNB was obtained from Pierce (Rockford, IL). TCEP and aminoethanethiol were obtained from Acros (Morris Plains, NJ). Cysteine was obtained from Alfa Aesar (Ward Hill, MA).
Preparation of ADCs
cAC10-vcMMAE with an average of 4 MMAE/mAb, referred to as E4 mixture, was prepared as follows. Methods A and D were used to make cAC10-vcMMAE with 2 MMAE/mAb, referred to as E2 mixture, with the indicated substitutions.
Method A: Limited DTT Reduction
cAC10 was treated with 3.25 molar equivalents of DTT (2.25 molar equivalents for E2 mixture) in 0.025 M sodium borate pH 8, 0.025 M NaCl, 1 mM DTPA for 2 h at 37 °C. The excess DTT was purified away from the partially reduced cAC10 in the following manner. The mixture was first diluted 5 fold with water and applied to a hydroxyapatite column (Macroprep ceramic type I 40 μm, BioRad, Hercules, CA) at a flow rate of 10 mL/min. The column size was 1 mL per 10 mg of cAC10. The column was previously equilibrated with 5 column volumes of 0.5 M sodium phosphate pH 7, 10 mM NaCl and 5 column volumes of 10 mM sodium phosphate pH 7, 10 mM NaCl. Following application, the column was washed with 5 column volumes of 10 mM sodium phosphate pH 7, 10 mM NaCl and then eluted with 100 mM sodium phosphate pH 7, 10 mM NaCl. DTPA was added to 1 mM following elution. The protein concentration was quantified using an absorbance value of 1.58 at 280 nm for a 1.0 mg/mL solution, and the molar concentration determined using a molecular weight of 148,449 g/mol. The concentration of mAb-cysteine thiols produced was determined by titrating with DTNB, typically resulting in 4.0 to 4.5 thiols/mAb when 3.25 molar equivalents of DTT were used.
Partially reduced cAC10 was alkylated with 1.1 molar equivalents of vcMMAE/mAb-cysteine thiol. The alkylation reaction was performed at 0 °C for 30 min. Cysteine (1 mM final) was used to quench any unreacted, excess vcMMAE. cAC10-vcMMAE was purified by hydroxyapatite chromatography as described above. Following elution, the buffer was changed to phosphate buffered saline (Invitrogen, Carlsbad, CA) using Amicon (Millipore, Bedford, MA) Ultrafree 30K cutoff spin concentration devices. The protein concentration was quantified using an absorbance value of 1.62 at 280 nm for a 1.0 mg/mL solution.
Method B: Limited TCEP Reduction
cAC10 was treated with 2.75 molar equivalents of TCEP in 0.025 M sodium borate pH 8, 0.025 M NaCl, 1 mM DTPA for 2 h at 37 °C. Without purification, the mixture was then cooled to 0 °C and partially reduced cAC10 alkylated with vcMMAE as described above. cAC10-vcMMAE was purified using PD-10 columns (Amersham Biosciences, Piscataway, NJ) equilibrated with phosphate buffered saline.
Method C: Reduction of cAC10 with Aminoethanethiol
cAC10 was reduced with aminoethanethiol as previously described for gamma globulin (23). The reduced mAb was alkylated with vcMMAE as described above.
Method D: DTNB Reoxidation of Fully Reduced cAC10
cAC10 was treated with a large excess (10 mM) of DTT in 0.025 M sodium borate pH 8, 0.025 M NaCl, 1 mM DTPA for 1 h at 37 °C and purified by hydroxyapatite chromatography as described above. The fully reduced cAC10 was cooled to 0 °C and then treated with 2.0 equivalents of DTNB (3.0 for E2 mixture) or 4,4’-dipyridyldithiol (from 10 mM stock solutions) with rapid mixing. This mixture was incubated at 0 °C for 20 min. Without further purification, partially reoxidized cAC10 was alkylated with vcMMAE as previously described and the resulting conjugate was purified by hydroxyapatite chromatography as described above.
cAC10 Reduction Time Course
cAC10 was reduced with 3.0 equivalents of DTT in 50 mM sodium phosphate pH 7.5 and 5 mM EDTA at 37 °C. At various times, samples were removed, quenched with an equal volume of ice-cold 200 mM sodium citrate pH 5, and purified using PD-10 columns equilibrated with phosphate buffered saline containing 5 mM EDTA. Reduced cAC10 was treated with vcMMAE as previously described and purified using PD-10 columns equilibrated with phosphate buffered saline.
Purification of E2, E4, and E6 by HIC
cAC10-vcMMAE was purified by HIC as previously described to yield ADCs with exactly 2, 4, or 6 MMAE/mAb (referred to as E2, E4, and E6, respectively) (21). Briefly, NaCl was added to cAC10-vcMMAE to give a final concentration of 2.0 M and this mixture applied to a phenyl HIC column. E2, E4, and E6 were eluted using step gradients of decreasing NaCl concentration. These purified fractions were analyzed by HIC-HPLC to determine drug loading purity as previously described (21, 24). HIC peaks corresponding to E2, E4, or E6 were identified as previously described: cAC10 and vcMMAE have distinct absorbance maxima (λmax = 280 and 248 nm, respectively) and overlaying the UV spectra of these peaks yields A248/280 ratios that increase with drug loading (21, 24). The HIC-HPLC method was also used to isolate small amounts of E2, E4, and E6.
Characterization of ADCs
The drug loading was estimated using the molar extinction coefficients at 248 and 280 nm of the mAb (9.413 × 104 and 2.3415 × 105 M−1 cm−1, respectively) and drug (1.5 × 103 and 1.59 × 104 M−1 cm−1, respectively) as previously described (21, 24). Size-exclusion HPLC and C12 reversed-phase HPLC were used as previously described (17) to determine the amount of aggregate and free vcMMAE, respectively, present in the ADCs.
Analysis of ADCs by Capillary Electrophoresis
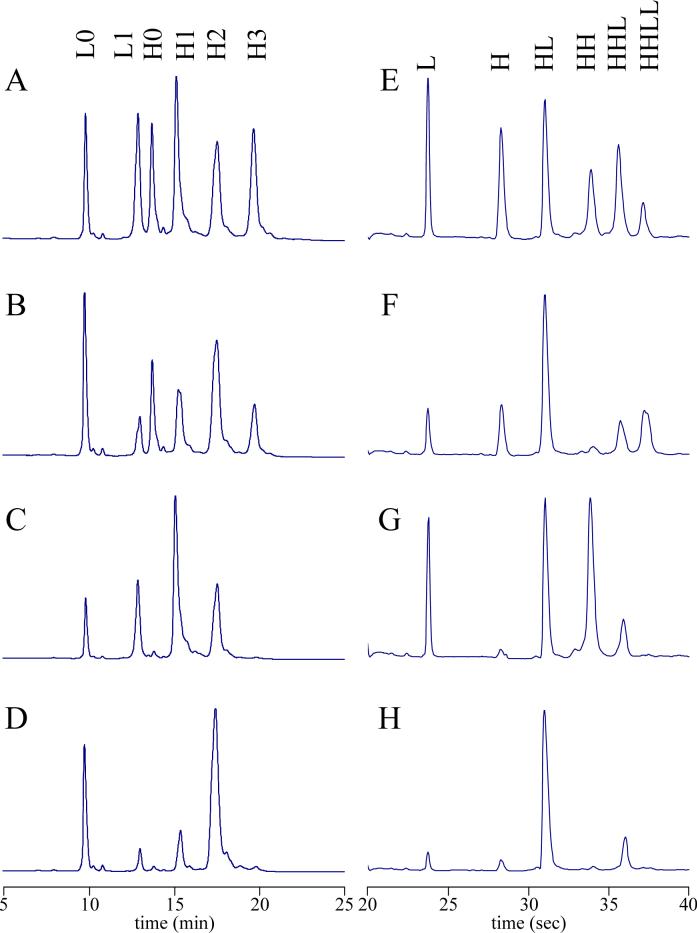
The ADCs were analyzed under denaturing but non-reducing conditions using an Agilent Bioanalyzer (Palo Alto, CA). A protein 200 chip was used under denaturing but non-reducing conditions as described by the manufacturer. Briefly, 4 μL of 1 mg/mL cAC10-vcMMAE was mixed with 2 μL of non-reducing loading buffer and heated to 100 °C for 5 min. Water (84 μL) was added and 6 μL of this mixture was loaded into each well of the chip. The molecular weights of peaks were determined using the molecular weight ladder provided and run concurrently, allowing assignment of L, H, HL, HH, HHL, and LHHL. Unconjugated cAC10 confirmed the assignment of LHHL, while E8 confirmed the assignment of L and H.
Analysis of ADCs by Reversed-Phase HPLC
ADCs were analyzed under denaturing and reducing conditions using a 2.1 × 150 mm, 8μ, 1000 Å PLRP-S column (Polymer Laboratories, Amherst, MA). The flow rate was 1 mL/min and the column temperature was 80 °C. Solvent A was 0.05% trifluoroacetic acid in water and solvent B was 0.04% trifluoroacetic acid in acetonitrile. The method consisted of the following: Isocratic 25% B for 3 min, a 25-min linear gradient to 50% B, a 2-min linear gradient to 95% B, a 1-min linear gradient to 25% B, and isocratic 25% B for 2 min. Injections were 10−20 μL of 1 mg/mL cAC10-vcMMAE previously reduced with 20 mM DTT at 37 °C for 15 min to cleave the remaining interchain disulfides. The number of moles and mole fraction of each chain was determined using the following molar extinction coefficients: 30,160 M−1 cm−1 (L0); 31,660 M−1 cm−1 (L1); 86,915 M−1 cm−1 (H0); 88,415 M−1 cm−1 (H1); 89,915 M−1 cm−1 (H2); 91,415 M−1 cm−1 (H3). Peak assignments were made with unconjugated cAC10 (L0 and H0) and E8 (L1 and H3). H1 and H2 were assigned by their elution time (eluting between H0 and H3) and UV spectra (the A248/280 ratio increases with drug loading).
Determination of Isomeric Distribution
The isomeric distribution for E2 and E6 was determined using solely reversed-phase HPLC data from the mole fractions determined above. For E2 isomer A, the mole fraction of L0 and H1 are equal, while for E2 isomer B, the mole fraction of L0, L1, H0, and H1 are equal. With the sum of the mole percent of L0, L1, H0, and H1 set to 100%, the percent isomer B can be determined as follows:
| (1) |
where %L1 = mole fraction of L1 and %H0 = mole fraction H0. The percent of isomer A is assumed to be 100 - %B.
Similarly, for E6 isomer A, the mole fraction of L1 and H2 are equal, while for E6 isomer B, the mole fractions of L0, L1, H2, and H3 are equal. With the sum of the mole percent of L0, L1, H2, and H3 set to 100%, the percent isomer B can be determined as follows:
| (2) |
where %L0 = mole fraction of L0 and %H3 = mole fraction H3. The percent of isomer A is assumed to be 100 - %B.
Due to more possible isomers, the percentages of the E4 isomers cannot be obtained solely from the reversed-phase HPLC data because no isomer gives a unique light and heavy chain pattern. The amount of E4 isomer A was first determined from capillary electrophoresis data using the HHL, HH, and HL peak areas as follows:
| (3) |
With the sum of the mole percent of L0, L1, H1, and H2 set to 100%, reversed-phase HPLC data was then used to solve for the remaining contribution of E4 isomers B and C using the following formulas:
| (4) |
| (5) |
where %H1 = mole fraction of H1, %L1 = mole fraction L1, %H2 = mole fraction H2, and %L0 = mole fraction L0.
Simulations
The simulated partial reduction of cAC10 was carried out using a series of coupled ordinary differential equations (25) with an integration algorithm (4th order Runge-Kutta) implemented as a VBA function in Excel (Microsoft Corp., Redmond, WA). The rate constants for reduction of heavy-light and heavy-heavy disulfides were equal. The simulation assumes 2 equivalent heavy-light and 2 equivalent heavy-heavy disulfides per mAb, with the relative rate constants for the reduction of the first and second of these equivalent disulfides being 2:1 due to the 2:1 stoichiometry (for example, when the first heavy-light disulfide is reduced, there are 2 heavy-light disulfides present, whereas when the second heavy-light disulfide is reduced, there is only 1 heavy-light disulfide present). The simulation used a concentration of reductant sufficient to reduce an average of 2 disulfides per mAb. Figures S1 and S2 show the mechanism and the differential equations used in the simulation.
Binding Studies
ADCs were labeled with 36 molar equivalents of europium N1-isothiocyanate (Perkin Elmer Biosciences, Wellesley MA) overnight at 4 °C in 50 mM Na2CO3, pH 9.0. Small molecular weight agents were removed by gel filtration on PD-10 columns equilibrated with Tris buffered saline (TBS, 50 mM Tris HCl, pH 8.0, 150 mM NaCl) followed by dialysis overnight in TBS at 4 °C. Conjugate concentration was determined by UV absorbance. Analysis of the europium content (by release into DELFIA enhancement solution, Perkin Elmer) indicated that there were 5 − 7 europium labels/mAb. Karpas 299 ALCL cells (100 μL/well at 1 × 106/mL in TBS + 0.5% FBS, 10 μM EDTA) were incubated with increasing dilutions of labeled ADC or parental cAC10 for 1 h at 4 °C on a plate rocker. Following incubation, the cells were washed 3 times in TBS, resuspended in 200 μL of enhancement solution, transferred to DELFIA yellow sided plates and incubated on an orbital shaker for 5 min at room temperature. Fluorescence was monitored using a Fusion HTS microplate reader (Perkin Elmer) using the time-resolved fluorescence mode using an excitation wavelength of 335 nm and emission wavelength set at 620 nm. The time delay between excitation and emission was 400 μsec. After reading, data were reduced and binding analyzed using the one-site binding model in GraphPad Prism version 4.01 for Windows (GraphPad Software, San Diego CA). Binding curves experiments were performed a minimum of three times to ensure accuracy.
Cytotoxicity Assays
The experiments were performed as previously described (17). Briefly, Karpas 299 cells were treated with ADCs for 92 h before adding resazurin and incubating an additional 4 h. Dye reduction was measured using a fluorescent plate reader.
In Vivo Studies
In vivo efficacy of ADCs in xenograft models of human CD30+ tumors in SCID mice and the maximum tolerated dose of ADCs in BALB/c mice were performed as previously described (21). Briefly, subcutaneous Karpas-299 tumors were grown in C.B.−17 SCID mice, with the test articles administered when tumors reached approximately 100 mm3 (length × width2)/2. Animals (7−10/group) were injected iv with the ADCs at the doses indicated. Animals were considered cured if they were tumor free 101 days post tumor implant. Groups were compared for statistical significance by pair wise treatment of Kaplan-Meier analysis using GraphPad Prism version 4.01. To evaluate the MTD, BALB/c mice (parent strain of the C.B.−17 SCID) were administered with ADCs. Animal weights were measured and clinical observations were recorded over a 14 day period. The MTD was assigned as the highest single dose administered to an animal that did not result in weight loss ≥20% or lead to any signs of distress.
Results
Reduction Methods
Two general strategies were used to prepare ADCs with an average of 4 MMAE/mAb (referred to as E4 mixture). The first involved limited reduction of the interchain disulfides of the chimeric mAb cAC10, an IgG1, followed by alkylation with maleimidocaproyl-val-cit-PABA-MMAE (vcMMAE). This two-step process led to the formation of cAC10-vcMMAE. About 3.25 and 2.75 molar equivalents of the strong reducing agents DTT and TCEP, respectively, were sufficient to cleave 2 interchain disulfide bonds, yielding an average of 4 cAC10 cysteines/mAb. Partial reduction of cAC10 could also be accomplished using the weaker reducing agent aminoethanethiol, which at pH 5 requires about 500 molar equivalents to yield 4 cAC10 cysteines/mAb. Unlike the thiol-based reducing agents, phosphines such as TCEP react poorly with maleimides, and the excess reducing agent does not have to be removed before adding vcMMAE.
A second general strategy for controlled drug substitution involves fully reducing cAC10 with DTT and then partially reoxidizing the resulting thiol groups with DTNB. This reoxidation process is very efficient, requiring 2.0 equivalents of DTNB to reoxidize 8 mAb cysteines back to 4. Treatment of this reoxidized mAb with a thiol such as cysteine does not liberate any bound thionitrobenzoic (TNB) acid, suggesting that the reoxidized cysteines are in the form of mAb disulfides rather than mixed TNB-cysteine disulfides. The remaining mAb cysteines can then be conjugated to vcMMAE.
Quantification of Drug Loading Levels
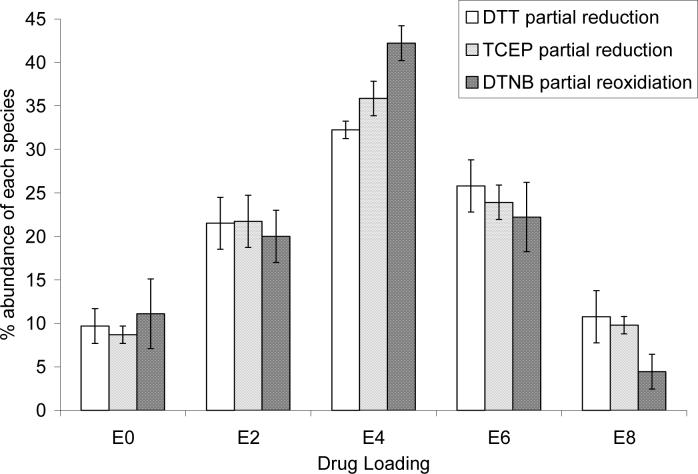
Figure 2 shows the normalized percentages of the even drug loaded species prepared by DTT partial reduction, TCEP partial reduction, and partial DTNB reoxidation as determined by HIC-HPLC. The DTNB partial reoxidation method yields a slightly higher percentage of E4 (42%) than the partial reduction methods (32% for DTT and 36% for TCEP). This comes at the expense of mainly E6 and E8, which total about 37% for DTT partial reduction and 34% for TCEP partial reduction, while only 27% for DTNB partial reoxidation.
2.
Composition of E4 mixture as a function of conjugation methodology. HICHPLC chromatograms were integrated to determine the abundance of E0, E2, E4, E6, and E8 in the mixtures. Values are ± standard deviation for 4 (DTT partial reduction), 3 (TCEP partial reduction), or 6 (DTNB partial reoxidation) separate experiments. The contributions from odd-loaded species, typically less than 10%, are not shown. The sum of E0, E2, E4, E6, and E8 was normalized to 100%. The simulated reduction assumes that the rate constants for reduction of the 4 interchain disulfides are equal for a random reduction process, and the sum of E0, E2, E4, E6, and E8 equals 100%.
Isomeric Populations
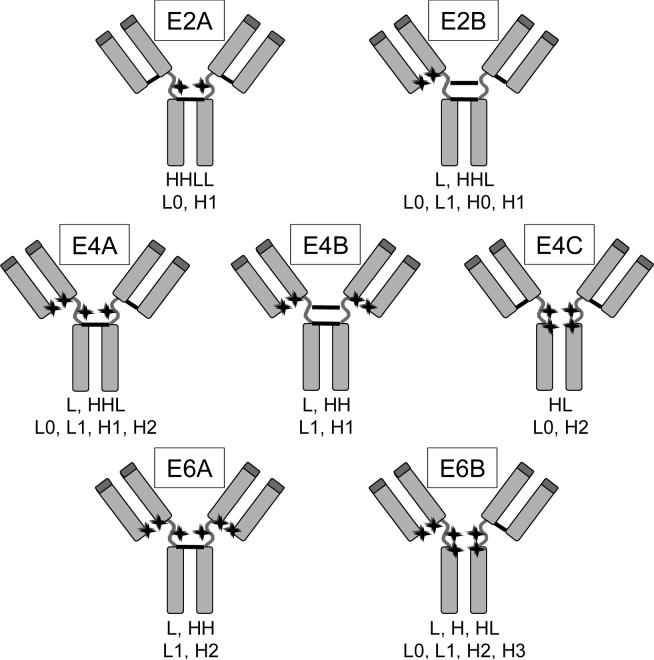
To determine the isomeric population of each of the drug loaded species, cAC10-vcMMAE prepared with various methods was first separated by preparative HIC, yielding E2, E4, and E6. The purities of these materials, with respect to their drug loading levels, was at least 95% as determined by analytical HIC-HPLC (21). The HIC-purified materials were then subjected a combination of reducing and non-reducing methods to assign the distribution of the drugs on the mAb. Reducing and denaturing reversed-phase HPLC was used to separate and quantify various light and heavy chain species. In this method, pretreatment of the ADC with an excess of DTT breaks the remaining interchain disulfides and allows separation of light chain with 0 or 1 drugs (L0 and L1) from heavy chain with 0, 1, 2, or 3 drugs (H0, H1, H2, and H3) (Figure 3, A-D). A second form of analysis involved non-reducing and denaturing capillary electrophoresis, allowing for the separation of 6 potential mAb species: L, H, HL, HH, HHL, and LHHL (Figure 3, E-H). Figure 4 shows the possible isomers of E2, E4, and E6 and the expected distribution of mAb fragments under denaturing conditions in both reducing and non-reducing environments. The isomeric populations of E2 and E6 were readily determined by reversed-phase HPLC since each isomer yields a unique fragment pattern. For example, isomer E2B yields L1 and H0 under denaturing and reducing conditions, while E2A yields L0 and H1. Under denaturing and non-reducing conditions, isomer E2A yields LHHL, while E2B yields L and HHL. For E4, the three potential isomers do not yield unique fragment patterns on reversed-phase HPLC or capillary electrophoresis alone, requiring that the combined results must be used to assign the isomeric population.
3.
Analytical characterization of ADCs. (A-D) Reversed-phase HPLC and (E-H) capillary electrophoresis were used to analyze (A, E) E4 mixture made by partial DTT reduction followed by MMAE conjugation; (B, F) E4 mixture made by DTNB partial reoxidation followed by MMAE conjugation; (C, G) E4 made by partial DTT reduction followed by MMAE conjugation and purified by preparative HIC ; and (D, H) E4 made by partial DTNB reoxidation followed by MMAE conjugation and purified by preparative HIC. HPLC injections were 20 μL of 1 mg/mL cAC10-vcMMAE treated with 20 mM DTT for 15 min at 37 °C. Separations were performed at 80 °C. Capillary electrophoresis was performed under non-reducing conditions.
4.
Potential isomers for E2, E4, and E6. Isomers with one heavy-heavy disulfide (2A, 4A, and 6A) can be further subdivided into isomers with either the first or the second disulfide intact, but because the analytical methods described herein cannot distinguish these subgroups, only one isomer is shown. Below the isomer are the chain compositions under denaturing conditions (first line, non-reducing; second line, reducing). For denaturing and non-reducing conditions, the possible species formed are L, H, HL, HH, HHL, and LHHL, where the remaining interchain disulfides link the indicated chains. For denaturing and reducing, the possible species formed are L0, L1, H0, H1, H2, and H3, in which the numbers indicate how many drug molecules are attached to the light or heavy chain. The locations of MMAE conjugation are indicated by stars. Unconjugated cAC10 (E0) and fully conjugated cAC10 (E8) are not shown.
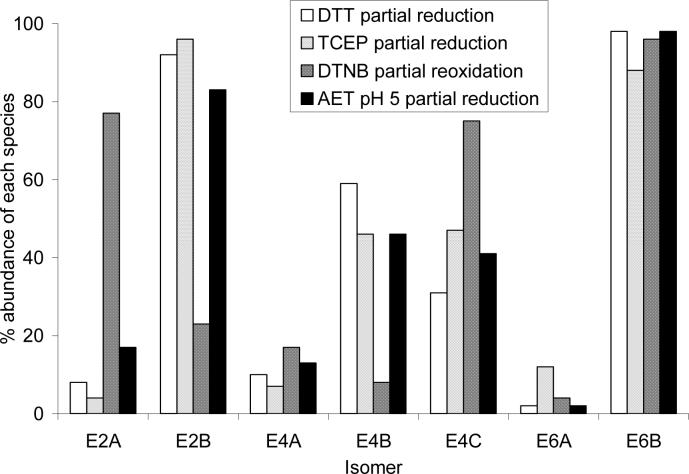
Figure 5 shows the percent composition for each of these isomers. Both partial reduction methods gave similar isomer populations. However, for E2, DTT partial reduction and DTNB partial reoxidation gave contrasting isomeric populations: the former method yielded 92% E2B, while the latter method yielded 77% E2A. Similarly, E4B is the predominant isomer for DTT partial reduction (59%), while E4C is the predominant isomer for DTNB partial reoxidation (75%). The chemistry used to prepare the reduced antibody plays a significant role in the location of the drugs in the resulting conjugate. Interestingly, both methods yield E6B as the predominant E6 isomer in very high yields (96−98%). There are also smaller differences in the distribution of isomers observed from the DTT and TCEP partial reduction methods, the largest of which can be seen in isomers E4B and E4C. DTT partial reduction yields about 2:1 E4B to E4C, while TCEP partial reduction yields about 1:1.
5.
Composition of isomeric population of purified E2, E4, and E6. The percent of the E2 and E6 isomers were determined from reversed-phase HPLC data using Equations 1 and 2, respectively. The percent of E4A was determined from capillary electrophoresis data using Equation 3 and then the percent of E4B and E4C were determined from reversed-phase HPLC data using Equations 4 and 5. For each drug loading level, the sum of the isomers is equal to 100%.
Simulated Partial Reduction
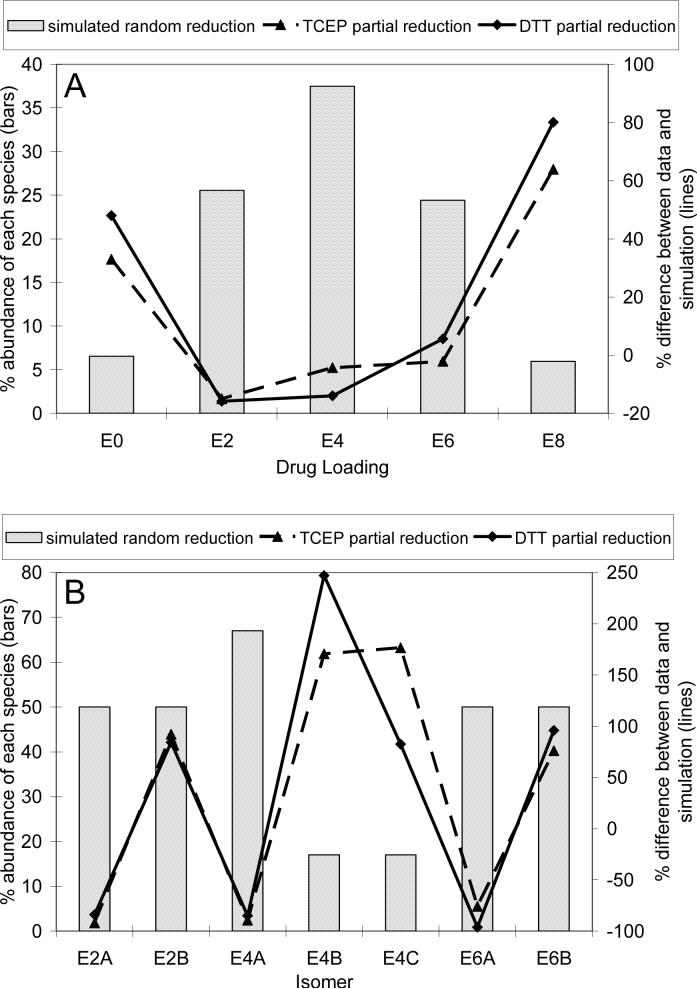
The observed drug loading levels and isomeric populations are significantly different for the partial reduction and partial reoxidation methods, which may indicate that there is some degree of selectivity in reducing and oxidizing mAb heavy-light and heavy-heavy disulfides. A simulation of cAC10 partial reduction was performed in which disulfides were allowed to be randomly reduced under conditions where the rate constants for the reduction of heavy-light and heavy-heavy disulfides were equal. Figure 6a shows the simulated drug loading levels for E0, E2, E4, E6, and E8 and the percent difference between these simulated values and the HIC-HPLC data for DTT and TCEP partial reduction. The percent difference varies systematically, in that the experimentally determined amounts of E0 and E8 were significantly larger than the simulated values, while the amounts of E2, E4, and E6 were slightly less than the simulated values. Figure 6b shows the simulated isomer populations and the percent difference between the simulation and the values determined from reversed-phase HPLC and capillary electrophoresis. The experimentally determined isomer abundances were as much as 250% more and up to nearly 100% less than the simulated values. No experimentally determined isomer abundance was within 75% of the simulated values. These results strongly suggest that the reduction is not a random process and the rate constants for reduction of individual cAC10 disulfides are widely different.
6.
Simulation of random reduction reaction. cAC10 reduction was simulated with equal rate constants for reduction of heavy-heavy and heavy-light disulfides. (A) The simulated drug loading abundance is shown with bars for the reduction of an average of 2 disulfides to yield an average of 4 antibody cysteines. The percent differences between the experimentally determined drug loading abundance from HIC-HPLC data for DTT and TCEP partial reduction shown in Figure 2 and the simulated values are shown in lines. (B) The simulated isomer distribution is shown in bars. The percent differences between the experimentally determined isomer abundances shown in Figure 5 and the simulated values are shown in lines. In both panels, a positive number indicates that the experimental data is greater than the simulated value, while a negative number indicates that the experimental data is lower than the simulated value.
DTT Reduction Time Course
Because of the long reduction time necessary to form partially reduced cAC10 with DTT, the isomeric population could change during the reduction reaction and using the end point of the reaction would not be representative of the overall reduction process. To address this, the isomeric compositions of the product ADCs were determined at various time intervals after the addition of the reducing agent. Reduction times of 10 to 120 min yielded drug loadings of 1.3 to 3.9 drugs/mAb. Table 1 shows that the isomeric composition does not change over the course of the reduction reaction. Thus, the reduction process is under kinetic rather than thermodynamic control, and using the end point of the reaction is appropriate.
Table 1.
Time course of isomer distribution for DTT partial reduction. Numbers given are in mole fraction. For each drug loading level, the sum of the isomers is equal to 100%.
| Time (min)a | Drugs/mAbb | E2Ac | E2Bc | E4Ad | E4Be | E4Ce | E6Af | E6Bf |
|---|---|---|---|---|---|---|---|---|
| 10 | 1.3 | 12 | 88 | 9 | 63 | 28 | N/D | N/D |
| 20 | 2.1 | 9 | 91 | 7 | 65 | 29 | 7 | 93 |
| 35 | 2.7 | 9 | 91 | 7 | 63 | 31 | 6 | 94 |
| 55 | 3.3 | 9 | 91 | 7 | 63 | 30 | 8 | 92 |
| 80 | 3.6 | 9 | 92 | 7 | 61 | 32 | 6 | 94 |
| 120 | 3.9 | 11 | 90 | 8 | 61 | 31 | 7 | 93 |
Reduction time. Once reduced, all antibodies were treated with vcMMAE for identical times.
Determined by HIC-HPLC.
Determined from reversed-phase HPLC data using Equation 1.
Determined from capillary electrophoresis data using Equation 3.
Determined from reversed-phase HPLC data using Equations 4 and 5.
Determined from reversed-phase HPLC data using Equation 2. N/D, not determined. At this time point, very little E6 was produced and this material was not sufficient for determining the isomer population.
Binding to CD30 Positive Cells
cAC10-vcMMAE conjugates were tested for their abilities to bind to CD30 positive Karpas 299 Anaplastic Large Cell Lymphoma cells. Table 2 shows that unmodified cAC10 and ADCs with 2 drugs/mAb bound with similar affinities, 2.0 to 3.6 nM. ADCs with 4 and 8 drugs/mAb bound slightly weaker, 6.5 to 7.8 nM. The chemistry used to prepare the ADCs did not significantly influence the ability of the ADCs to bind to the cells.
Table 2.
In vitro binding and cytotoxicity of cAC10-vcMMAE.
| ADC | Binding Kd (nM)a | Karpas 299 IC50 (ng/mL)b |
|---|---|---|
| E0 (cAC10) | 2.70 ± 1.91 | N/D |
| E2 mixture DTT | N/D | 11.4 ± 2.4 |
| E2 DTT | 3.57 ± 2.41 | 13.8 ± 3.6 |
| E2 mixture DTNB | N/D | 11.7 ± 4.5 |
| E2 DTNB | 2.02 ± 1.22 | 13.2 ± 2.7 |
| E4 mixture DTT | N/D | 3.4 ± 1.2 |
| E4 DTT | 7.76 ± 3.93 | 4.8 ± 0.7 |
| E4 mixture DTNB | N/D | 5.0 ± 0.0 |
| E4 DTNB | 7.69 ± 4.42 | 4.3 ± 0.9 |
| E8 | 6.53 ± 3.09 | 2.7 ± 0.2 |
Binding to Karpas 299, determined from 4−7 independent measurement ± standard deviation. N/D, not determined.
In vitro cytotoxicity, in ng cAC10/mL, determined from 3 independent measurements ± standard deviation. N/D, not determined. cAC10 alone displays poor activity against Karpas 299.
In Vitro Cytotoxicity
Mixtures with an average of 2 and 4 drugs/mAb as well as HIC purified E2 and E4 were investigated for their in vitro activities on Karpas 299 cells. Table 2 shows the ADCs with 2 and 4 drugs/mAb have IC50 values from 11.4 to 13.8 ng/mL and 3.4 to 5.0 ng/mL, respectively. E8 gave the lowest value of 2.7 ng/mL and unconjugated cAC10 showed no effect. Thus, cytotoxic activity increases with increased drug substitution, but is not dependent on ADC isomeric composition.
In Vivo Toxicity and Therapeutic Efficacy
MTD values in immunocompetent BALB/c mice were determined by iv ADC injections. As previously described, the MTD of E8 was 50 mg/kg (21). Significantly more of the ADCs could be administered with lower drug loading. The best tolerated ADC was E2 made by DTT partial reduction, which was devoid of toxicity at 250 mg/kg, the highest dose tested. The MTD of E4 prepared by the DTNB partial reoxidation method was 120 mg/kg, which was slightly less toxic than the corresponding E4 ADC prepared by DTT partial reduction (MTD of 100 mg/kg). Consistent with in vitro cytotoxicity, the number of drugs/mAb had a much more pronounced influence on toxicity than the placement of the drug on the mAb.
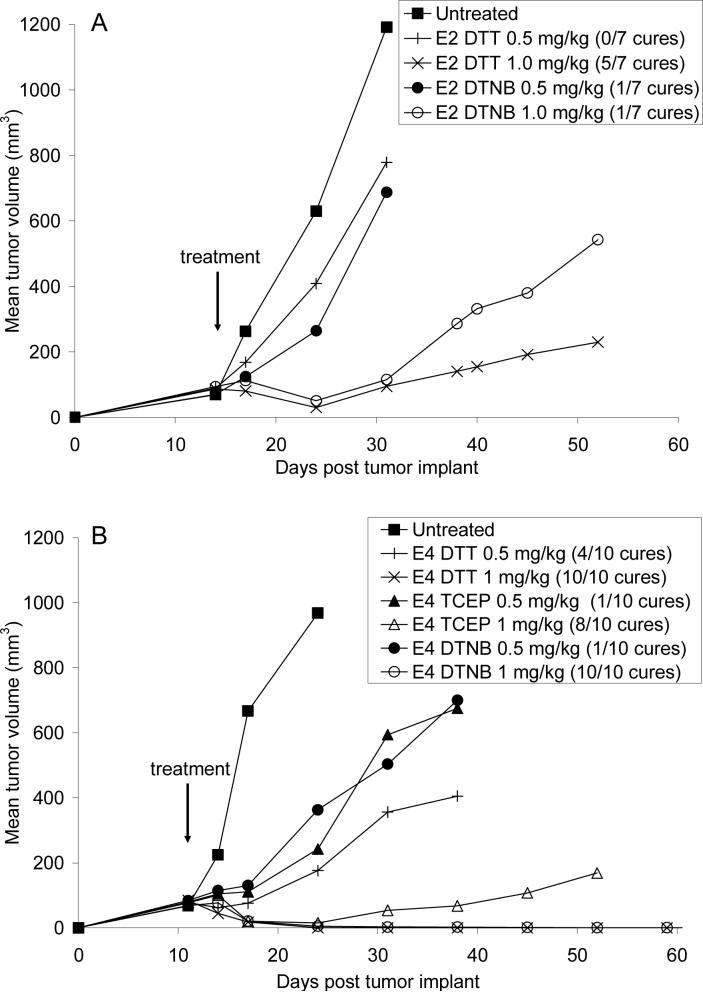
Therapeutic efficacy experiments were performed in SCID mice with subcutaneous Karpas 299 tumors. Single doses of the ADCs were given 11 (E4) or 14 (E2) days after tumor implant, when the tumors had reached approximately 100 mm3. Figure 7a shows that for E2 made by DTT partial reduction and DTNB partial reoxidation methods, 1.0 mg/kg of ADC both gave delays in tumor growth. Although E2 made by DTT partial reduction gave 5 cures out of 7 while E2 made by DTNB partial reoxidation provided 1 cure out of 7, Kaplan-Meier analysis for survival showed that there was not a statistical difference between these 2 chemistries (P=0.08). However, since the P value is trending towards significance (0.05<P<0.1), further studies are required with larger group sizes to determine if these chemistries yield differences in efficacy. The E4 ADCs which were made by DTT partial reduction, TCEP partial reduction, and DTNB partial reoxidation gave delays in tumor growth at both 0.5 and 1.0 mg/kg (Figure 7b). Nearly identical cure rates were obtained at 1 mg/kg for all 3 production methods (8 to 10 cures out of 10), suggesting that the production methods yielded ADCs with similar efficacies (P=0.15 for E4 made by TCEP partial reduction compared to E4 made by either DTT partial reduction or DTNB partial reoxidation).
7.
Single dose efficacy of E2 and E4. SCID mice with Karpas 299 tumors were treated with single doses of (A) E2 made by DTT partial reduction and DTNB partial reoxidation and (B) E4 made by DTT partial reduction, TCEP partial reduction, and DTNB partial reoxidation. Animals were considered cured if they were tumor free 101 days post tumor implant.
Discussion
We have demonstrated for the first time that IgG1 interchain disulfide bonds can be distinguished by chemical reduction and oxidization methods. This was made possible using the drug MMAE to fix the disulfide bonding pattern by alkylating free cysteines, allowing separation of the drug loading levels by HIC, and facilitating the analysis of the isomeric populations.
As shown in Figure 3, the production method significantly affects the location of the drugs, suggesting that the mAb disulfides can be selectively reduced. Partial DTT reduction yields predominantly isomer E2B, which results from reduction of one of the heavy-light chain disulfides. Conjugates consisting of E4 were comprised mainly of isomer E4B, which results from the reduction of both heavy-light chain disulfides. The main E6 species detected was isomer E6B, which results from reduction of both heavy-heavy chain disulfides and one heavy-light chain disulfide, while isomers with one heavy-heavy chain disulfide reduced were in the extreme minority. On the other hand, partial DTNB reoxidation yields almost the opposite result for E2 and E4 isomers, with predominantly isomers E2A and E4C, where one heavy-heavy chain disulfide is intact, and the same result for E6. We have observed qualitatively similar preferences of the isomer populations from partial reduction and partial reoxidiation of the humanized anti-Her2 IgG1 Herceptin (data not shown).
Previous investigations of mAb disulfide biochemistry did not demonstrate whether there was selective reduction of particular mAb interchain disulfides, in part because of the inability to fractionate the reduction reaction into subpopulations together with the low resolution of the analytical techniques employed. SDS-PAGE was used extensively (26-28), but this technique could not resolve and quantify the 9 possible isomers present in a mAb reduction reaction. Figure 3 shows that the E4 mixtures prepared by partial DTT reduction and partial DTNB reoxidation are difficult to distinguish, while HIC-purified E4 prepared by these chemistries are readily distinguished. In the past, analytical ultracentrifugation was also used to characterize mAb interchain disulfide reduction using aminoethanethiol under acidic conditions, and this data was used to suggest that exclusively heavy-heavy disulfides can be cleaved under these conditions (29-31). These data are even lower resolution than SDS-PAGE and cannot provide molecular characterization of the generated species. Our data are in contrast to this report, since partial reduction of cAC10 by aminoethanethiol at pH 5 results in a mixture of E4B and E4C present in nearly a 1:1 ratio (Figure 5), rather than only E4C, which would result from exclusive heavy-heavy disulfide reduction. The analytical technologies used in the previous reports (29-31) were evidently not sensitive enough to support the conclusions drawn.
The isomeric populations that result from partial DTT reduction demonstrate that IgG1 heavy-heavy disulfide bond cleavage is a cooperative process. Very little of any of the isomers with one heavy-heavy disulfide bond intact are observed, even at very early time points during the reduction (Table 1). The E2 isomer formed in greater than 90% abundance (E2B) has one heavy-light disulfide bond cleaved, while the predominant E4 isomer (E4B) has both of the heavy-light chain disulfides cleaved.
The simulated partial reduction with equal rate constants for heavy-light and heavy-heavy disulfide reduction confirm that the reduction of cAC10 is not a random process. The drug loading distribution and isomer distribution are significantly different between the simulation and experimental data, suggesting that a more complex model with unequal rate constants is much more appropriate in describing the cAC10 reduction process. We are currently performing additional simulations with other models to determine the rate constants that describe cAC10 reduction. These simulations will enable us to determine the relative rate constants of reduction of the heavy-light versus heavy-heavy disulfides, as well as the rate constants of reduction of the first versus the second heavy-light and heavy-heavy disulfides. Simulations may also be able to explain the small observed differences between DTT and TCEP partial reduction, with these two reducing agents potentially having different absolute reducing strengths as well as differences in their rate constants for reduction of individual disulfides.
Inspection of X-ray crystal structures (PDB structures include 1HZH and 1IGT) (20, 32, 33) show the region around these heavy-heavy disulfides to be highly flexible, and electron density is often absent. These disulfide bonds may be responsible for any local structure, and release of one may open up this region, allowing the second to be more exposed to solvent and reducing agents. The cleavage of these disulfides is not expected to cause large changes in the overall protein structure (20).
The partial DTNB reoxidation process preferentially re-forms the heavy-light chain disulfides, as was seen previously for air reoxidation (26). The E6 isomer formed in greater than 90% abundance (E6B) has one heavy-light disulfide, and the predominant E4 isomer (E4C) has both heavy-light disulfides. Unlike DTT reduction, reformation of the heavy-heavy disulfides does not appear to be cooperative, in that the predominant E2 isomer has just one heavy-heavy disulfide. Reformation of heavy-heavy disulfides appears to be merely kinetically slower than the heavy-light disulfides. If cleavage of the heavy-heavy disulfides releases the local structure and the previously paired cysteines can move away from each other, the kinetics of heavy-heavy disulfide bond formation will be slow. The heavy-light disulfide is probably not responsible for local structure or relative positioning of the heavy and light chains, and the unpaired cysteines will remain in close proximity and proper orientation for disulfide bond formation.
The chemistries described herein enable the generation of highly site-specific conjugates. For cAC10-vcMMAE, the effect of the location of the drugs on biological activity is modest, with the only significant difference being the MTD values of E4 made by DTNB partial reoxidation being higher than E4 made by DTT partial reduction. It is likely that other biologically interesting molecules such as fluorophores and fluorescence resonance energy transfer reagents may benefit from site-specific conjugation, and this is an area that we are currently investigating. The ability to control the molecular composition in ADCs by chemical means complements recombinant strategies, and allows for the generation of well defined ADCs with potent biological activities.
Supplementary Material
Acknowledgement
We thank Angela Merges, Sarah Hall, and Jamie Andreyka for technical assistance. This work was supported in part by Grant R44 CA088583-02A1 from the National Institutes of Health.
Abbreviations
- ADC
mAb drug conjugate
- DTNB
5,5’-dithiobis(2-nitrobenzoic acid)
- DTPA
diethyeletriaminepentaacetic acid
- DTT
dithiothreitol
- E0, E2, E4, E6, E8, cAC10
0, 2, 4, 6, 8 MMAE per mAb, respectively
- H0, H1, H2, H3, heavy chain
0, 1, 2, 3 MMAE, respectively
- HIC
hydrophobic interaction chromatography
- L0, L1, light chain
0, 1 MMAE, respectively
- mAb
monoclonal antibody
- MMAE
monomethyl auristatin E
- MTD
maximum tolerated dose
- TCEP
tris(2-carboxyethyl)phosphine
- vcMMAE
maleimidocaproyl-valine-citruline-PABA-MMAE
References
- 1.Senter PD, Springer CJ. Selective activation of anticancer prodrugs by monoclonal antibody-enzyme conjugates. Adv Drug Deliv Rev. 2001;53:247–64. doi: 10.1016/s0169-409x(01)00206-x. [DOI] [PubMed] [Google Scholar]
- 2.Sharkey RM, Goldenberg DM. Perspectives on cancer therapy with radiolabeled monoclonal antibodies. J Nucl Med. 2005;46(Suppl 1):115S–27S. [PubMed] [Google Scholar]
- 3.Trail PA, King HD, Dubowchik GM. Monoclonal antibody drug immunoconjugates for targeted treatment of cancer. Cancer Immunol Immunother. 2003;52:328–37. doi: 10.1007/s00262-002-0352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohdiewicz PJ. Indium-111 satumomab pendetide: the first FDA-approved monoclonal antibody for tumor imaging. J Nucl Med Technol. 1998;26:155–63. quiz 170−1. [PubMed] [Google Scholar]
- 5.Van de Wiele C, Revets H, Mertens N. Radioimmunoimaging. Advances and prospects. Q J Nucl Med Mol Imaging. 2004;48:317–25. [PubMed] [Google Scholar]
- 6.Porstmann T, Kiessig ST. Enzyme immunoassay techniques. An overview. J Immunol Methods. 1992;150:5–21. doi: 10.1016/0022-1759(92)90061-w. [DOI] [PubMed] [Google Scholar]
- 7.Lichlyter DJ, Grant SA, Soykan O. Development of a novel FRET immunosensor technique. Biosens Bioelectron. 2003;19:219–26. doi: 10.1016/s0956-5663(03)00215-x. [DOI] [PubMed] [Google Scholar]
- 8.Wisdom GB. Conjugation of antibodies to fluorescein or rhodamine. Methods Mol Biol. 2005;295:131–4. doi: 10.1385/1-59259-873-0:131. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson BM, Young NM, Oomen RP. Characterisation of residues in antibody binding sites by chemical modification of surface-adsorbed protein combined with enzyme immunoassay. J Immunol Methods. 1993;158:215–27. doi: 10.1016/0022-1759(93)90217-u. [DOI] [PubMed] [Google Scholar]
- 10.Wines BD, Easterbrook-Smith SB. Carbodiimide crosslinking of human C1q and rabbit IgG. Mol Immunol. 1990;27:221–6. doi: 10.1016/0161-5890(90)90133-k. [DOI] [PubMed] [Google Scholar]
- 11.Casey JL, Coley AM, Tilley LM, Foley M. Green fluorescent antibodies: novel in vitro tools. Protein Eng. 2000;13:445–52. doi: 10.1093/protein/13.6.445. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi N, Kipriyanov S, Fuchs P, Welschof M, Dorsam H, Little M. A single expression system for the display, purification and conjugation of single-chain antibodies. Gene. 1995;160:129–30. doi: 10.1016/0378-1119(95)00218-u. [DOI] [PubMed] [Google Scholar]
- 13.Schneider P. Production of recombinant TRAIL and TRAIL receptor: Fc chimeric proteins. Methods Enzymol. 2000;322:325–45. doi: 10.1016/s0076-6879(00)22031-4. [DOI] [PubMed] [Google Scholar]
- 14.Siemers NO, Kerr DE, Yarnold S, Stebbins MR, Vrudhula VM, Hellstrom I, Hellstrom KE, Senter PD. Construction, expression, and activities of L49-sFv-beta-lactamase, a single-chain antibody fusion protein for anticancer prodrug activation. Bioconjug Chem. 1997;8:510–9. doi: 10.1021/bc9700751. [DOI] [PubMed] [Google Scholar]
- 15.Rodwell JD, Alvarez VL, Lee C, Lopes AD, Goers JW, King HD, Powsner HJ, McKearn TJ. Site-specific covalent modification of monoclonal antibodies: in vitro and in vivo evaluations. Proc Natl Acad Sci U S A. 1986;83:2632–6. doi: 10.1073/pnas.83.8.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang IN, Lin JN, Andrade JD, Herron JN. Photoaffinity labeling of antibodies for applications in homogeneous fluoroimmunoassays. Anal Chem. 1995;67:959–66. doi: 10.1021/ac00101a025. [DOI] [PubMed] [Google Scholar]
- 17.Doronina SO, Toki BE, Torgov MY, Mendelsohn BA, Cerveny CG, Chace DF, DeBlanc RL, Gearing RP, Bovee TD, Siegall CB, Francisco JA, Wahl AF, Meyer DL, Senter PD. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat Biotechnol. 2003;21:778–84. doi: 10.1038/nbt832. [DOI] [PubMed] [Google Scholar]
- 18.Trail PA, Willner D, Lasch SJ, Henderson AJ, Hofstead S, Casazza AM, Firestone RA, Hellstrom I, Hellstrom KE. Cure of xenografted human carcinomas by BR96-doxorubicin immunoconjugates. Science. 1993;261:212–5. doi: 10.1126/science.8327892. [DOI] [PubMed] [Google Scholar]
- 19.Francisco JA, Cerveny CG, Meyer DL, Mixan BJ, Klussman K, Chace DF, Rejniak SX, Gordon KA, DeBlanc R, Toki BE, Law CL, Doronina SO, Siegall CB, Senter PD, Wahl AF. cAC10-vcMMAE, an anti-CD30-monomethyl auristatin E conjugate with potent and selective antitumor activity. Blood. 2003;102:1458–65. doi: 10.1182/blood-2003-01-0039. [DOI] [PubMed] [Google Scholar]
- 20.Saphire EO, Stanfield RL, Crispin MD, Parren PW, Rudd PM, Dwek RA, Burton DR, Wilson IA. Contrasting IgG structures reveal extreme asymmetry and flexibility. J Mol Biol. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- 21.Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, Kissler KM, Bernhardt SX, Kopcha AK, Zabinski RF, Meyer DL, Francisco JA. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10:7063–70. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 22.Wahl AF, Klussman K, Thompson JD, Chen JH, Francisco LV, Risdon G, Chace DF, Siegall CB, Francisco JA. The anti-CD30 monoclonal antibody SGN-30 promotes growth arrest and DNA fragmentation in vitro and affects antitumor activity in models of Hodgkin's disease. Cancer Res. 2002;62:3736–42. [PubMed] [Google Scholar]
- 23.Mandy WJ, Nisonoff A. Effect of Reduction of Several Disulfide Bonds on the Properties and Recombination of Univalent Fragments of Rabbit Antibody. Journal of Biological Chemistry. 1963;238:206–213. [Google Scholar]
- 24.Sanderson RJ, Hering MA, James SF, Sun MM, Doronina SO, Siadak AW, Senter PD, Wahl AF. In vivo drug-linker stability of an anti-CD30 dipeptide-linked auristatin immunoconjugate. Clin Cancer Res. 2005;11:843–52. [PubMed] [Google Scholar]
- 25.Olson JS. Numerical analysis of kinetic ligand binding data. Methods Enzymol. 1981;76:652–67. doi: 10.1016/0076-6879(81)76149-4. [DOI] [PubMed] [Google Scholar]
- 26.Sears DW, Beychok S. Acquisition of the covalent quaternary structure of an immunoglobulin G molecule. Theoretical reoxidation models. Biochemistry. 1977;16:2026–31. doi: 10.1021/bi00628a041. [DOI] [PubMed] [Google Scholar]
- 27.Sears DW, Mohrer J, Beychok S. Relative susceptibilities of the interchain disulfides of an immunoglobulin G molecule to reduction by dithiothreitol. Biochemistry. 1977;16:2031–5. doi: 10.1021/bi00628a042. [DOI] [PubMed] [Google Scholar]
- 28.Virella G, Parkhouse RM. Sensitivity to reduction of human immunoglobulin G of different heavy chain sub-classes. Immunochemistry. 1973;10:213–7. doi: 10.1016/0019-2791(73)90197-3. [DOI] [PubMed] [Google Scholar]
- 29.Hermanson G. Bioconjugate Techniques. Academic Press; San Diego: 1996. [Google Scholar]
- 30.Kato K, Hamaguchi Y, Fukui H, Ishikawa E. Enzyme-linked immunoassay. Conjugation of rabbit anti-(human immunoglobulin G) antibody with beta-D-galactosidase from Escherichia coli and its use for human immunoglobulin G assay. Eur J Biochem. 1976;62:285–92. doi: 10.1111/j.1432-1033.1976.tb10159.x. [DOI] [PubMed] [Google Scholar]
- 31.Palmer JL, Nisonoff A, Vanholde KE. Dissociation of Rabbit Gamma Globulin into Subunits by Reduction and Acidification. Proc Natl Acad Sci U S A. 1963;50:314–21. doi: 10.1073/pnas.50.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36:1581–97. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- 33.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–9. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.