Abstract
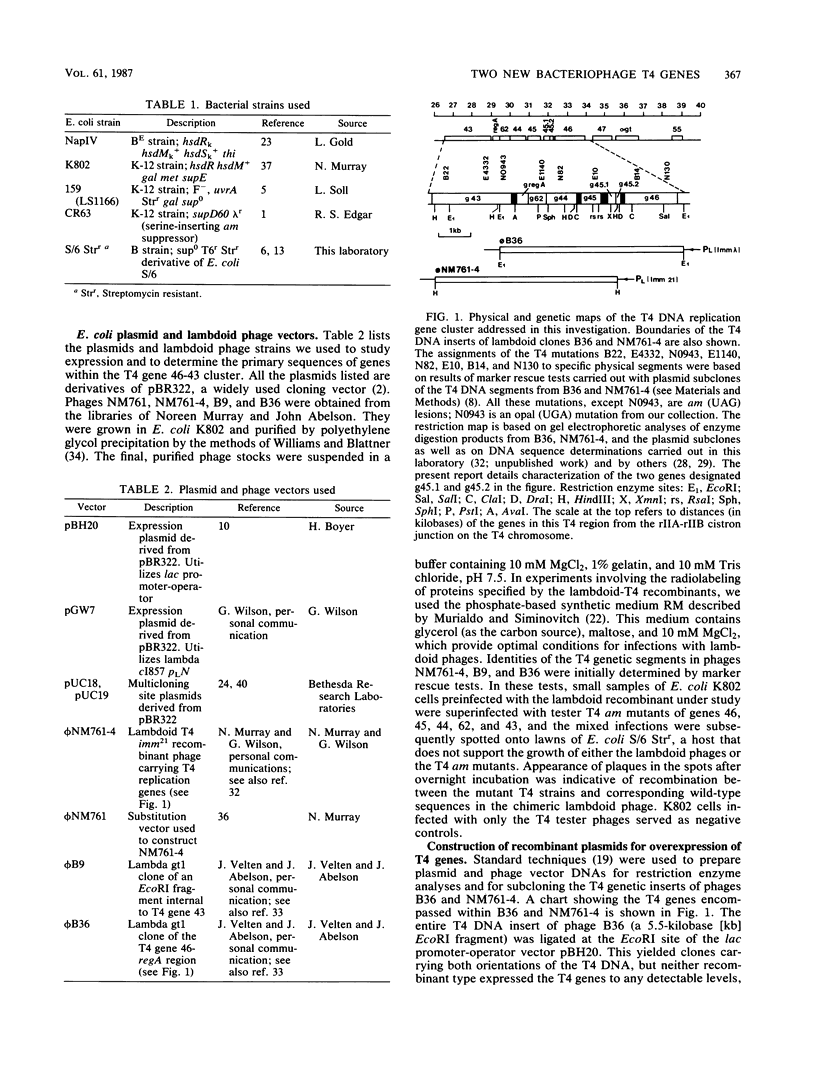
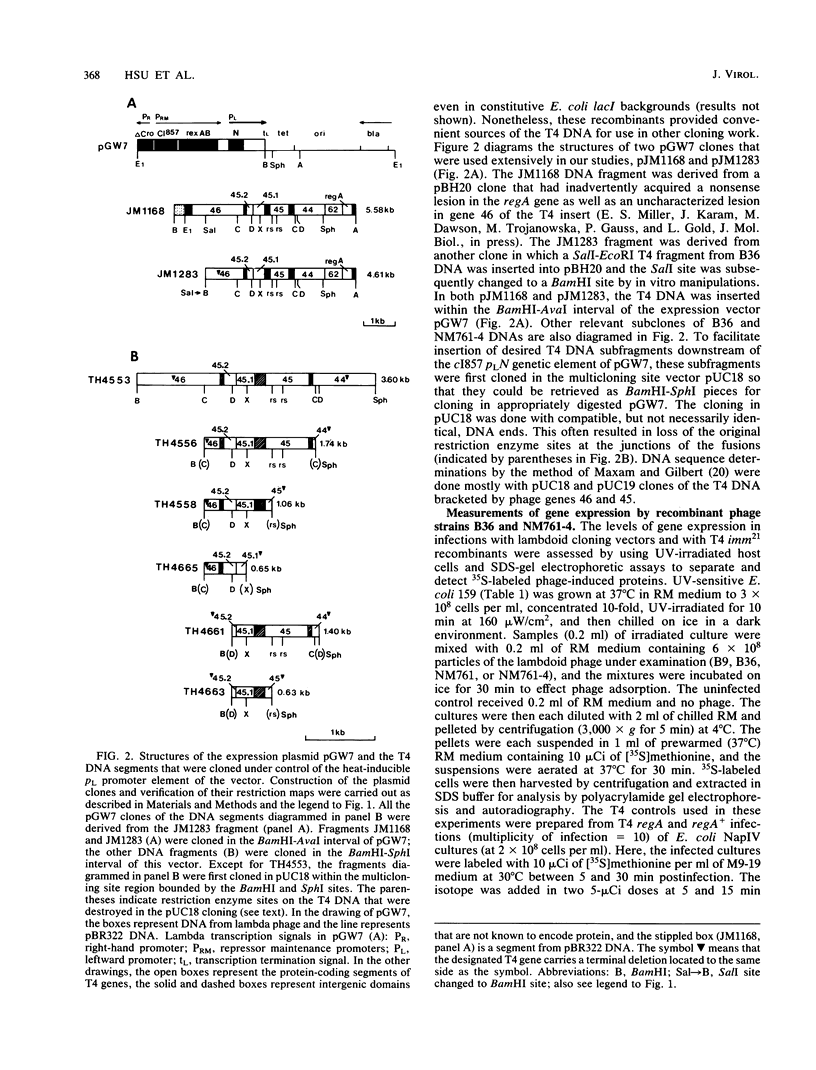
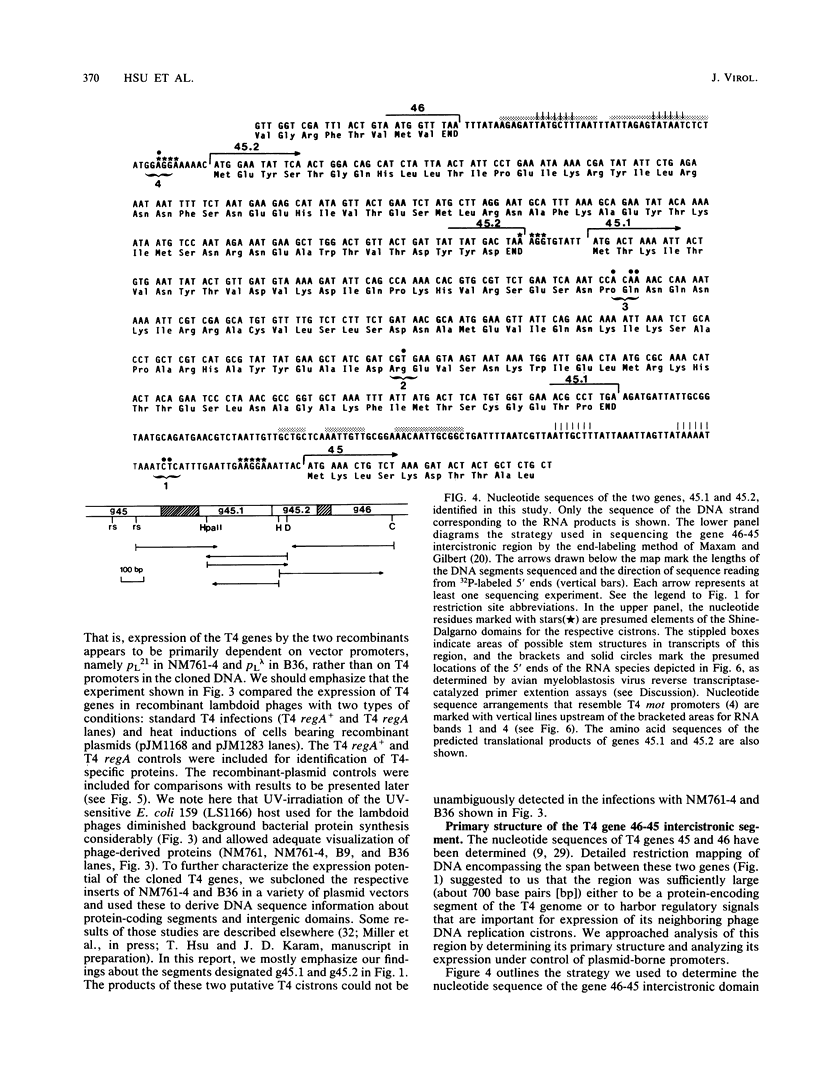
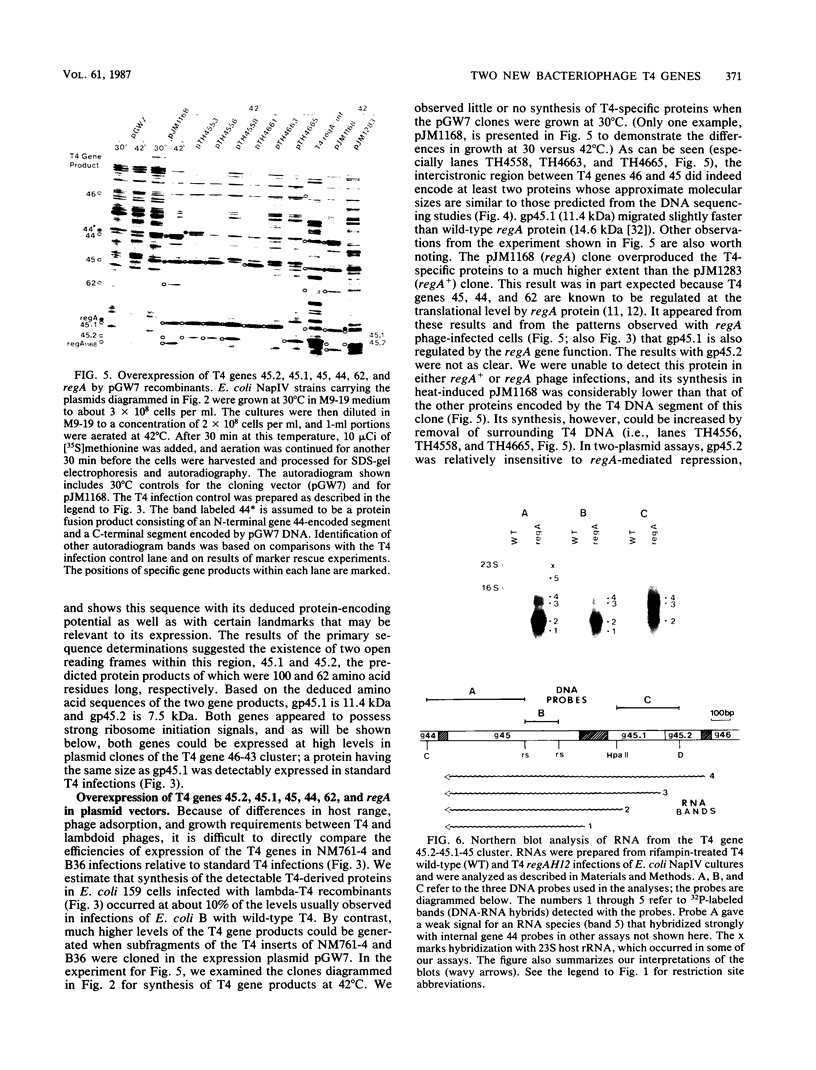
We have identified two bacteriophage T4 genes, 45.1 and 45.2, that map in the intergenic space between phage replication genes 46 (which encodes a recombination initiation protein) and 45 (which encodes a bifunctional protein required in replication and transcription). The existence of genes 45.1 and 45.2 had not been previously recognized by mutation analysis of the T4 genome. We cloned the T4 gene 45.1/45.2 segment, determined its nucleotide sequence, and expressed its two reading frames at high levels in bacterial plasmids. The results predicted molecular weights of 11,400 (100 amino acids) for gp45.1 and 7,500 (62 amino acids) for gp45.2. We also determined that in T4-infected Escherichia coli, genes 45.1 and 45.2 are cotranscribed with their distal neighbor, gene 45, by at least one mode of transcription. In an accompanying report (K. P. Williams, G. A. Kassavetis, F. S. Esch, and E. P. Geiduschek, J. Virol. 61:600-603, 1987), it is shown that the product of gene 45.1 is the so-called T4-induced 15K protein, an RNA polymerase-binding protein of unknown role in phage development. Possibly, T4 genes 45.2, 45.1, and 45 constitute an operon for host RNA polymerase-binding phage proteins. Jointly with Williams et al., we propose the term rpb (RNA polymerase-binding) to refer to T4 genes whose products bind to the host RNA polymerase and have adopted the name rpbA for T4 gene 45.1.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- APPLEYARD R. K., MCGREGOR J. F., BAIRD K. M. Mutation to extended host range and the occurrence of phenotypic mixing in the temperate coliphage lambda. Virology. 1956 Aug;2(4):565–574. doi: 10.1016/0042-6822(56)90012-5. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: requirements for late messenger synthesis. J Mol Biol. 1968 Apr 28;33(2):339–362. doi: 10.1016/0022-2836(68)90193-9. [DOI] [PubMed] [Google Scholar]
- Buchwald M., Murialdo H., Siminovitch L. The morphogenesis of bacteriophage lambda. II. Identification of the principal structural proteins. Virology. 1970 Oct;42(2):390–400. doi: 10.1016/0042-6822(70)90282-5. [DOI] [PubMed] [Google Scholar]
- Doermann A H, Hill M B. Genetic Structure of Bacteriophage T4 as Described by Recombination Studies of Factors Influencing Plaque Morphology. Genetics. 1953 Jan;38(1):79–90. doi: 10.1093/genetics/38.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald W. L., Karam J. D. Expression of a DNA replication gene cluster in bacteriophage T4: genetic linkage and the control of gene product interactions. Genetics. 1984 Aug;107(4):537–549. doi: 10.1093/genetics/107.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram H., Rüger W. Genes 55, alpha gt, 47 and 46 of bacteriophage T4: the genomic organization as deduced by sequence analysis. EMBO J. 1985 Jan;4(1):257–264. doi: 10.1002/j.1460-2075.1985.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura K., Hirose T., Crea R., Riggs A. D., Heyneker H. L., Bolivar F., Boyer H. W. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977 Dec 9;198(4321):1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- Karam J. D., O'Donnell P. V. Suppression of amber mutations of bacteriophage T4 gene 43 (DNA polymerase) by translational ambiguity. J Virol. 1973 Jun;11(6):933–945. doi: 10.1128/jvi.11.6.933-945.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam J., Bowles M., Leach M. Expression of bacteriophage T4 genes 45, 44, and 62. I. Discoordinate synthesis of the T4 45- and 44-proteins. Virology. 1979 Apr 15;94(1):192–203. doi: 10.1016/0042-6822(79)90449-5. [DOI] [PubMed] [Google Scholar]
- Karam J., McCulley C., Leach M. Genetic control of mRNA decay in T4 phage-infected Escherichia coli. Virology. 1977 Feb;76(2):685–700. doi: 10.1016/0042-6822(77)90251-3. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. J., Huang W. M. Identification of the origins of T4 DNA replication. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7248–7252. doi: 10.1073/pnas.79.23.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luder A., Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Dimitrov M., Goldfarb A. Initiation of transcription by bacteriophage T4-modified RNA polymerase independently of host sigma factor. J Mol Biol. 1985 Sep 5;185(1):83–91. doi: 10.1016/0022-2836(85)90184-6. [DOI] [PubMed] [Google Scholar]
- Malik S., Goldfarb A. The effect of a bacteriophage T4-induced polypeptide on host RNA polymerase interaction with promoters. J Biol Chem. 1984 Nov 10;259(21):13292–13297. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ratner D. Letter to the editor: Bacteriophage T4 transcriptional control gene 55 codes for a protein bound to Escherichia coli RNA polymerase. J Mol Biol. 1974 Nov 15;89(4):803–807. doi: 10.1016/0022-2836(74)90054-0. [DOI] [PubMed] [Google Scholar]
- Ratner D. The interaction bacterial and phage proteins with immobilized Escherichia coli RNA polymerase. J Mol Biol. 1974 Sep 15;88(2):373–383. doi: 10.1016/0022-2836(74)90488-4. [DOI] [PubMed] [Google Scholar]
- Spicer E. K., Noble J. A., Nossal N. G., Konigsberg W. H., Williams K. R. Bacteriophage T4 gene 45. Sequences of the structural gene and its protein product. J Biol Chem. 1982 Aug 10;257(15):8972–8979. [PubMed] [Google Scholar]
- Stevens A. New small polypeptides associated with DNA-dependent RNA polymerase of Escherichia coli after infection with bacteriophage T4. Proc Natl Acad Sci U S A. 1972 Mar;69(3):603–607. doi: 10.1073/pnas.69.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A., Rhoton J. C. Characterization of an inhibitor causing potassium chloride sensitivity of an RNA polymerase from T4 phage-infected Escherichia coli. Biochemistry. 1975 Nov 18;14(23):5074–5079. doi: 10.1021/bi00694a007. [DOI] [PubMed] [Google Scholar]
- Trojanowska M., Miller E. S., Karam J., Stormo G., Gold L. The bacteriophage T4 regA gene: primary sequence of a translational repressor. Nucleic Acids Res. 1984 Aug 10;12(15):5979–5993. doi: 10.1093/nar/12.15.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten J., Fukada K., Abelson J. In vitro construction of bacteriophage lambda and plasmid DNA molecules containing DNA fragments from bacteriophage T4. Gene. 1976;1(1):93–106. doi: 10.1016/0378-1119(76)90009-3. [DOI] [PubMed] [Google Scholar]
- Williams B. G., Blattner F. R. Construction and characterization of the hybrid bacteriophage lambda Charon vectors for DNA cloning. J Virol. 1979 Feb;29(2):555–575. doi: 10.1128/jvi.29.2.555-575.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. P., Kassavetis G. A., Esch F. S., Geiduschek E. P. Identification of the gene encoding an RNA polymerase-binding protein of bacteriophage T4. J Virol. 1987 Feb;61(2):597–599. doi: 10.1128/jvi.61.2.597-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Molecular cloning of the DNA ligase gene from bacteriophage T4. I. Characterisation of the recombinants. J Mol Biol. 1979 Aug 15;132(3):471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Wu R., Geiduschek E. P. The role of replication proteins in the regulation of bacteriophage T4 transcription. I. Gene 45 and hydroxymethyl-C-containing DNA. J Mol Biol. 1975 Aug 25;96(4):513–538. doi: 10.1016/0022-2836(75)90137-0. [DOI] [PubMed] [Google Scholar]
- Wu R., Geiduschek E. P. The role of replication proteins in the regulation of bacteriophage T4 transcription. II. Gene 45 and late transcription uncoupled from replication. J Mol Biol. 1975 Aug 25;96(4):539–562. doi: 10.1016/0022-2836(75)90138-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]





