Abstract
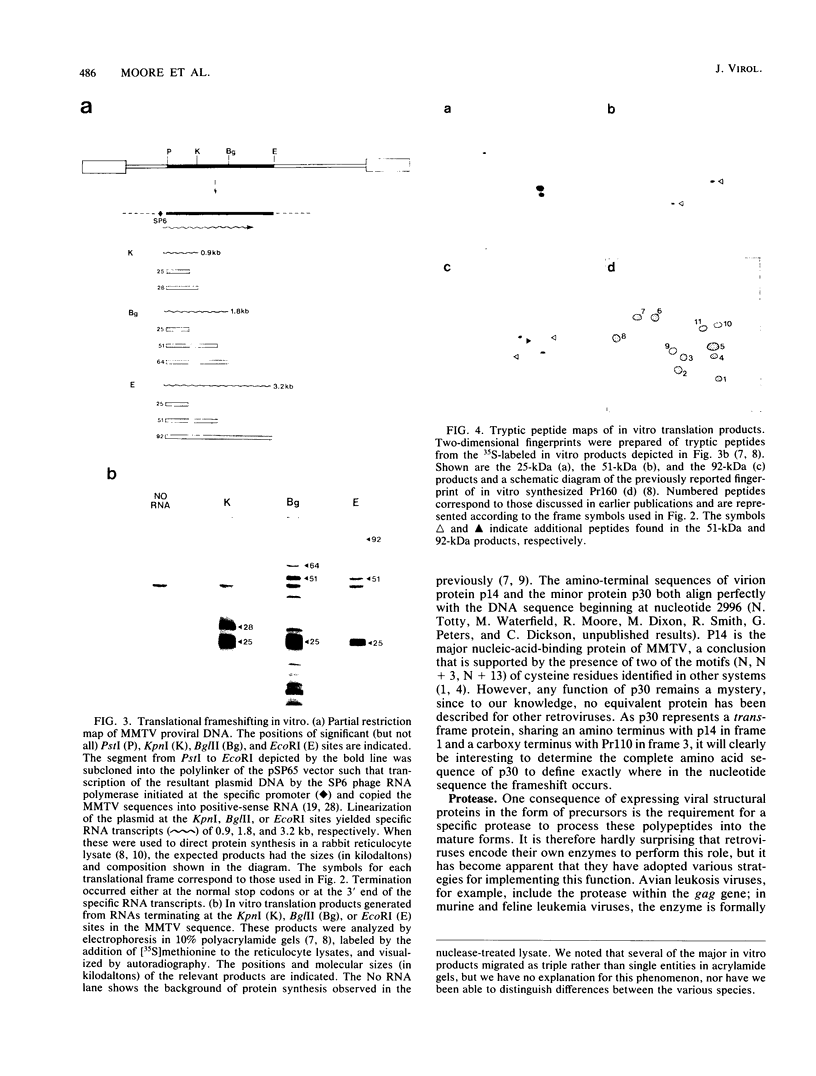
We sequenced two recombinant DNA clones constituting a single provirus of the milk-transmitted mouse mammary tumor virus characteristic of BR6 mice. The complete provirus is 9,901 base pairs long, flanked by 6 base-pair duplications of cellular DNA at the site of integration. Five extensive blocks of open reading frame corresponding to the gag gene, the presumed protease, the pol and env genes, and the open reading frame orf within the long terminal repeat of the provirus were readily discernible. Translation of gag, protease, and pol involved three different translational reading frames to produce the three overlapping polyprotein precursors Pr77, Pr110, and Pr160 found in virus-infected cells. Synthesis of the reverse transcriptase and endonuclease therefore required two separate frameshifts to suppress the termination codons at the ends of the Pr77 and Pr110 domains. Direct evidence is presented for translational readthrough of both stop codons in an in vitro protein synthesis system.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arthur L. O., Long C. W., Smith G. H., Fine D. L. Immunological characterization of the low-molecular-weight DNA binding protein of mouse mammary tumor virus. Int J Cancer. 1978 Oct 15;22(4):433–440. doi: 10.1002/ijc.2910220411. [DOI] [PubMed] [Google Scholar]
- Brookes S., Placzek M., Moore R., Dixon M., Dickson C., Peters G. Insertion elements and transitions in cloned mouse mammary tumour virus DNA: further delineation of the poison sequences. Nucleic Acids Res. 1986 Nov 11;14(21):8231–8245. doi: 10.1093/nar/14.21.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetti E., Diggelmann H. Cloned mouse mammary tumor virus DNA is biologically active in transfected mouse cells and its expression is stimulated by glucocorticoid hormones. Cell. 1981 Feb;23(2):335–345. doi: 10.1016/0092-8674(81)90129-x. [DOI] [PubMed] [Google Scholar]
- Chiu I. M., Callahan R., Tronick S. R., Schlom J., Aaronson S. A. Major pol gene progenitors in the evolution of oncoviruses. Science. 1984 Jan 27;223(4634):364–370. doi: 10.1126/science.6197754. [DOI] [PubMed] [Google Scholar]
- Copeland T. D., Oroszlan S., Kalyanaraman V. S., Sarngadharan M. G., Gallo R. C. Complete amino acid sequence of human T-cell leukemia virus structural protein p15. FEBS Lett. 1983 Oct 17;162(2):390–395. doi: 10.1016/0014-5793(83)80793-5. [DOI] [PubMed] [Google Scholar]
- Crawford S., Goff S. P. A deletion mutation in the 5' part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985 Mar;53(3):899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deen K. C., Sweet R. W. Murine mammary tumor virus pol-related sequences in human DNA: characterization and sequence comparison with the complete murine mammary tumor virus pol gene. J Virol. 1986 Feb;57(2):422–432. doi: 10.1128/jvi.57.2.422-432.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Atterwill M. Composition, arrangement and cleavage of the mouse mammary tumor virus polyprotein precursor Pr77gag and p110gag. Cell. 1979 Aug;17(4):1003–1012. doi: 10.1016/0092-8674(79)90339-8. [DOI] [PubMed] [Google Scholar]
- Dickson C., Peters G. Protein-coding potential of mouse mammary tumor virus genome RNA as examined by in vitro translation. J Virol. 1981 Jan;37(1):36–47. doi: 10.1128/jvi.37.1.36-47.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Peters G. Proteins encoded by mouse mammary tumour virus. Curr Top Microbiol Immunol. 1983;106:1–34. doi: 10.1007/978-3-642-69357-1_1. [DOI] [PubMed] [Google Scholar]
- Dickson C., Smith R., Peters G. In vitro synthesis of polypeptides encoded by the long terminal repeat region of mouse mammary tumour virus DNA. Nature. 1981 Jun 11;291(5815):511–513. doi: 10.1038/291511a0. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Huang A. L., Hager G. L. Regulatory and coding potential of the mouse mammary tumor virus long terminal redundancy. J Virol. 1981 Jan;37(1):226–238. doi: 10.1128/jvi.37.1.226-238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOULDS L. Mammary tumours in hybrid mice; the presence and transmission of the mammary tumour agent. Br J Cancer. 1949 Jun;3(2):230–239. doi: 10.1038/bjc.1949.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Buetti E., Firzlaff J., Pearson K., Diggelmann H. Nucleotide sequence of the 5' noncoding region and part of the gag gene of mouse mammary tumor virus; identification of the 5' splicing site for subgenomic mRNAs. Nucleic Acids Res. 1983 Oct 25;11(20):6943–6955. doi: 10.1093/nar/11.20.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Pearson K., Buetti E., Diggelmann H. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1982;1(1):3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D., Quinn T., Hippenmeyer P. J., Oroszlan S. Structural characterization of the avian retrovirus reverse transcriptase and endonuclease domains. J Biol Chem. 1985 Jul 15;260(14):8243–8249. [PubMed] [Google Scholar]
- Henderson L. E., Sowder R., Smythers G., Oroszlan S. Terminal amino acid sequences and proteolytic cleavage sites of mouse mammary tumor virus env gene products. J Virol. 1983 Oct;48(1):314–319. doi: 10.1128/jvi.48.1.314-319.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984 Feb;49(2):471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Kennedy N., Knedlitschek G., Groner B., Hynes N. E., Herrlich P., Michalides R., van Ooyen A. J. Long terminal repeats of endogenous mouse mammary tumour virus contain a long open reading frame which extends into adjacent sequences. Nature. 1982 Feb 18;295(5850):622–624. doi: 10.1038/295622a0. [DOI] [PubMed] [Google Scholar]
- Klemenz R., Reinhardt M., Diggelmann H. Sequence determination of the 3' end of mouse mammary tumor virus RNA. Mol Biol Rep. 1981 May 22;7(1-3):123–126. doi: 10.1007/BF00778742. [DOI] [PubMed] [Google Scholar]
- Kohno T., Roth J. R. A Salmonella frameshift suppressor that acts at runs of A residues in the messenger RNA. J Mol Biol. 1978 Nov 25;126(1):37–52. doi: 10.1016/0022-2836(78)90278-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L. I., Goff S. P. Reverse transcription of retroviral genomes: mutations in the terminal repeat sequences. J Virol. 1985 Feb;53(2):447–455. doi: 10.1128/jvi.53.2.447-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G., Varmus H. E. Frameshift and intragenic suppressor mutations in a Rous sarcoma provirus suggest src encodes two proteins. Cell. 1983 Mar;32(3):871–879. doi: 10.1016/0092-8674(83)90072-7. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. H., Long C. A., Vaidya A. B., Sheffield J. B., Dion A. S., Lasfargues E. Y. Mammary tumor viruses. Adv Cancer Res. 1979;29:347–418. doi: 10.1016/s0065-230x(08)60850-7. [DOI] [PubMed] [Google Scholar]
- Moore R., Casey G., Brookes S., Dixon M., Peters G., Dickson C. Sequence, topography and protein coding potential of mouse int-2: a putative oncogene activated by mouse mammary tumour virus. EMBO J. 1986 May;5(5):919–924. doi: 10.1002/j.1460-2075.1986.tb04304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984 Mar;36(3):673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Temin H. M. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Peters G., Glover C. tRNA's and priming of RNA-directed DNA synthesis in mouse mammary tumor virus. J Virol. 1980 Jul;35(1):31–40. doi: 10.1128/jvi.35.1.31-40.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Placzek M., Brookes S., Kozak C., Smith R., Dickson C. Characterization, chromosome assignment, and segregation analysis of endogenous proviral units of mouse mammary tumor virus. J Virol. 1986 Sep;59(3):535–544. doi: 10.1128/jvi.59.3.535-544.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson L., Andersson P., Olshevsky U., Weinberg R., Baltimore D., Gesteland R. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978 Jan;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- Redmond S. M., Dickson C. Sequence and expression of the mouse mammary tumour virus env gene. EMBO J. 1983;2(1):125–131. doi: 10.1002/j.1460-2075.1983.tb01393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice N. R., Stephens R. M., Burny A., Gilden R. V. The gag and pol genes of bovine leukemia virus: nucleotide sequence and analysis. Virology. 1985 Apr 30;142(2):357–377. doi: 10.1016/0042-6822(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Ringold G. M. Regulation of mouse mammary tumor virus gene expression by glucocorticoid hormones. Curr Top Microbiol Immunol. 1983;106:79–103. doi: 10.1007/978-3-642-69357-1_4. [DOI] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A. M., Oroszlan S. In vivo modification of retroviral gag gene-encoded polyproteins by myristic acid. J Virol. 1983 May;46(2):355–361. doi: 10.1128/jvi.46.2.355-361.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Hirayama Y., Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Takahashi Y., Shimizu N., Gojobori T., Golde D. W., Chen I. S., Miwa M., Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proc Natl Acad Sci U S A. 1985 May;82(10):3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985 Aug;42(1):369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Barker C., Hunter E., Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986 May 9;45(3):375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Sorge J., Hughes S. H. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982 Aug;43(2):482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucker D. S., Ross S. R., Yamamoto K. R. Mammary tumor virus DNA contains sequences required for its hormone-regulated transcription. Cell. 1981 Dec;27(2 Pt 1):257–266. doi: 10.1016/0092-8674(81)90409-8. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Temin H. M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5' long terminal repeat and the start of the gag gene. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B. Molecular model of ribosome frameshifting. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5797–5801. doi: 10.1073/pnas.81.18.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. A., Butel J. S., Medina D., Cardiff R. D., Hager G. L. Transcription of mouse mammary tumor virus: identification of a candidate mRNA for the long terminal repeat gene product. J Virol. 1983 Apr;46(1):42–49. doi: 10.1128/jvi.46.1.42-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Oroszlan S. Translational readthrough of an amber termination codon during synthesis of feline leukemia virus protease. J Virol. 1985 Sep;55(3):870–873. doi: 10.1128/jvi.55.3.870-873.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaka Y., Katoh I., Copeland T. D., Smythers G. W., Oroszlan S. Bovine leukemia virus protease: purification, chemical analysis, and in vitro processing of gag precursor polyproteins. J Virol. 1986 Mar;57(3):826–832. doi: 10.1128/jvi.57.3.826-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooyen A. J., Michalides R. J., Nusse R. Structural analysis of a 1.7-kilobase mouse mammary tumor virus-specific RNA. J Virol. 1983 May;46(2):362–370. doi: 10.1128/jvi.46.2.362-370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]






