Abstract
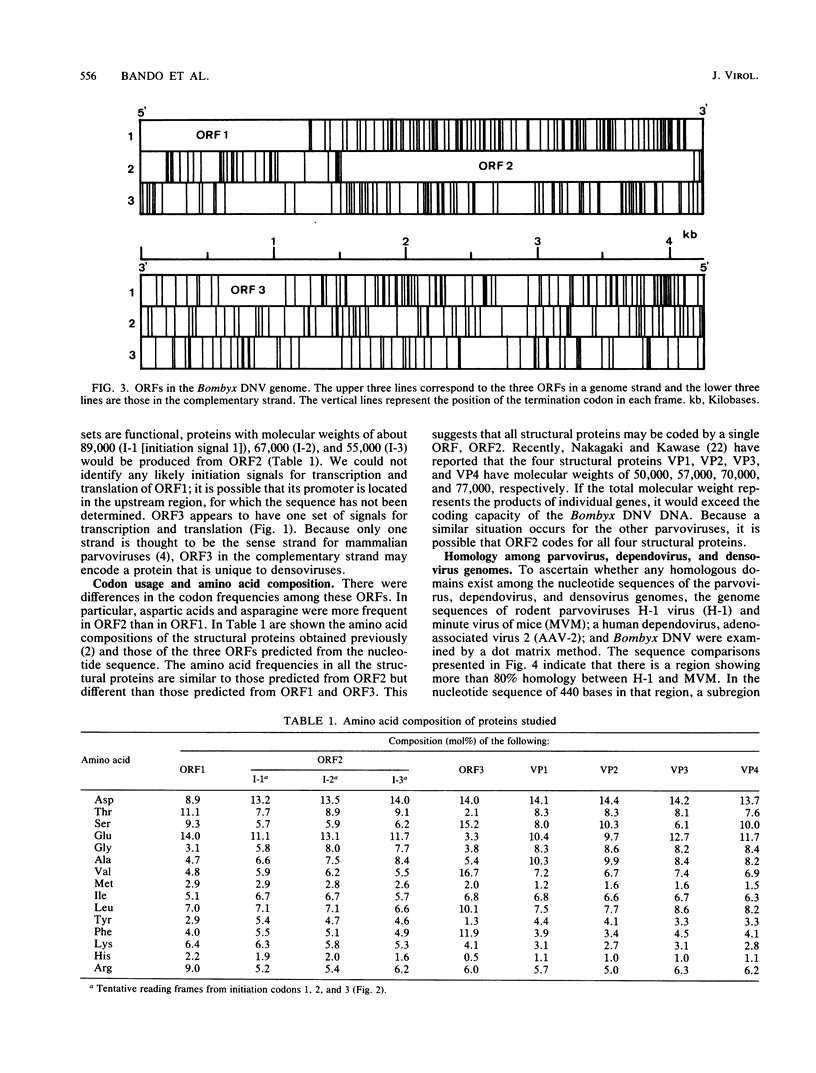
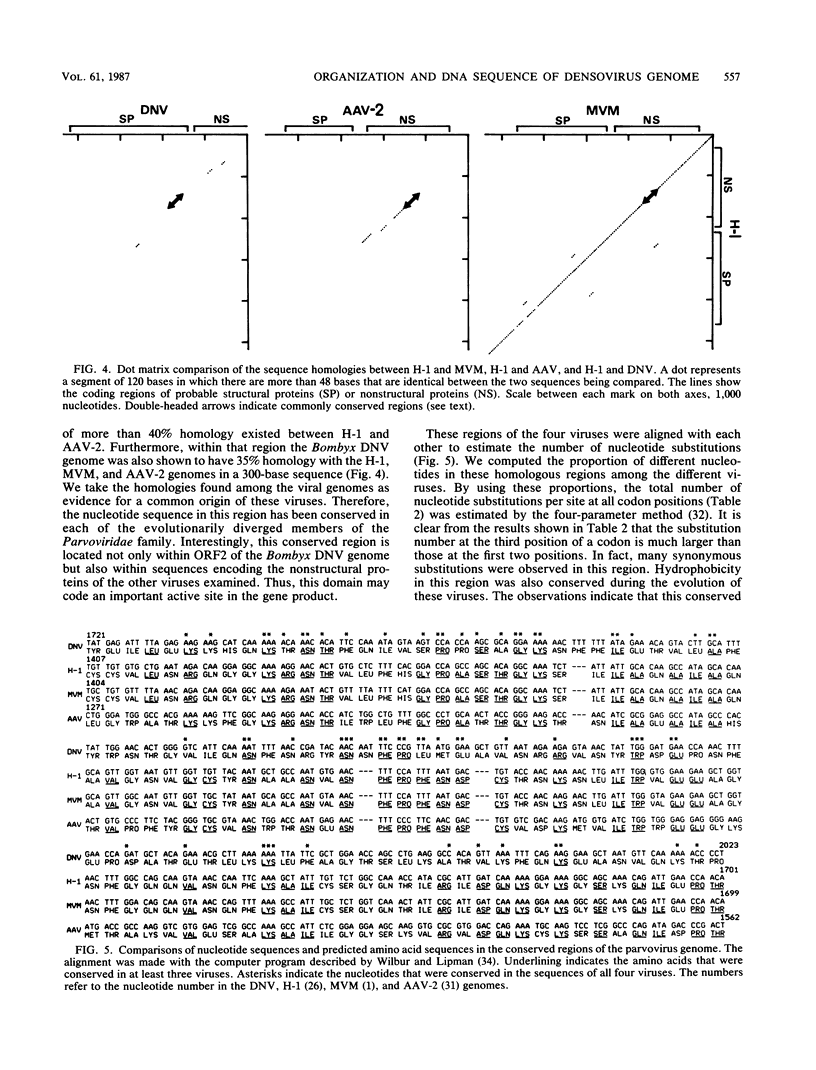
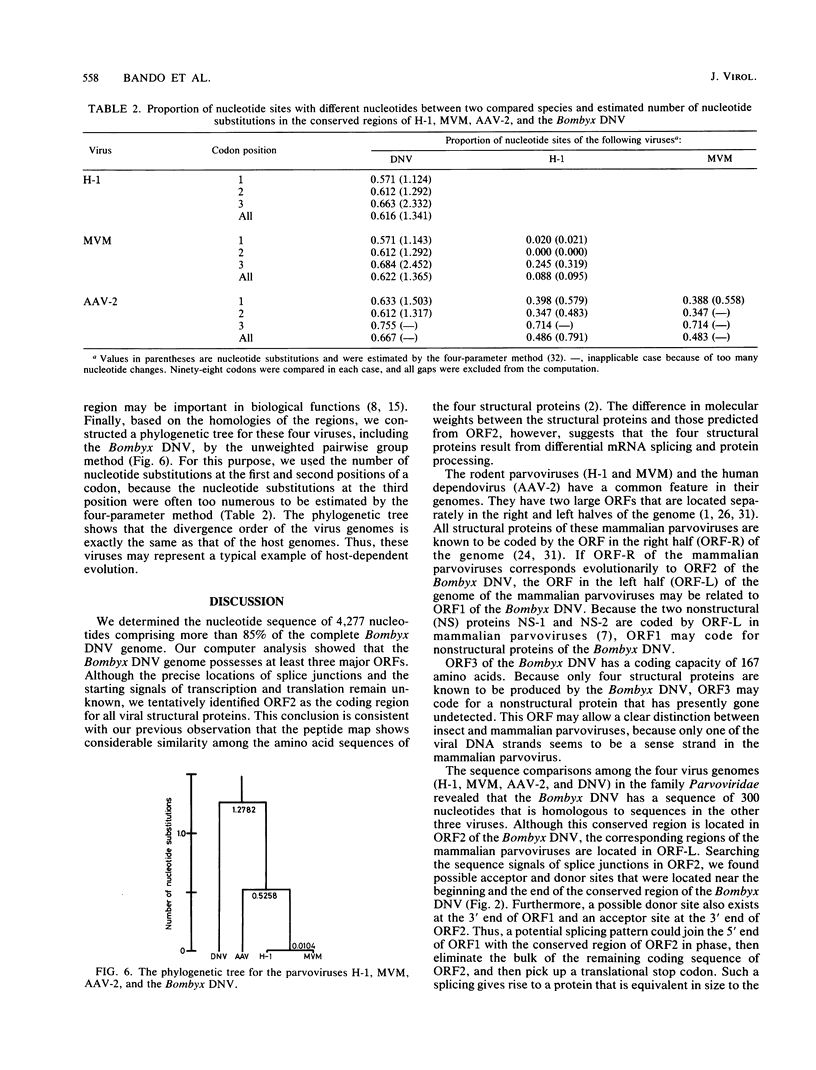
The genome structure of a densovirus from a silkworm was determined by sequencing more than 85% of the complete genome DNA. This is the first report of the genome organization of an insect parvovirus deduced from the DNA sequence. In the viral genome, two large open reading frames designated 1 and 2 and one smaller open reading frame designated 3 were identified. The first two open reading frames shared the same strand, while the third was found in the complementary sequence. Computer analysis suggested that open reading frame 2 may encode all four structural proteins. The genome organization and a part of the nucleotide sequence were conserved among the insect densovirus, rodent parvoviruses, and a human dependovirus. These viruses may have diverged from a common ancestor.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astell C. R., Thomson M., Merchlinsky M., Ward D. C. The complete DNA sequence of minute virus of mice, an autonomous parvovirus. Nucleic Acids Res. 1983 Feb 25;11(4):999–1018. doi: 10.1093/nar/11.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando H., Kondo N., Kawase S. Molecular homology among the structural proteins of densonucleosis virus from silkworm, Bombyx mori. Arch Virol. 1984;80(2-3):209–218. doi: 10.1007/BF01310660. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. K., Hoggan M. D., Hauswirth W. W., Berns K. I. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J Virol. 1980 Feb;33(2):739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classification and nomenclature of viruses. Fourth report of the International Committee on Taxonomy of Viruses. Intervirology. 1982;17(1-3):1–199. doi: 10.1159/000149278. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Cotmore S. F., Sturzenbecker L. J., Tattersall P. The autonomous parvovirus MVM encodes two nonstructural proteins in addition to its capsid polypeptides. Virology. 1983 Sep;129(2):333–343. doi: 10.1016/0042-6822(83)90172-1. [DOI] [PubMed] [Google Scholar]
- Go M., Miyazawa S. Relationship between mutability, polarity and exteriority of amino acid residues in protein evolution. Int J Pept Protein Res. 1980 Mar;15(3):211–224. doi: 10.1111/j.1399-3011.1980.tb02570.x. [DOI] [PubMed] [Google Scholar]
- Gojobori T., Yokoyama S. Rates of evolution of the retroviral oncogene of Moloney murine sarcoma virus and of its cellular homologues. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4198–4201. doi: 10.1073/pnas.82.12.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H., Shiroki K., Shimojo H. Establishment and characterization of KB cell lines latently infected with adeno-associated virus type 1. Virology. 1977 Oct 1;82(1):84–92. doi: 10.1016/0042-6822(77)90034-4. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977 May 19;267(5608):275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kurstak E., Vago C. Transmission du virus de la densonuCL'EOSE PAR LE PARASITISME D'un Hyménoptère. Rev Can Biol. 1967 Dec;26(4):311–316. [PubMed] [Google Scholar]
- Merchlinsky M. J., Tattersall P. J., Leary J. J., Cotmore S. F., Gardiner E. M., Ward D. C. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J Virol. 1983 Jul;47(1):227–232. doi: 10.1128/jvi.47.1.227-232.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Paradiso P. R., Williams K. R., Costantino R. L. Mapping of the amino terminus of the H-1 parvovirus major capsid protein. J Virol. 1984 Oct;52(1):77–81. doi: 10.1128/jvi.52.1.77-81.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd, Paradiso P. R. Parvovirus genome: nucleotide sequence of H-1 and mapping of its genes by hybrid-arrested translation. J Virol. 1983 Jan;45(1):173–184. doi: 10.1128/jvi.45.1.173-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd trans-Activation of parvovirus P38 promoter by the 76K noncapsid protein. J Virol. 1985 Sep;55(3):886–889. doi: 10.1128/jvi.55.3.886-889.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Maruyama T., Arrand J. R., Griffin B. E. Host-dependent evolution of three papova viruses. Nature. 1980 May 15;285(5761):165–167. doi: 10.1038/285165a0. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Lusby E. W., Berns K. I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983 Feb;45(2):555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Kimura M. A model of evolutionary base substitutions and its application with special reference to rapid change of pseudogenes. Genetics. 1981 Jul;98(3):641–657. doi: 10.1093/genetics/98.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]


