Abstract
Eukaryotic cells actively block entry into mitosis in the presence of DNA damage or incompletely replicated DNA. This response is mediated by signal transduction cascades called cell cycle checkpoints. We show here that the human checkpoint control protein hRAD9 physically associates with two other checkpoint control proteins, hRAD1 and hHUS1. Furthermore, hRAD1 and hHUS1 themselves interact, analogously to their fission yeast homologues Rad1 and Hus1. We also show that hRAD9 is present in multiple phosphorylation forms in vivo. These phosphorylated forms are present in tissue culture cells that have not been exposed to exogenous sources of DNA damage, but it remains possible that endogenous damage or naturally occurring replication intermediates cause the observed phosphorylation. Finally, we show that hRAD9 is a nuclear protein, indicating that in this signal transduction pathway, hRAD9 is physically proximal to the upstream (DNA damage) signal rather than to the downstream, cytoplasmic, cell cycle machinery.
INTRODUCTION
The eukaryotic cell cycle consists of a number of tightly regulated events whose precise order ensures that the important tasks of DNA replication and cell division occur with high fidelity. Cells maintain the order of these events by making later events dependent on the successful completion of earlier events. This dependency is enforced by cellular mechanisms called checkpoints (Weinert and Hartwell, 1988, 1990). The DNA damage (G2) and DNA replication (S-phase) checkpoints arrest eukaryotic cells at the G2/M transition in the presence of damaged or incompletely replicated DNA, respectively (Weinert and Hartwell, 1988, 1990; Enoch and Nurse, 1990; Enoch et al., 1992; al-Khodairy and Carr, 1992; al-Khodairy et al., 1994; Rowley et al., 1992). This arrest provides time for the cell to repair damage or complete replication before entry into mitosis.
Various lines of evidence support a model for G2 checkpoint regulation in which the ultimate event is phosphorylation of the tyrosine 15 residue of the cyclin-dependent kinase Cdc2 (Enoch and Nurse, 1990; O’Connell et al., 1997; Rhind et al., 1997). Phosphorylation of this residue is regulated primarily by the Cdc25 phosphatase and the Wee1 protein kinase, and the activity of these enzymes is regulated in turn by the kinases Chk1 and Cds1, respectively (Walworth et al., 1993; Furnari et al., 1997). Chk1 is only required for the DNA damage checkpoint (Walworth et al., 1993) and functions by phosphorylating and inhibiting Cdc25, thereby preventing Cdc2 dephosphorylation and mitotic entry (Furnari et al., 1997). When the S-phase checkpoint is triggered, activation of Cds1 results in activating phosphorylation of Wee1, which then results in inhibitory phosphorylation of Cdc2 (Boddy et al., 1998). Although the mechanistic detail involved in the G2 checkpoints upstream of these proteins is unclear, it is known that a group of six proteins in fission yeast are required for both G2 and S-phase checkpoint control. These proteins are Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1 and are collectively termed the checkpoint rad proteins (al-Khodairy and Carr, 1992; al-Khodairy et al., 1994; Enoch et al., 1992; Rowley et al., 1992). Evidence that these genes are all critical components of both the damage and replication checkpoints is based on observations that the checkpoint rad mutants, unlike wild-type cells, do not block mitotic entry in response to DNA-damaging agents or transient inhibition of DNA synthesis (al-Khodairy and Carr, 1992; al-Khodairy et al., 1994; Enoch et al., 1992; Rowley et al., 1992). The checkpoint rads are placed upstream of the Cdc2 regulators in the emerging checkpoint signal transduction pathway, because the checkpoint-induced phosphorylation of the Chk1 and Cds1 kinases is dependent on the presence of all of the checkpoint rad proteins (Walworth and Bernards, 1996; Lindsay et al., 1998). More recently, it was shown that Rad1 and Hus1 form a stable complex that is dependent on Rad9, suggesting that these three proteins may exist in a three-way complex in fission yeast (Kostrub et al., 1998).
Many of the genes involved in the G2 checkpoint pathways are conserved between humans and yeast (Table 1). Human homologues of all of the fission yeast checkpoint Rad proteins, with the exception of Rad26, have been identified, suggesting that the fission yeast G2 checkpoint signaling mechanism may be similar to that of humans (Cimprich et al., 1996; Lieberman et al., 1996; Kostrub et al., 1998; Parker et al., 1998a; Udell et al., 1998). In vitro evidence has suggested that the human homologues of fission yeast Chk1 and Cds1 phosphorylate and inhibit Cdc25C in response to DNA damage (Sanchez et al., 1997; Matsuoka et al., 1998). Furthermore, this response is dependent on ATM, a human homologue of fission yeast Rad3 (Savitsky et al., 1995a,b). Therefore, the human equivalents of the checkpoint rads appear to be functioning upstream of the Cdc2 regulatory machinery, as they do in fission yeast.
Table 1.
Summary of G2 checkpoint proteins in yeasts and humans
| S. pombe | S. cerevisiae | Homo sapiens |
|---|---|---|
| Rad1 | RAD17 | hRAD1 |
| Rad3 | MEC1 | ATR/FRP1 |
| Rad9 | DDC1 | hRAD9 |
| Rad17 | RAD24 | hRAD17 |
| Rad26 | ||
| Hus1 | hHUS1 | |
| Chk1 | CHK1 | Chk1 |
| Cds1 | RAD53 | Chk2 |
Here, we identify further conservation between the fission yeast and human G2 checkpoints by demonstrating that human homologues of Schizosaccharomyces pombe checkpoint rads hRAD1, hRAD9, and hHUS1 physically interact with one another in vivo. We also show that endogenous hRAD9 is phosphorylated and that it localizes primarily to the nucleus in unperturbed HeLa and HaCaT cells.
MATERIALS AND METHODS
Yeast Two-Hybrid Library Screen
hRAD9 cDNA was subcloned from pBluescript (Stratagene, La Jolla, CA) into the SmaI and SalI restriction sites of the GAL4 DNA binding domain pGBT9 vector (Clontech, Palo Alto, CA). pGBT9-hRAD9 was then transformed into the budding yeast strain HF7c (Feilotter et al., 1994) according to the manufacturer’s instructions. Transformants were plated on synthetic dropout (SD) media minus tryptophan (6.7 g/l Difco [Detroit, MI] yeast nitrogen base without amino acids, 2% glucose, 0.62 g/l Bio 101 [La Jolla, CA] complete supplement mixture minus histidine (−his), leucine (−leu), and tryptophan (−trp), 20 mg/l histidine, 100 mg/l leucine, and 20 g/l Difco Bacto-Agar). To ensure that the hRAD9 GAL4 DNA binding domain hybrid construct alone did not activate the HIS3 and/or lacZ reporter genes, colonies were streaked onto the SD agar −trp, −his and tested for β-galactosidase activity using a filter assay described in the Clontech manual. One HF7c colony harboring the pGBT9-hRAD9 vector was picked into 150 ml of SD − trp liquid media and grown to saturation for 2 d at 30°C. The saturated culture was then diluted by adding 1 l of YTD (10 g/l yeast extract, 20 g/l tryptone, and 20 g/l dextrose) and grown to an OD600 of 0.5. These yeast were then transformed, as described by the manufacturer, with 0.5 mg of a directionally cloned HeLa cDNA library in the pGAD-GH GAL4 activation domain vector (Clontech). The transformants were plated on 44 15-cm plates containing SD agar −trp, −leu, −his and incubated at 30°C. To determine the efficiency of the library transformation, serial dilutions of a small aliquot of the transformed yeast were plated on SD agar −trp, −leu. After 10 d, ∼500 colonies grew larger than background on the triple dropout plates. These colonies were subcultured onto SD agar −trp, −leu −his and 5 mM 3-aminotriazole (3-AT) and incubated at 30°C for 2 d, after which 15 positive clones were identified. Plasmid DNA was then prepared from 15 saturated liquid cultures essentially as described by the manufacturer (Clontech). XL1-Blue competent bacteria were then transformed with this DNA and plated on Luria–Bertani agar containing ampicillin. Inserts in pGAD-GH were sequenced using fluorescently labeled SK primer and an automated sequencer (Applied Biosystems, Foster City, CA). DNA sequence analysis was performed using the BLAST algorithm (Altschul et al., 1990).
For analysis of individual interactions among hRAD9, hRAD1, and hHUS1, HF7c were simultaneously cotransformed with pGBT9-hHUS1 and pGAD-hRAD1, pGBT9-hRAD9 and pGAD-hHUS1, and pGBT9-hRAD9 and pGAD-hRAD1, as described by the manufacturer (Clontech). Cotransformants were plated on SD agar −trp, −leu and incubated at 30°C for 2–3 d. As negative controls, pGBT9 fusion constructs were cotransformed with empty pGAD-GH vector, and pGAD fusion constructs were cotransformed with empty pGBT9 vector. As a positive control, a p53-DNA binding domain fusion construct was cotransformed with a pSV40 T antigen-activation domain fusion construct. A single isolated colony from each plate was streaked onto both SD agar −trp, −leu and SD agar −trp −leu −his and 5mM 3-AT and grown at 30°C for 2–3 d.
hRAD9 Polyclonal Antibody Preparation and Purification
hRAD9 cDNA was PCR cloned into the SmaI and BamHI restriction sites of the pGEX1 bacterial expression vector (Pharmacia, Piscataway, NJ). An hRAD9-GST fusion protein was then expressed in Escherichia coli and affinity purified on glutathione-Sepharose (Pharmacia) according to previously described methods (Frangioni and Neel, 1993). α-hRAD9 polyclonal chicken antibodies were generated against this hRAD9 fusion protein (RCH antibodies).
Ten milligrams of purified GST were batch adsorbed to 2 ml of glutathione-Sepharose for 2 h at 4°C. Sepharose was washed with 40 vol of PBS. Two milliliters of antibody supernatant were batch adsorbed with the GST-bound glutathione-Sepharose overnight. Sepharose was subjected to centrifugation, and the supernatant was harvested.
Thirty-five micrograms of purified GST-hRAD9 protein were subjected to electrophoresis through a 10% acrylamide gel and then electroblotted onto a nitrocellulose membrane. The protein band was visualized by Ponceau S staining, and the band was excised and cut into small pieces with a scalpel. Membrane pieces were blocked overnight in 1% casein in PBS and 0.1% Tween 20 (PBST) at 4°C in a microfuge tube. The membrane was then washed three times for 5 min each in PBST. One milliliter of precleared antibody supernatant was added to the membrane pieces and rocked at 4°C for 4 h. The supernatant was removed, and the membrane was washed two times rapidly and once for 15 min with PBST. The tube was centrifuged briefly, and all traces of the wash were removed. The antibody was eluted from membrane with 300 μl of 0.2 M glycine, pH 2.8. A second elution with 100 μl of glycine was pooled with the first, and the antibody supernatant was neutralized with 0.2 vol of 1 M Tris, pH 8.0.
Coimmunoprecipitation Experiments
Coimmunoprecipitations used the myc and flag epitope tags, and for simplicity, proteins expressed with these tags are denoted by a subscript m or f, respectively. hRAD1 cDNA was amplified by PCR and cloned into the XbaI and EcoRI restriction sites of the mammalian expression vector pyDF31 (a gift from Dr. David LeBrun, Queen’s University, Department of Pathology), in frame with one copy of the flag epitope. hRAD9 cDNA was PCR cloned into the XbaI and XhoI restriction sites of the pCS2-MT, a mammalian expression vector with six copies of the myc epitope (Rupp et al., 1994; Turner and Weintraub, 1994). A hHUS1-myc fusion construct was generated by PCR amplifying hHUS1 cDNA and cloning it into the pCS2-MT vector. The constructs used to express the negative controls HLFf and FerΔNm constructs were gifts of Dr. David LeBrun and Dr. Peter Greer (Queen’s University, Cancer Research Laboratories), respectively.
COS-1 cells that were ∼50% confluent in 10-cm tissue culture plates were transiently cotransfected with 24 μg each of the indicated constructs, using Lipofectin reagent (Sigma, St. Louis, MO) according to the manufacturer’s instructions. Cells were then washed twice with 10 ml of sterile PBS, and 10 ml of complete Dulbecco’s modified Eagle’s medium were added (Dulbecco’s modified Eagle’s medium and 10% FBS). Transfected cells were cultured at 37°C in a 5% CO2 atmosphere for 48 h. Cells were lysed directly on the plate in mammalian cell lysis solution (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% NP-40, 1 mM Na3VO4, 1 mM PMSF, 20 μg/ml aprotinin, and 10 μg/ml leupeptin). Lysates were passed through 18- and then 23-gauge syringes several times to shear genomic DNA, incubated on ice for 30 min, and centrifuged at 16,000 × g to remove any insoluble material. Each cotransfected cell lysate was split into two equal portions. To one set lysates were precleared with 35 μl of α-immunoglobulin y (IgY) agarose (Promega, Madison, WI) on a Nutator (Becton Dickinson, Oakville, Canada) at 4°C for 45 min and immunoprecipitated with polyclonal chicken α-hRAD9 antibodies on a Nutator at 4°C for 1 h. These immune complexes were collected on 35 μl of α-IgY agarose (Promega) at 4°C for 1 h. To the other set lysates were precleared with 10 μl of protein G-Sepharose (Pharmacia) and immunoprecipitated with ∼1 μg of α-myc 9E10 mouse monoclonal antibody. These immune complexes were collected on 10 μl of protein G-Sepharose at 4°C for 1 h. Both the α-myc and α-hRAD9 immunoprecipitated complexes were collected by centrifugation at 500 × g, washed four times with PBS, and incubated at 100°C for 5 min in 50 μl of SDS-PAGE sample buffer (New England Biolabs, Beverly, MA). After centrifugation at 16,000 × g for 20 min, 10 μl of each supernatant was electrophoresed through a single 6% acrylamide gel. Protein was transferred to nitrocellulose (0.2 μm pore size; Xymotech, Toronto, Canada) which was blocked in 5% MPBST (PBS, 5% nonfat milk powder, and 0.1% Tween 20) at room temperature for 2 h and then probed with α-myc 9E10 mouse monoclonal antibody. After extensive washing in PBST, HRP-conjugated anti-mouse secondary antibody was added, and the membrane was incubated for 45 min at room temperature. Protein antigens were detected by chemiluminescence using the ECL detection system (Amersham, Arlington Heights, IL), followed by exposure to x-ray film (Eastman Kodak, Rochester, NY).
For the hRAD1/hHUS1 and hRAD9/hRAD1 coimmunoprecipitations, the same methods as described for the hRAD9/hHUS1 coimmunoprecipitations were used, with the following exceptions. All lysates were precleared with 10 μl of protein G-Sepharose (Pharmacia) on a Nutator at 4°C for 45 min. Either α-myc 9E10 monoclonal antibody or α-flag M2 monoclonal antibody was used for immunoprecipitation. Samples were size fractionated on 10% polyacrylamide gels. Immunoblotting was carried out using α-myc 9E10 mouse monoclonal or α-flag M2 monoclonal antibody, as indicated.
Calf Intestinal Phosphatase (CIP) Treatments
COS-1 cells were transfected with 24 μg of pCS2-MT-hRAD9 as described previously. Two days after the transfection, cells were harvested and immunoprecipitated with α-myc monoclonal antibody as before. After collecting the immune complexes on protein G-Sepharose, beads were washed four times with PBS and resuspended in 200 μl of NEB buffer 3 (50 mM Tris-HCl, 10 mM MgCl2, 100 mM NaCl, and 1 mM DTT) and 1% SDS. Protein was removed from the Sepharose beads by heating at 100°C for 5 min followed by centrifugation at 16,000 × g. Twenty microliters of the supernatant were then treated with 30 U of calf intestinal alkaline phosphatase (Promega) in 1× NEB buffer 3 in the presence or absence of 2 mM sodium orthovanadate (Na3VO4) for 30 min at 37°C. To sufficiently dilute the SDS in the sample, the total volume of the reactions was 200 μl. Both reactions, along with 20 μl of untreated immunoprecipitate, were made up to 1 ml with PBS and reimmunoprecipitated with α-myc monoclonal antibody, electrophoresed through 6% acrylamide, and immunoblotted with α-myc monoclonal antibody essentially as above.
Endogenous hRAD9 protein was immunoprecipitated from ∼9 × 106 HeLa cells with polyclonal chicken α-hRAD9 antibodies essentially as described above. The phosphatase procedure followed was identical to that for exogenous hRAD9m, except samples were electrophoresed through 8% acrylamide and immunoprecipitated and immunoblotted with α-hRAD9 antibodies.
hRAD9 Immunofluorescence
HaCaT or HeLa cells were seeded on coverslips for 1 h (HeLa) or overnight (HaCaT) at 37°C in 5% CO2. Cells were washed twice with PBS and fixed with 10% paraformaldehyde for 10 min at room temperature. Fixed cells were washed twice more with PBS, covered with methanol, and incubated at −20°C for 20 min. Cells were rinsed twice and then washed for 30 min in PBST. PBST and 1% normal goat serum (NGS) were used to block cells at room temperature for 1 h. Incubation in polyclonal α-hRAD9 chicken antibodies in PBST and 1% NGS for 1 h at room temperature was followed by two rinses and one 30-min wash in PBST. Cells were then incubated in Alexa 488 goat anti-chicken secondary antibody (Molecular Probes, Eugene, OR) and diluted to 10 μg/ml in PBST and 1% NGS for 1 h at room temperature. After two rinses with PBST and two 10-min washes in PBS, cells were treated with 200 μg/ml RNase A in 1% PBS for 1 h at 37°C. After two rinses and two 5-min washes in PBST, nuclei were stained with 2 μg/ml propidium iodide in PBS for 5 min at room temperature. Cells were rinsed twice and washed once for 10 min with PBST. Coverslips were mounted on glass slides and visualized using a Meridian Instruments (Lansing, MI) Insight Plus confocal microscope. Images were captured from a cooled Meridian video with a Matrox 1280 frame grabber (Matrox Electronic Systems, Dorval, Quebec, Canada) and pseudocolored and saved using MCID M4 software (Imaging Research, St. Catherines, Ontario, Canada).
RESULTS
hRAD9 and hHUS1 Physically Interact
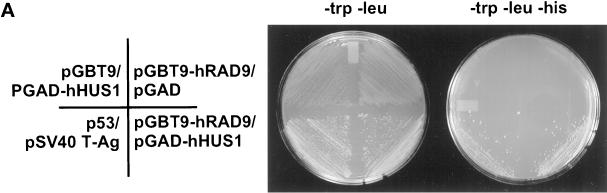
We set about to identify proteins interacting with hRAD9 using a two-hybrid screen. We screened 5.5 × 106 total transformants and from these identified 15 primary positive clones, each of which was viable on triple dropout medium in the presence of 5 mM 3-AT. Eleven of the 15 isolates contained hHUS1 cDNA sequences. Two approaches were taken, to substantiate the interaction we observed between hRAD9 and hHUS1 in the two-hybrid screen. First, GAL4 fusion constructs for hRAD9 and hHUS1 were retransformed into Saccharomyces cerevisiae HF7c, and the two-hybrid interaction was confirmed (Figure 1A). pGBT9-hRAD9 and pGAD-hHUS1, encoding the entire hHUS1 cDNA sequence, were transformed individually and together into HF7c. In the individual transformations, empty vector of the complementary plasmid was cotransformed. Positive control plasmids fusing p53 and SV40 T antigen to the GAL4 DNA binding and transactivation domains, respectively, were also cotransformed. Cotransformants were selected on double (−leu, −trp) dropout media and then subcultured onto triple (−leu, −trp, −his) dropout media to verify interactions. Neither pGBT9-hRAD9 nor pGAD-hHUS1 could drive expression of the HIS3 reporter gene (Figure 1A, upper two quadrants). Only when the hRAD9 and hHUS1 fusions were cotransformed together were HIS+ colonies isolated (Figure 1A, lower right panel), indicating that interaction between the two proteins is required for reconstitution of the GAL4 transcriptional regulator.
Figure 1.
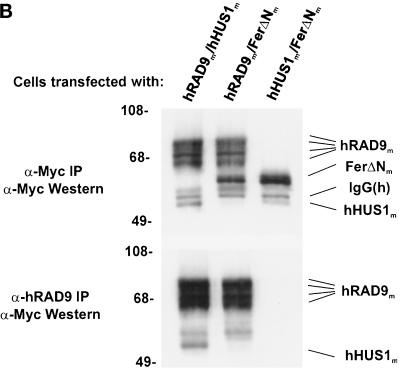
hRAD9 and hHUS1 physically interact. (A) S. cerevisiae strain HF7c was transformed with the indicated GAL4 fusion plasmids and plated on media selecting for cotransformants. Single colonies were subcultured onto selective media in the presence (left) or absence (right) of histidine. Growth in the absence of histidine is indicative of a protein–protein interaction, as demonstrated by the p53/SV40 T-Ag positive control (lower left quadrant). When pGBT9-hRAD9 and pGAD-hHUS1 were cotransformed separately with the corresponding empty vector, no growth on triple dropout was observed (upper two quadrants). Expression of both hRAD9 and hHUS1 GAL4 fusions was required for viability in the absence of histidine (bottom right quadrant). (B) COS-1 cells were transiently cotransfected with constructs expressing hRAD9m and hHUS1m, hRAD9m and FerΔNm, or hHUS1m and FerΔNm, as indicated. After harvesting, lysates were immunoprecipitated with α-myc 9E10 monoclonal antibody (upper panel) or chicken α-hRAD9 polyclonal antibodies (lower panel) and in both cases were immunoblotted with α-myc 9E10 monoclonal antibody. Specific coimmunoprecipitation of hRAD9m and hHUS1m is observed in lane 1 of the lower panel.
The second approach we took to study this potential interaction was to coimmunoprecipitate hRAD9 and hHUS1 proteins exogenously expressed in COS-1 cells (Figure 1B). Both hRAD9 and hHUS1 cDNA were subcloned into the pCS2-MT mammalian expression vector. This vector placed six copies of the myc epitope at the C terminus of hRAD9 and hHUS1. pCS2-MT-hRAD9 and pCS2-MT-hHUS1 were cotransfected into COS-1 cells. Cotransfections with pCS2-MT-FerΔN were also performed to ensure that the coimmunoprecipitation of hHUS1 with hRAD9 was not the result of nonspecific interactions involving the myc epitope tag. Cell lysates were immunoprecipitated with α-myc 9E10 monoclonal antibody or α-hRAD9 polyclonal chicken antibodies. Immunoprecipitates were then size fractionated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with α-myc monoclonal antibody. hHUS1 coimmunoprecipitated with hRAD9 when cell lysates were incubated with α-hRAD9 antibodies (Figure 1B, α-hRAD9 IP/α-Myc Western, lane 1). Although hHUS1m was exogenously expressed at similar levels in both pCS2-MT-hHUS1–transfected cells (Figure 1B, α-Myc IP/α-Myc Western, lanes 1 and 3), in the absence of hRAD9m, hHUS1m did not immunoprecipitate with polyclonal α-hRAD9 antibodies. In this and other coimmunoprecipitation experiments described below, the relative expression levels of the epitope tagged-proteins were constant between experimental and control lanes, indicating that the observed interactions were not simply due to overexpression of the proteins.
hHUS1 and hRAD1 Physically Interact
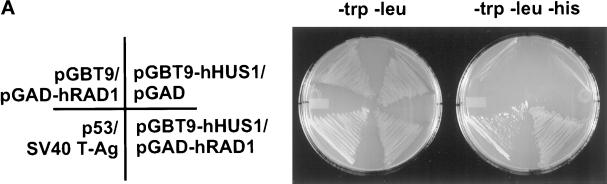
Because an Hus1–Rad1 interaction had been previously described in S. pombe (Kostrub et al., 1998), we investigated whether a similar interaction existed between hHUS1 and hRAD1. We cotransformed pGBT9-hHUS1 and pGAD-hRAD1 into HF7c and looked for activation of the HIS3 reporter gene by subculturing cotransformants on triple dropout media (Figure 2A). The same positive and negative controls were used as before. Although neither fusion plasmid on its own was sufficient for growth in the absence of histidine, cotransformation of pGBT9-hHUS1 and pGAD-hRAD1 resulted in viable HIS+ cotransformants. This suggested that a specific interaction existed between hHUS1 and hRAD1.
Figure 2.
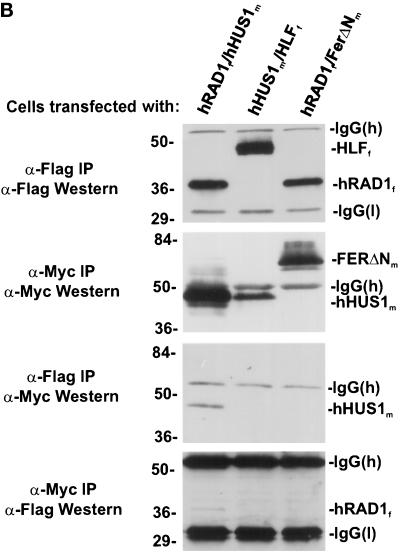
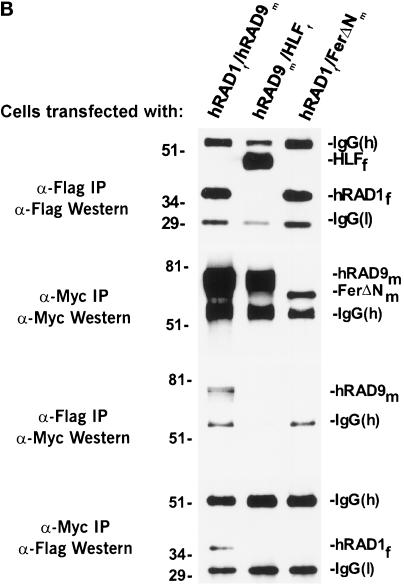
hRAD1 and hHUS1 interact specifically. (A) S cerevisiae strain HF7c was cotransformed with the indicated GAL4 fusion plasmids and plated on media selecting for cotransformants. Single colonies were then subcultured onto selective media in the presence (left) or absence (right) of histidine. Similar to the positive control (lower left quadrant), cotransformation of pGBT9-hHUS1 and pGAD-hRAD1 resulted in growth in the absence of histidine (bottom right quadrant). When either construct was cotransformed separately with the corresponding empty GAL4 vector, no growth on −trp, −leu, −his media was observed (upper two quadrants). (B) COS-1 cells were transiently cotransfected with constructs expressing hRAD1f and hHUS1m, HLFf and hHUS1m, or hRAD1f and FerΔNm, as indicated. After harvest, lysates were immunoprecipitated with α-flag M2 monoclonal antibody or α-myc 9E10 monoclonal antibody. Each of these immunoprecipitations were then subjected to immunoblotting with either α-myc or α-flag antibodies, as indicated on the left. Specific coimmunoprecipitation of hRAD1f and hHUS1m is observed in lane 1 of each of the bottom two panels.
To confirm this hypothesis, exogenously expressed hRAD1 and hHUS1 were coimmunoprecipitated in COS-1 cells (Figure 2B) using flag epitope-tagged hRAD1 and myc-epitope tagged hHUS1. HLFf and FerΔNm were included as negative controls to ensure the specificity of the interaction. Cells were cotransfected as indicated with hRAD1f/hHUS1m, HLFf/hHUS1m, or hRAD1f/FerΔNm. The cells were harvested 48 h after transfection, lysed, and immunoprecipitated with either α-flag M2 monoclonal antibody or α-myc 9E10 monoclonal antibody. Two aliquots from each sample were electrophoresed through two identical polyacrylamide gels, one of which was used for an α-flag Western blot and the other for an α-myc Western blot. Although exogenous hRAD1f protein levels were approximately equivalent in both pyDF31-hRAD1 transfections (Figure 2B, α-Flag IP/α-Flag Western), hRAD1f immunoprecipitated only with hHUS1m and not FerΔNm (Figure 2B, α-Myc IP/α-Flag Western). Similarly, hHUS1m immunoprecipitated with hRAD1f but not HLFf (Figure 2B, α-Flag IP/α-Myc Western). Together, these results verify the existence of a specific physical interaction between hHUS1 and hRAD1.
hRAD9 and hRAD1 Physically Interact
Having observed the two interactions described above, it seemed logical to explore the association status of hRAD9 and hRAD1. Using the pGBT9-hRAD9 and pGAD-hRAD1 GAL4 fusion constructs, we repeated the yeast two-hybrid experiment described above (Figure 3A). Despite growth of the p53/pSV40 T-Ag–positive control (Figure 3A, lower left quadrant), coexpression of hRAD9 and hRAD1 fusions failed to assemble a functional GAL4 and hence did not produce viable yeast in the absence of histidine (Figure 3A, lower right quadrant). Therefore, although the yeast two-hybrid system demonstrated interactions between hHUS1 and hRAD9, and hHUS1 and hRAD1, it showed no interaction between hRAD9 and hRAD1.
Figure 3.
hRAD9 and hRAD1 coimmunoprecipitate (A) S. cerevisiae strain HF7c was cotransformed with the indicated GAL4 fusion plasmids and plated on media selecting for cotransformants. Single colonies were then subcultured onto selective media in the presence (left panel) or absence (right panel) of histidine. Although the p53/SV40 T-Ag positive control grew in the absence of histidine (lower left quadrant), expression of both hRAD9 and hRAD1 GAL4 fusions did not result in the formation of HIS+ yeast colonies (bottom right quadrant). Therefore, no interaction was detectable between these two proteins. (B) COS-1 were transiently cotransfected with constructs expressing hRAD1f and hRAD9m, HLFf and hRAD9m, or hRAD1f and FerΔNm, as indicated. After harvest, lysates were immunoprecipitated with α-flag M2 monoclonal antibody or α-myc 9E10 monoclonal antibody. Each immunoprecipitation was then immunoblotted with α-myc or α-flag antibodies, as indicated on the left. Specific coimmunoprecipitation of hRAD1f and hRAD9m is observed in lane 1 of each of the bottom two panels.
We went on to examine the ability of hRAD9 and hRAD1 to coimmunoprecipitate in COS-1 cells (Figure 3B). hRAD9m and hRAD1f were exogenously expressed either together or separately with HLFf or FerΔNm, as described above. Neither hRAD1 nor hRAD9 protein expression levels varied significantly between different transfection (Figure 3B, α-Flag IP/α-Flag Western and α-Myc IP/α-Myc Western, respectively). Contrary to the yeast two-hybrid data, hRAD9m, but not FerΔNm, immunoprecipitated with hRAD1f (Figure 3B, α-Flag IP/α-Myc Western), and hRAD1f, but not HLFf, immunoprecipitated with hRAD9m (Figure 3B, α-Myc IP/α-Flag Western). Therefore, although hRAD1 and hRAD9 show no interaction in the two-hybrid system, they do specifically coimmunoprecipitate with each other when exogenously expressed in COS-1 cells.
hRAD9 Is Phosphorylated in Undamaged Cells
From the earliest immunoprecipitations we performed using antibodies directed against either native or epitope-tagged hRAD9 (Figures 1 and 3), we noted three discrete bands, the smallest of which corresponded approximately to the predicted size of hRAD9 (Lieberman et al., 1996). To test our hypothesis that these multiple bands were the result of phosphorylation, we examined the effect of phosphatase treatment on hRAD9’s migration through acrylamide. COS-1 cells were transfected with our hRAD9m-expressing construct and harvested 48 h later. The cells were lysed and immunoprecipitated with 9E10 monoclonal antibody directed against the myc epitope. Immunoprecipitates were either untreated or treated with CIP in the presence or absence of sodium orthovanadate. Samples were then subjected to SDS PAGE, transferred to nitrocellulose, and immunoblotted with α-myc monoclonal antibodies. Figure 4A, lane 1, shows the multiple banding pattern of hRAD9m in immunoblots, similar to that seen in Figures 1 and 3. Treatment with CIP causes the slower-migrating bands to disappear, leaving only the fastest form. This effect can be alleviated by the phosphatase competitor sodium orthovanadate, confirming that the slower-migrating bands are the result of multiple phosphorylation states of hRAD9m.
Figure 4.
hRAD9 is phosphorylated in undamaged cells. (A) COS-1 cells transiently transfected with the construct expressing hRAD9m. After harvest, lysates were immunoprecipitated with α-myc 9E10 monoclonal antibody. Fractions of the immunoprecipitate were either treated or not with CIP in the presence or absence of orthovanadate (VO4). Samples were then subjected to Western analysis using antibody directed against the myc epitope. CIP treatment resulted in elimination of slower-migrating forms of hRAD9m, an effect that was not observed when VO4 was present. (B) Logarithmically growing HeLa cells were harvested and subjected to Western analysis using polyclonal α-hRAD9 antibodies. As in A, samples also treated with CIP in the presence or absence of vanadate, as indicated.
We have also demonstrated that the phosphorylation of hRAD9m is due neither to the overexpression of the protein in COS-1 cells nor to the myc epitope tag. We did this using polyclonal chicken antibodies directed against hRAD9 that are of sufficient sensitivity and specificity to detect endogenous hRAD9 in HeLa cells. Essentially the same experiment as above was performed, with the exception that endogenous hRAD9 was detected by polyclonal α-hRAD9 antibodies. In this case, only a single slower-migrating band was observed (Figure 4B, lane 1). Also, by contrast with the overexpressed hRAD9 from COS-1 cells, most of the hRAD9 in HeLa cells are phosphorylated. The hRAD9 can be converted to the faster-migrating, dephosphorylated form by treatment with CIP, and this reaction is sensitive to the phosphatase inhibitor sodium orthovanadate (Figure 4B, lanes 2 and 3), indicating that endogenous hRAD9 is phosphorylated in HeLa cells.
hRAD9 Is a Nuclear Protein
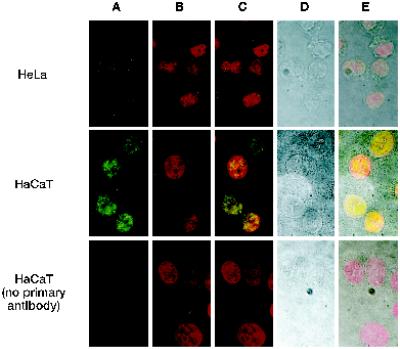
To determine where hRAD9 localizes in the cell, we used immunofluorescence with a fluorescent secondary antibody directed against the polyclonal α-hRAD9 chicken antibodies. These hRAD9 antibodies are able to specifically detect endogenous hRAD9, as evidenced by Figure 4B. The location of the Alexa 488 goat α-chicken secondary antibody is represented in green in Figure 5A. The specificity of the secondary antibody is demonstrated by the absence of signal in the absence of primary α-hRAD9 antibodies (Figure 5A; bottom row). Propidium iodide staining was used to determine the location of the nucleus (Figure 5B), and the images from Figure 5, A and B, are superimposed in Figure 5C. Corresponding light microscope images are presented in Figure 5D and superimposed with the fluorescent staining in Figure 5E. The cellular membranes are clearly visible in the HeLa cells, and hRAD9 staining is confined to the nucleus. Similarly, in the confluent HaCaT cells, all hRAD9 staining is nuclear. In both cases the staining is punctate.
Figure 5.
hRAD9 is located in the nucleus. Confocal immunofluorescence and light microscopy was performed on <50% confluent HeLa cells and 90% confluent HaCaT cells. Cells were fixed and probed with α-hRAD9 chicken polyclonal antibodies, followed by a fluorescently labeled anti-chicken IgY secondary antibody (A). DNA was visualized by staining with propidium iodide (B). Images from A and B were superimposed (C). The cellular borders of the HeLa and confluent HaCaT cells were visualized by light microscopy (D), and the light and fluorescent images were superimposed (E).
DISCUSSION
We have demonstrated three interactions between three human checkpoint rad proteins, hRAD1, hRAD9, and hHUS1. In all cases, these interactions were substantiated using both the yeast two-hybrid system and by coimmunoprecipitation, except for hRAD1 and hRAD9, which did not interact in the yeast two-hybrid system but did coimmunoprecipitate when exogenously expressed in COS-1 cells. The original observation that led to this work was that hRAD9 interacted with hHUS1 in a two-hybrid screen. Eleven of 15 interactions isolated in the screen that used hRAD9 as bait were hHUS1. It is interesting to note that hRAD1 was not among the remaining isolates, which are still being characterized. However, the observation that hRAD1 and hRAD9 show no interaction in the two-hybrid system had been made previously (Parker et al., 1998b). It now appears that this interaction may be dependent on factors that are absent in budding yeast, because hRAD1 and hRAD9 specifically coimmunoprecipitate in COS-1 cells. Such factors may include a budding yeast equivalent of hHUS1, the existence of which seems unlikely considering that no homologues have been identified based on sequence. Alternatively, the N-terminal GAL4 domain of the fusion proteins may result in a conformational change that prevents association of these two proteins. This hypothesis is supported by our observation that reversing the orientation of the hRAD9/hHUS1 and hRAD1/hHUS1 GAL4 fusions abolishes the HIS3 reporter gene activation (St. Onge and Udell, unpublished results). Furthermore, an N-terminal myc-tagged version of fission yeast hus1 has been shown to function as a dominant negative allele (Kostrub et al., 1997). Future use of dominant negative fusions involving human proteins could prove invaluable in uncovering the mechanistic details involved in checkpoint signaling.
It has been shown in fission yeast that Hus1 and Rad1 interact, and that this interaction is dependent on the presence of Rad9, because interaction does not occur in a rad9-null background (Kostrub et al., 1998). Our data offer strong evidence that such a complex also exists in humans, although it may be assembled differently. Although we have only demonstrated pair-wise interactions between the three human checkpoint proteins, the simplest explanation of this and the yeast data together is that a three-way complex exists among hRAD1, hRAD9, and hHUS1. We cannot rule out the possibility that the observed hRAD1–hHUS1 interactions described here are bridged by DDC1, the S. cerevisiae homologue of hRAD9 (Longhese et al., 1997; Paciotti et al., 1998), or by and endogenous monkey homologue of hRAD9 in COS-1 cells. Such evidence will ultimately have to be achieved using hRAD9-null cell lines. Furthermore, with the highly similar phenotypes observed in all of the fission yeast checkpoint rad mutants, and considering recent data demonstrating an interaction between hRAD1 and hRAD17 (Parker et al., 1998b), and that ATR, a human homologue of fission yeast Rad3, exists predominantly as part of a high-molecular-weight complex (Wright et al., 1998), the potential for a multiprotein complex involving all of the checkpoint rad proteins must not be overlooked.
We have also shown that both exogenous and endogenous hRAD9 are phosphorylated at multiple sites. Considering that S. cerevisiae DDC1 and S. pombe Hus1 both appear to be phosphorylated in response to DNA damage (Kostrub et al., 1997; Paciotti et al., 1998), phosphorylation is an integral component of checkpoint signaling. To determine whether checkpoint activation affects hRAD9 phosphorylation, we investigated whether γ radiation or hydroxyurea could induce a change in the migration pattern of endogenous hRAD9 on a Western blot. Neither a 4-Gy dose of γ radiation nor incubation in 0.1 mM hydroxyurea for up to 24 h affected the migration of endogenous hRAD9 from HaCaT cells, although hRAD9 is already highly phosphorylated in these cells. We cannot rule out the possibility that ongoing replication or the presence of endogenous DNA damage may be inducing hRAD9 phosphorylation in the absence of exogenous signals. It is worth noting that phosphorylation is not an absolute requirement for association of hRAD9 and hRAD1, because hRAD1 immunoprecipitation will coimmunoprecipitate all forms of hRAD9 (Figure 3).
Finally, we have investigated the subcellular localization of hRAD9, and we have shown that hRAD9 is a nuclear protein (Figure 5). This observation was not a foregone conclusion, because the start of the checkpoint signal transduction pathway is nuclear (DNA damage), whereas the end is cytoplasmic (the cell cycle machinery). Unlike hRAD1, which has been shown to be present mainly in a diffuse pattern in the nucleus (Freire et al., 1998), the staining pattern of hRAD9 within the nucleus shows discrete areas of intense staining. It will be interesting to further characterize the nature of these foci, including determining what other proteins are present, and whether DNA synthesis, either replicative or unscheduled, is occurring in these regions.
The reason for the current intense interest in cell cycle checkpoint control is the association of defects in checkpoint control with human cancers. Genomic instability is a common feature accompanying checkpoint loss, regardless of which checkpoint is compromised, and whether the cell is subjected to exogenous stresses (Weinert and Hartwell, 1990; Livingstone et al., 1992; Yin et al., 1992). A great deal of evidence now links genomic instability with the multistep origin of human cancer (Loeb, 1991; Loeb and Christians, 1996; Hartwell, 1992; Meyn, 1995; Smith and Fornace, 1995; Thrash-Bingham et al., 1995; Tlsty et al., 1995; Perucho, 1996). The number of checkpoint control genes that act as tumor suppressors under normal circumstances is growing and currently includes p53 (Malkin et al., 1990; Kastan et al., 1992; Kuerbitz et al., 1992), ATM (Savitsky et al., 1995a,b; Xu and Baltimore, 1996), BLM (Ellis et al., 1995; Davey et al., 1998), and hBUB1 (Cahill et al., 1998). Although none of the checkpoint rad proteins has yet been shown to act as a tumor suppressor, those that have been mapped all localize to regions associated with loss of heterozygosity in tumors, which is indicative of the presence of tumor-suppressing genes (Lieberman et al., 1996; Parker et al., 1998a,b). Also, genomic instability has been associated with G2 checkpoint deficiency in budding yeast rad9 mutants (Weinert and Hartwell, 1990). Ultimately, the work reported here will shed light on the mechanistic details of how genomic stability is maintained by the G2 and S-phase checkpoints.
ACKNOWLEDGMENTS
We thank Dr. Peter Greer and Dr. David LeBrun for providing plasmids used in this work. We also thank Jennifer Pelley and Dennis Kim for technical assistance and Lee Fraser and Deborah Greer for critically reading the manuscript. This work was supported by Medical Research Council of Canada grant MT-14352 and National Institutes of Health grant ES07940-01A1 (to S.D.). S.D. is a Cancer Care Ontario scientist. R.P.S.O. is the recipient of a Queen’s University graduate award. C.M.U. is the recipient of US Army Breast Cancer research studentship DAMD17-98-1-8080. The confocal microscope used in this work is partially supported by Medical Research Council of Canada equipment maintenance grant MT-7827.
REFERENCES
- al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Shin RB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey S, Han CS, Ramer SA, Klassen JC, Jacobson A, Eisenberger A, Hopkins KM, Lieberman HB, Freyer GA. Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom’s syndrome disease gene. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes & Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Freire R, Murguia JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP. Human and mouse homologs of Schizosaccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: linkage to checkpoint control and mammalian meiosis. Genes & Dev. 1998;12:2560–2573. doi: 10.1101/gad.12.16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, al-Khodairy F, Ghazizadeh H, Carr AM, Enoch T. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol Gen Genet. 1997;254:389–399. doi: 10.1007/pl00008606. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, Knudsen K, Subramani S, Enoch T, Kostrub CF, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman HB, Hopkins KM, Nass M, Demetrick D, Davey S. A human homolog of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc Natl Acad Sci USA. 1996;93:13890–13895. doi: 10.1073/pnas.93.24.13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tlsty TD. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- Loeb LA, Christians FC. Multiple mutations in human cancers. Mutat Res. 1996;350:279–286. doi: 10.1016/0027-5107(95)00117-4. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Meyn MS. Ataxia-telangiectasia and cellular responses to DNA damage. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Oostveen I, Yon J, Verhasselt P, Luyten WH. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998a;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Verhasselt P, Luyten WH. Identification of a human homologue of the Schizosaccharomyces pombe rad17+ checkpoint gene. J Biol Chem. 1998b;273:18340–18346. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- Perucho M. Cancer of the microsatellite mutator phenotype. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes & Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes & Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995a;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Sfes S, Jagle DA, Zio Y, Sartiel A, Collins FS, Shiloh Y, Rotman G. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet. 1995b;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- Smith ML, Fornace AJ., Jr Genomic instability and the role of p53 mutations in cancer cells. Curr Opin Oncol. 1995;7:69–75. [PubMed] [Google Scholar]
- Thrash-Bingham CA, Salazar H, Freed JJ, Greenberg RE, Tartof KD. Genomic alterations and instabilities in renal cell carcinomas and their relationship to tumor pathology. Cancer Res. 1995;55:6189–6195. [PubMed] [Google Scholar]
- Tlsty TD, Briot A, Gualberto A, Hall I, Hess S, Hixon M, Kuppuswamy D, Romanov S, Sage M, White A. Genomic instability and cancer. Mutat Res. 1995;337:1–7. doi: 10.1016/0921-8777(95)00016-d. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Udell CM, Lee SK, Davey S. HRAD1 and MRAD1 encode mammalian homologues of the fission yeast rad1+ cell cycle checkpoint control gene. Nucleic Acids Res. 1998;26:3971–3978. doi: 10.1093/nar/26.17.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to. cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes & Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]