Abstract
Screening of a library derived from primary human endothelial cells revealed a novel human isoform of vesicle-associated membrane protein-1 (VAMP-1), a protein involved in the targeting and/or fusion of transport vesicles to their target membrane. We have termed this novel isoform VAMP-1B and designated the previously described isoform VAMP-1A. VAMP-1B appears to be an alternatively spliced form of VAMP-1. A similar rat splice variant of VAMP-1 (also termed VAMP-1B) has recently been reported. Five different cultured cell lines, from different lineages, all contained VAMP-1B but little or no detectable VAMP-1A mRNA, as assessed by PCR. In contrast, brain mRNA contained VAMP-1A but no VAMP-1B. The VAMP-1B sequence encodes a protein identical to VAMP-1A except for the carboxy-terminal five amino acids. VAMP-1 is anchored in the vesicle membrane by a carboxy-terminal hydrophobic sequence. In VAMP-1A the hydrophobic anchor is followed by a single threonine, which is the carboxy-terminal amino acid. In VAMP-1B the predicted hydrophobic membrane anchor is shortened by four amino acids, and the hydrophobic sequence is immediately followed by three charged amino acids, arginine-arginine-aspartic acid. Transfection of human endothelial cells with epitope-tagged VAMP-1B demonstrated that VAMP-1B was targeted to mitochondria whereas VAMP-1A was localized to the plasma membrane and endosome-like structures. Analysis of C-terminal mutations of VAMP-1B demonstrated that mitochondrial targeting depends both on the addition of positive charge at the C terminus and a shortened hydrophobic membrane anchor. These data suggest that mitochondria may be integrated, at least at a mechanistic level, to the vesicular trafficking pathways that govern protein movement between other organelles of the cell.
INTRODUCTION
A group of membrane-bound proteins, known collectively as soluble N-ethylmaleidmide-sensitive factor attachment protein (SNAP) receptors (SNAREs),1 have garnered intense scrutiny because of their involvement in vesicular trafficking (reviewed by Rothman, 1994; and Pfeffer, 1996). One set of SNAREs (v- or vesicle-SNAREs) are embedded in the membrane of transport vesicles, and another set (t- or target-SNAREs) are embedded in the membrane to which the vesicles are targeted. Genetic studies in yeast and biochemical studies in both yeast and mammalian cells have demonstrated that these proteins are indispensable for transport vesicle fusion. Particularly compelling is the demonstration that clostridial neurotoxins with endometalloprotease activity inhibit regulated secretion by specifically cleaving SNARE proteins (Neimann et al., 1994). v-SNAREs directly bind to t-SNAREs in interactions that show pairwise specificity. For example, the well characterized v-SNAREs, vesicle associated membrane protein (VAMP) isoforms 1 and 2 (also known as synaptobrevins 1 and 2; for the purpose of clarity these proteins will be referred to as VAMPs for the remainder of this report), bind in vitro to the t-SNARE syntaxin isoforms 1 and 4, but not syntaxin isoforms 2 or 3 (Calakos et al., 1994). Furthermore, SNAREs are specifically localized to different organelles in the cell. In yeast and mammalian cells both v- and t-SNAREs have been identified that are localized to transport vesicles and target membranes associated with the endoplasmic reticulum (Newman et al., 1990; Banfield et al., 1994; Hay et al., 1996), Golgi (Hardwick and Pelham, 1992; Hay et al., 1996; Nagahama et al., 1996), and plasma membrane (Aalto et al., 1993; Bennett, et al., 1993) among other sites. These two attributes, specificity in binding and specificity in localization, have given rise to the “SNARE hypothesis” (Sollner et al., 1993), which states, in its most simple form, that the interaction between v- and t-SNAREs governs the targeting of transport vesicles to the appropriate target membrane. A targeting role for the SNARES is, however, the subject of debate. For example, the consequence of inactivating either syntaxin or VAMP is the accumulation of secretory vesicles at the target membrane (Hunt et al., 1994; Broadie et al., 1995), consistent with a role for the SNAREs in fusion, but clouding the issue of SNARE involvement in vesicle targeting.
The localization of the SNARE proteins to specific membranes is thought to be intrinsic to their mechanistic role and has been directly shown to be essential for function (Regazzi et al., 1996). Both the syntaxin- and VAMP-related SNAREs are anchored by carboxy-terminal hydrophobic anchors (another important but unrelated t-SNARE, SNAP-25, is anchored by palmitoylation). Membrane insertion of this class of membrane protein appears to be posttranslational (reviewed in Kutay et al., 1993). However, at least for VAMP-2, the protein is not inserted directly into transport vesicles, but instead is inserted into the membrane of the endoplasmic reticulum, by a mechanism distinct from the signal recognition particle-mediated pathway, and routed through the Golgi apparatus for incorporation into vesicles (Kutay et al., 1995). The mechanism directing VAMP-2 to synaptic vesicles relies on an amphipathic helix (helix-1) in the conserved cytoplasmic domain of the protein (Grote et al., 1995). When this helix is disrupted, VAMP-2 is localized predominantly in endosomes, which therefore may be considered a “default” localization for this protein. For other carboxy-terminal–anchored proteins, the localization sequences within the proteins are, by and large, less well defined, although both transmembrane and cytosolic portions of these proteins have been implicated (Kutay et al., 1993; Banfield et al., 1994; Yang et al., 1997). The role of the structure of the signal-anchor sequence in targeting a v-SNARE has been studied in detail in yeast (Rayner and Pelham, 1997). The protein studied, Ufe1p, is a v-SNARE localized at steady state to the endoplasmic reticulum. In the case of Ufe1p, the length of the hydrophobic anchor is critically important to retention in the endothelial reticulum (ER). Lengthening the Ufe1p anchor by four hydrophobic residues results in relocalization to a endosome-like compartment, and further lengthening by two additional amino acids leads to plasma membrane localization. Interestingly, a sequence-specific retention in the ER was also demonstrated, indicating that proteins within the plane of the membrane interact with Ufe1p to retain it in the ER.
We undertook screening of an endothelial cell library for SNARE proteins to better understand regulated secretion in those cells. This screen uncovered the presence of VAMP-2 and SNAP-25 (our unpublished data). In addition, as reported here, we have found an isoform of VAMP-1 that, relative to the previously reported VAMP-1 sequence, has an altered carboxy-terminal tail. This alteration results in the creation of a mitochondrial targeting sequence.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, all reagents were from commercial sources and of reagent-grade or higher.
Antibodies
Anti-FLAG epitope monoclonal M2 was purchased from Kodak Imaging Systems (Rochester, NY). Polyclonal rabbit anti-Tom20 (Hanson et al., 1996) was a generous gift of Dr. Nicholas Hoogenraad (La Trobe University, Bundoora, Victoria, Australia). Biotinylated swine anti-rabbit antibody was from DAKO (Glostrup, Denmark). Streptavidin conjugated to Tri-color was purchased from Caltag Laboratories (South San Francisco, CA). FITC-conjugated goat anti-mouse was from Silenus (Hawthorne, Victoria, Australia).
Cells
Human umbilical vein endothelial cells (HUVEC) were isolated and cultured on gelatin-coated flasks as previously described (Wall et al., 1978). Cells were generally used at passage 3. For transfection and immunofluorescence, cells were plated into eight-well chamber slides (Nalge Nunc International, Naperville, IL) coated with fibronectin, 50 μg/ml (Boehringer-Mannheim, Indianapolis, IN) at 1.25 × 104 cells/well. Primary fibroblasts were isolated from umbilical cord by a similar procedure. Human peripheral blood neutrophils were isolated from healthy donors by dextran density gradient sedimentation followed by hypotonic lysis of erythrocytes. Jurkat cells were cultured in RPMI + 10% FCS and HepG2 cells were cultured in DMEM + 10% FCS.
Immunofluorescence
Endothelial cells were washed once with PBS and then fixed for 10 min at room temperature in 4% paraformaldehyde in PBS. Cells were then washed three times with PBS/0.1% Triton X-100 (PBS/Triton) and permeabilized for 10 min in PBS/Triton. Primary antibodies were then added in PBS/Triton containing 3% BSA and incubated for 1 h followed by three washes with PBS/Triton. Secondary and, where appropriate, tertiary antibodies, diluted in PBS/Triton/BSA, were added for 1 h followed by three washes with PBS/Triton. Coverslips were then added over a mounting solution of 2% n-propylgallate in glycerol. M2 was generally used at 3.6 μg/ml. Anti-Tom20 was diluted 1:200. M2 staining was visualized with FITC anti-mouse diluted 1:100. Anti-Tom20 staining was visualized with biotinylated anti-rabbit (diluted 1:300), and then streptavidin Tri-color (diluted 1:50). Confocal microscopy was performed on an Olympus BH2 microscope (Tokyo, Japan) (40× objective) equipped with a Bio-Rad MRC-600 argon laser confocal system (Bio-Rad, Richmond, CA) with the A1 and A2 filter blocks (excitation at 514 nm, A-1 emission filter, 525–555 bandpass; A-2 emission filter, 600 longpass).
Human brain RNA was the generous gift of Drs. Hugh Reid and Claude Bernard (La Trobe University, Bundoora, Victoria, Australia). RNA from tissue culture cells was prepared as described previously (Chomczynski and Sacchi, 1987).
Cloning of VAMP-1B
A HUVEC cDNA library was constructed from poly A+ mRNA isolated from first-passage HUVEC plated at low density (0.5 × 105 cells/3.5-cm diameter plate) and reverse transcribed using the ZAP Express cDNA synthesis kit (Stratagene, La Jolla, CA) according to the manufacture’s instructions. The resulting cDNA was then cloned into the pBK Phagemid vector (Stratagene) according to the manufacturer’s instructions (Roberts, unpublished). Plaques (5 × 105) were screened with a sequence spanning the VAMP-1 coding sequence generated by PCR from human brain DNA (see below). The probe was hybridized in 0.5% nonfat dry milk/5× SSC/0.5% SDS/100 μg/ml salmon sperm DNA/50% formamide at 42°C overnight and then washed three times in 0.1% SDS/0.1× SSC at 65°C. After three rounds of plaque purification, the sequences were subjected to in vivo excision into pBK-CMV for sequencing. Clones were sequenced from both ends using the Ready Reaction Cycle Sequencing kit (Perkin Elmer-Cetus, Branchburg, NJ) on an ABI 373 DNA sequencer (Perkin Elmer-Cetus).
PCR Conditions
cDNAs were produced using avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI) according to the manufacturer’s instructions. In all cases, hot-start PCR was used. VAMP-1A was amplified from human brain cDNA using primers derived from the rat VAMP-1 sequence (5′-primer, 5′-GATCGAATTCAAATGCTGCTCCAGCTC-3′, 3′-primer, 5′-GACTGATTCCTCAAGTAAAAATGTAGATT AC-3′). Long-range PCR was performed using a Taq (Perkin Elmer-Cetus) to Pfu (Stratagene) ratio of 80:1 under buffer and nucleotide conditions as described previously (Barnes, 1994). PCR conditions were 95°C (2 min) [95°C (1 min), 53°C (1 min), 70°C(2 min)] × 36 and 72°C (5 min), 4°C utilizing a PTC-100 thermal cycler (MJ Research, Watertown, MA). For PCR analysis of VAMP-1A and -1B mRNA content of cells in culture and human brain, similar PCR conditions were used except that extension was at 72°C. Primers spanning the complete coding sequence of the corresponding proteins were used: VAMP-1A, 5′-GACTGAATTCAAATGTCTGCTCCAGCTC-3′ (5′-primer); 5′-GACTGAATTCTTTCAAGTAAAAAAAGTAGATTAC-3′ (3′-primer); VAMP-1B, same 5′-primer as for VAMP-1A; 5′-GACTGAATTCAATCAGTCCCGCCTAACAAT-3′ (3′-primer); VAMP-2, 5′-GACTGAATTCAAATGTCTGCTACCGCTG-3′ (5′-primer); 5′-GACTGAATTCATTTAAGAGCTGAAGTAAACT-3′ (3′ primer); GAPDH, 5′-ACCACCATGGAGAAGGCTGG-3′ (5′ primer); 5′-CTCAGTGTAGCCCAGGATGC-3′ (3′-primer). After electrophoresis of the PCR products on 2% agarose gels, the products were hybridized to Hybond-N (Amersham, Buckinghamshire, England) nylon membranes and probed for VAMP-1A, 1B, and VAMP-2 under conditions identical to those used for screening the endothelial cell cDNA library. For VAMP-1A and -1B, the probe used was the complete VAMP-1B clone (Figure 1). For VAMP-2 the sequence was a 900-base pair (bp) fragment of human VAMP-2 cloned from the endothelial cell library, including the complete coding sequence, 63 bp upstream of the start site, and 474 bp of 3′-untranslated sequence.
Figure 1.
DNA and amino acid sequence comparison between the novel VAMP isoform VAMP-1B and VAMP-1A. (A) Comparison of cDNA sequence of clones encoding VAMP-1B and VAMP-1A (deduced from the genomic sequence). Solid arrows indicate limits of the coding sequences, and the open arrow indicates a splice junction at the 3′-end of the coding sequence. The VAMP-1B cDNA sequence has been deposited in GenBank with acccession number AF060538. (B) Carboxy-terminal hydrophobic amino acid sequences of VAMP-1A and human and rat VAMP-1B predicted from the corresponding cDNAs. Also shown are the sequences of BCL-2, monoamine oxidases A and B, and metaxin, four proteins anchored in mitochondria membranes by carboxy-terminal hydrophobic sequences. The hydrophobic sequences are underlined, and charged amino acids are indicated.
Construction of Expression Vectors Containing FLAG Epitope-Tagged VAMP-1A and VAMP-1B
The FLAG epitope was added in-frame to the N terminus of both VAMP-1A and VAMP-1B by PCR amplification of the cloned sequences (using conditions identical to those described above) with primers introducing an NcoI site and a consensus Kozak sequence at the 5′-end and an SpeI site at the 3′-end of the new coding sequence. The VAMP-1A primers used were: 5′-AGTCCACCATGGACTACAAGGACGACGATGACAAGTCTGCTCCAGCTCAGCCACC-3′ (5′-primer) and 5′-ACGTACTAGTAGTTCATTTTTTCATCTAATGTT ATTG-3′ (3′-primer). The 5′-primer used to amplify the VAMP-1B sequence was identical to the VAMP-1A 5′-primer. The 3′-primer used was 5′-ACGTACTAGTCTT CAGTCCCGCCTTACAATAAC-3′. The restriction fragments were cloned into the NcoI/Xba site of p-act (transcription driven by the β-actin promoter) (Nishina et al., 1989).
Transfection of HUVEC
HUVEC at passage 3 were plated in six-well trays at 1.25 × 105 cells/well overnight. Lipofectin (6 μl) (GIBCO-BRL, Grand Island, NY) was incubated in 100 μl Opti-MEM (GIBCO-BRL) for 45 min and then mixed with 1 μg of DNA in 100 μl Opti-MEM. After 15 min this mixture was diluted with 800 μl Opti-MEM. Cells were washed with warm Opti-MEM, and 300 μl of the transfection mix were added to each well. The cells were incubated for 3 h at 37°C, the transfection mix was removed and replaced with complete medium, and the cells were incubated for 48 h before preparation for immunofluorescence microscopy.
RESULTS
Cloning of a Novel Isoform of VAMP-1 from a Human Endothelial Cell Library
In the course of identifying the so-called SNARE proteins in vascular endothelial cells, an endothelial cell library was probed with a sequence derived from human VAMP-1. Twenty-seven clones were identified on initial screening. Seven plaques were isolated and purified. Sequencing revealed that three clones represent VAMP-2 sequences (our unpublished data). The remaining four clones were identical and contained a sequence with partial identity to VAMP-1. The 5′-portion of this sequence was virtually identical to the sequence of human neuronal VAMP-1 cDNA (Figure 1A). The single base deviation at the extreme 5′-end of the sequence (base 8) may be due to sequence ambiguity or polymorphism. The endothelial cell VAMP-1 sequence completely diverged from the previously derived sequence at the extreme 3′-end of the coding sequence. The VAMP-1 mRNA is encoded by five exons (Archer et al., 1990). The sequence divergence between the VAMP-1 sequence cloned here and the previously described neuronal VAMP-1 comes precisely at the boundary between exons IV and V. Exon V encodes the C-terminal five amino acids of the previously described VAMP-1. Therefore, the sequence divergence described here alters the very carboxy terminus of the protein (Figure 1B). Because of the sequence identity at the 5′-end of these sequences, even in the 5′-untranslated region, the fact that the divergence comes at a known splice junction, and because only a single VAMP-1 gene has been detected in human genomic DNA (Archer et al., 1990), we presume that this new sequence represents an alternatively spliced form of VAMP-1. We have therefore termed the originally derived sequence as VAMP-1A and the newly described isoform as VAMP-1B. A similar rat splice variant of VAMP-1 (also termed VAMP-1B) has been described recently (Mandic et al., 1997). The 3′-region of this sequence also shows strong homology to an uncharacterized cDNA fragment isolated from murine spleen by differential display (Ivanova and Belyavsky, 1995).
VAMP-1B Is Widely Distributed among Cell Lines
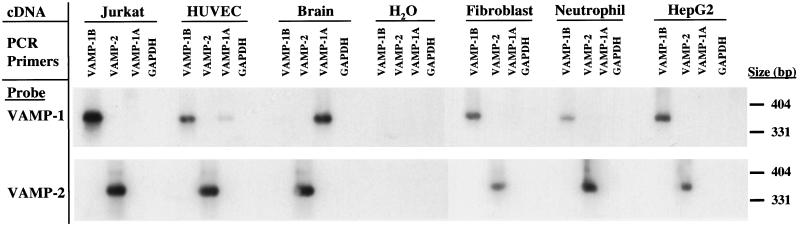
To determine whether VAMP-1B is restricted to endothelial cells or has a broader distribution, the presence of VAMP-1B mRNA was determined in a number of cell types by RT-PCR analysis. The presence of VAMP-1A and VAMP-2 message was also determined. Primers were designed that allowed amplification of the complete coding sequences of the VAMPs. In many cases the reaction products were poorly visualized on ethidium bromide-stained gels; therefore, the amplified bands were detected by Southern blotting using cDNA probes that either detect both VAMP-1A and VAMP-1B (derived from the complete VAMP-1B coding sequence) or VAMP-2. VAMP-1B was detected in cDNA from Jurkat cells, HUVEC, fibroblasts, neutrophils, and HepG2 cells (Figure 2). VAMP-1B was also detected in HeLa cell mRNA (not shown). The products were all of the predicted size (363 bp). In contrast, no VAMP-1A was detected in the cultured cells, except a variable weak band in endothelial cells. Human brain RNA produced an easily detectable amplification product with the VAMP-1A primers, as expected. All cDNAs tested amplified VAMP-2 sequences of the correct size (363 bp).
Figure 2.
PCR analysis of the distribution of VAMP-1A, VAMP-1B, and VAMP-2 mRNA. mRNA from Jurkat cells, human endothelial cells (HUVEC), human brain, primary fibroblasts, human neutrophils, and HepG2 cells was prepared and reversed transcribed. The cDNAs and a water control (no cDNA) were then amplified using primers specific for either VAMP-1A, VAMP-1B, VAMP-2, or GAPDH (see MATERIALS AND METHODS). The resulting reactions were separated on agarose gels, blotted to nylon membranes, and then visualized in a Southern blot with probes derived from the complete coding sequences for VAMP-1B (top) or VAMP-2 (bottom). The VAMP-1B probe used here is identical to the VAMP-1A sequence over 339 of 347 bases, and so readily recognizes the VAMP-1A sequence.
Preliminary Northern blotting was performed on RNA from cell lines and human tissues using probes derived from the divergent 3′-untranslated regions of VAMP-1A and VAMP-1B (our unpublished results). This probe hybridized to a band of ∼3 kilobases (kb) in mRNA from heart, placenta, lung, liver, skeletal muscle, kidney, pancreas, and brain. The signal was particurly strong in liver and pancreas. In addition, a weak, ∼1-kb band hybridized to the probe in mRNA from cells in culture but not in brain mRNA. The size of this message is identical to that of a potential murine VAMP-1B homologue isolated by differential display (Ivanova and Belyavsky, 1995). In brain, a strongly hybridizing 2.8-kb band was detected with the VAMP-1B probe, as well as a VAMP-1A probe. This is the reported size of the VAMP-1A message (Elferink et al., 1989). The analysis of Mandic and colleagues indicates that in rat the VAMP-1B message contains the VAMP-1A exon in its 3′- untranslated sequence. This presumably explains the cross-hybridization between the 3′-probe used here and the VAMP-1A message in brain.
VAMP-1B Contains a Mitochondrial Targeting Sequence
In VAMP-1A, exon V encodes the carboxy-terminal five amino acids, consisting of the last four hydrophobic amino acids of the membrane anchor, and a C-terminal threonine. The alternative exon in VAMP-1B replaces these five amino acids by two basic and one acidic amino acid (Figure 1B). This therefore shortens the hydrophobic membrane anchor sequence by four amino acids relative to the sequence of VAMP-1A and, possibly more importantly, adds charge to the carboxy terminus of the protein. This creates a motif that is broadly shared by other proteins with carboxy-terminal membrane anchors that are targeted to mitochondria, a hydrophobic anchor flanked by positively charged residues.
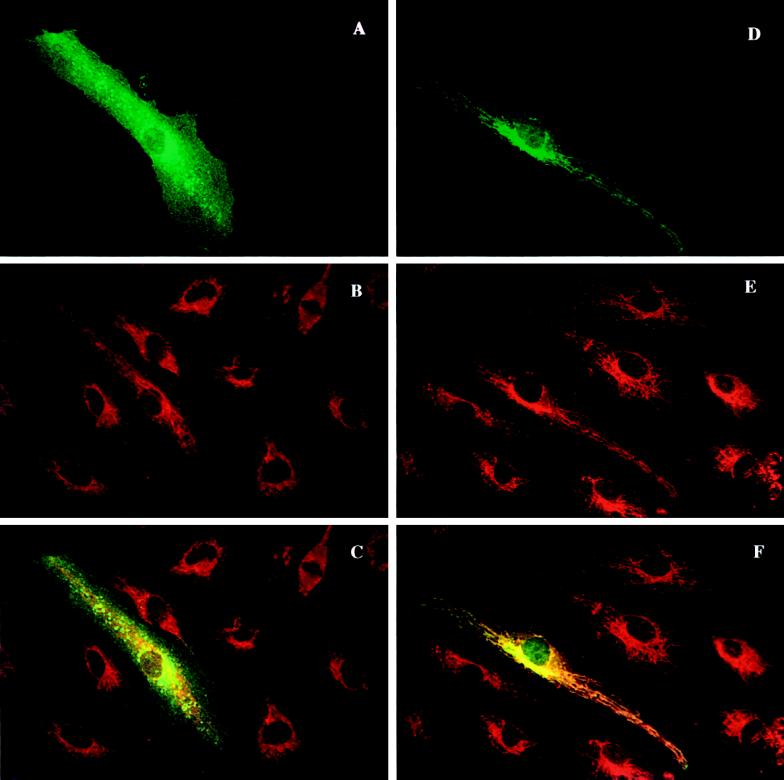
To test whether the sequence difference between VAMP-1A and VAMP-1B affects protein localization, constructs encoding these proteins were transfected into endothelial cells. The FLAG epitope was introduced into the N terminus of the VAMPs to create an antigenic marker. Amino-terminal modifications have been shown to be benign to function and localization of several SNARE proteins (Banfield et al., 1994; Hay et al., 1996; Nagahama et al., 1996). These constructs were then visualized by immunofluorescence microscopy (Figure 3). The cells were counterstained with an antibody directed against the mitochondrial outer membrane protein Tom20 (Hanson et al., 1996). The staining of VAMP-1A consisted of a diffuse surface labeling as well as punctate perinuclear staining (Figure 3A). The punctate staining resembles that of endosomes as assessed by fluorescent dextran uptake (our unpublished data). The endosomal distribution is not surprising given that endosomes are a “default” destination of VAMP-2 when VAMP-2 is not targeted to regulated secretory vesicles (Grote et al., 1995). What is somewhat surprising is that VAMP-1A was not detected in the secretory granules of endothelial cells, the Weibel-Palade body. The mitochondrial marker Tom20 did not show any appreciable colocalization with VAMP-1A (Figure 3, B and C). The distribution of VAMP-1B was strikingly different. VAMP-1B was localized to an array of fibrillar organelles that emanated from a perinuclear concentration. The staining of mitochondria with anti-Tom20 extensively overlapped that of VAMP-1B (Figure 3, E and F). This staining pattern was observed with cells expressing VAMP-1B at a variety of levels, including those with the lowest visible staining. In some transfected cells, particularly those exhibiting very high expression levels, there was some epitope-tagged VAMP-1B staining on the plasma membrane in addition to the fibrillar staining.
Figure 3.
VAMP-1B, but not VAMP-1A, colocalizes with mitochondria in transiently transfected cells. Plasmids were constructed containing sequences encoding the FLAG eptiope fused to either VAMP-1A or VAMP-1B at the N terminus of the coding sequences. The plasmids were transfected into human endothelial cells, and the FLAG epitope and mitochondria were visualized 48 h later. (A–C) VAMP-1A-FLAG transfectants. (D–F) VAMP-1B-FLAG transfectants. (A and D) M-2 Anti-FLAG epitope. (B and E) Anti-Tom20 (mitochondria). (C and F) Merged images.
Mitochondrial Targeting of VAMP-1B Depends Both on a Shortened Hydrophobic Membrane Anchor and the Addition of Positive Charge at the Carboxy Terminus
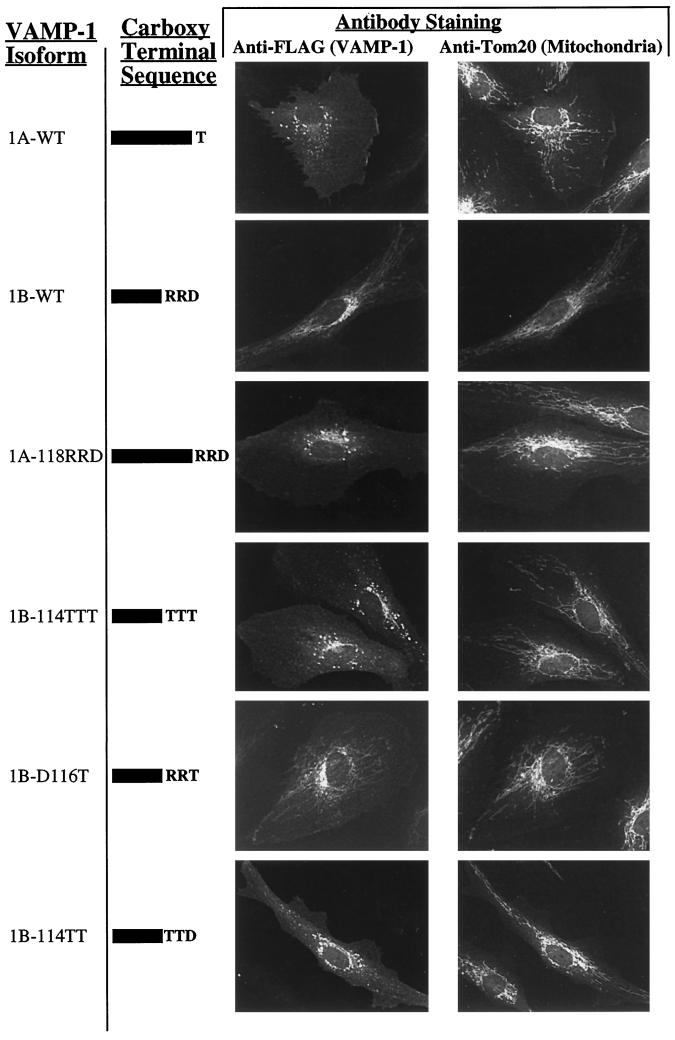
The sequence of VAMP-1B is altered, relative to VAMP-1A, by a shortened hydrophobic membrane anchor and the addition of charged residues to the carboxy terminus. To determine which aspects of this alteration lead to mitochondrial targeting, several mutations were introduced into the carboxy-terminal sequence of Flag-VAMP-1 and transfected into endothelial cells (Figure 4). To test whether it was the additional charge that governed mitochondrial targeting, the −RRD sequence at the terminus of VAMP-1B was used to replace the threonine in VAMP-1A, thus retaining the length of the VAMP-1A hydrophobic anchor but introducing the charged VAMP-1B sequence (Figure 4, mutant 1A-118RRD). This mutant form localized identically to the unmodified VAMP-1A. Conversely, to test whether a relatively short membrane anchor was sufficient for mitochondrial targeting, the charged residues in VAMP-1B were replaced by polar, but uncharged, residues (threonine), thus retaining the shorter hydrophobic anchor but eliminating the carboxy-terminal charge (Figure 4, mutant 1B-114TTT). In this case, the mutant construct was not distributed to mitochondria, but was localized to plasma membrane and endosome-like structures. These data indicate that the mitochondrial targeting of VAMP-1B requires both carboxy-terminal charged residues and a relatively short hydrophobic anchor. Positively charged residues are sufficient for mitochondrial targeting since replacement of the C-terminal apartate by threonine retains mitochondrial targeting (Figure 4, mutant 1B-D116T), whereas the the aspartate by itself, when the arginines are replaced by threonine, is not sufficient for mitochondrial targeting (Figure 4, mutant 1B-114TT). The localization of VAMP-1B mutations replacing either arginine-114 or arginine-115 with threonine, thus producing a net neutral charge at the C terminus, were also tested (our unpublished results). These proteins had a mixed distribution with some mitochondrial staining and some plasma membrane/endosome-like staining. The distribution of staining was variable from cell to cell in the same culture, such that in some cells localization to mitochondria was seen to predominate and in other cells localization to the plasma membrane/endosome was favored.
Figure 4.
Mitochondrial targeting of VAMP-1B is determined by a combination of a shortened hydrophobic anchor and positive charge at the carboxy terminus. Plasmids encoding VAMP-1 sequences with the FLAG epitope fused to the N terminus were transfected into human endothelial cells, and cells were stained for immunofluorescence with antibodies to either the FLAG epitope (VAMP protein) or Tom20 (mitochondria) after 24 h. In addition to wild type VAMPs 1A and 1B, the constructs included replacement of the carboxy-terminal threonine of VAMP-1A by Arg-Arg-Asp (1A-118RRD), replacement of the carboxy-terminal Arg-Arg-Asp of VAMP-1B by a triple threonine (1B-114TTT), replacement of the carboxy-terminal Asp of VAMP-1B by threonine (1B-116T), and replacement of Arg114 and Arg115 of VAMP-1B by threonines (1B-114TT).
DISCUSSION
We describe here the characterization of a novel splice-isoform of VAMP-1. This isoform, which we have termed VAMP-1B, results from the use of an alternative exon at the extreme 3′-end of the protein-coding region. A similar splice isoform was recently reported in rat (Mandic et al., 1997). In rat the exon encoding VAMP-1B is located in between exon 4 and exon 5, the latter of which encodes the carboxy-terminal five amino acids of VAMP-1A. We presume this organization is retained in the human sequence since the increased message size of VAMP-1B over VAMP-1A is consistent between rat and human. The mRNA for VAMP-1B is found in all cultured cell types that we have examined, although the levels of mRNA appear to be low. The expression of VAMP-1A and VAMP-1B may be, to a certain extent, mutually exclusive. In human brain cDNA, VAMP-1A was easily detectable, but no VAMP-1B was detected. In contrast, cultured cells all contain VAMP-1B but not VAMP-1A (except for a weak signal in vascular endothelial cells). From these data it appears that VAMP-1B is a widely distributed isoform, and VAMP-1A is restricted to specialized cells. A wide tissue distribution was also noted in rat (Mandic et al., 1997). It should be noted, however, that VAMP-1A message has been detected in a variety of tissues in rat, although the cell type distribution within those tissues has not been examined in detail (Rossetto et al., 1996). In addition to VAMP-1B, VAMP-2 mRNA was detected in all cell types examined, including cells that do not possess a known regulated secretory pathway, such as fibroblasts, HepG2 cells, and Jurkat cells. This may indicate that VAMP-2, best characterized for its involvement in regulated secretion, may well serve an alternate, more general role in all cells.
The carboxy-terminal amino acid sequence of VAMP-1 is changed by the alternative splicing in VAMP-1B. VAMPs 1–3 all terminate in either serine or threonine immediately adjacent to the hydrophobic transmembrane segment. In contrast, VAMP-1B contains three charged amino acids, and, in addition, the hydrophobic segment is shortened by four amino acids. The result of this change is to direct VAMP-1B to mitochondria in transfected cells. It should be noted that the low levels of endogenous VAMP-1B protein in cultured cells have made detection of the endogenous protein difficult. Therefore, we have not been able to confirm that endogenous VAMP-1B is localized to mitochondria, and this localization must be considered tentative. However, considering that there are many examples of transfected, epitope-tagged SNARE proteins correctly identifying the localization of the endogenous counterparts (Banfield et al., 1994; Grote et al., 1995; Hay et al., 1996; Nagahama et al., 1996; Yang et al., 1997), we are confident that the mitochondrial localization of the transfected protein reflects the localization of the endogenous protein.
The motif created by the alternative splicing, a hydrophobic carboxy-terminal sequence flanked by positive charges, is shared by several other carboxy-terminal–anchored mitochondrial proteins (Figure 2B). A similar basic-hydrophobic-basic motif is found in carboxy-terminal–anchored proteins that are directed to other sites. These include BOS1 (ER), the rat homologue of BET1 (Golgi), and syntaxin 2 (plasma membrane). One distinction between these proteins and those localized to mitochondria is the length of the hydrophobic sequence. With the exception of metaxin, the mitochondrially targeted proteins have hydrophobic segments that are 17 residues in length or shorter, whereas BOS1, rat BET1, and syntaxin 1 each contain hydrophobic segments that are 20 residues or longer. The differential localization of VAMPs 1A and 1B provide an ideal system for testing the sequence requirements for mitochondrial targeting of signal-anchored proteins, since VAMPs 1A and 1B differ by a small, discrete sequence change. We have found that a shortened hydrophobic sequence alone is not sufficient for mitochondrial targeting. Positively charged residues at the carboxy terminus (presumably intermembrane space side) of the sequence are also required for mitochondrial targeting. Comparison of the human and rat VAMP-1B protein sequences substantiates the importance of these positive charges (Figure 1B). These sequences diverge in exact sequence at the end of the hydrophobic sequence, yet both forms contain two positively charged residues. This finding mirrors in vitro studies of the mitochondrial targeting of an N-terminal signal-anchor sequence from Tom70, which is thought to be targeted to the mitochondrial membrane by the same mechanism that targets proteins with C-terminal signal anchors (McBride et al., 1992). In those studies it was shown that whereas the hydrophobic core was sufficient for targeting, the efficiency of targeting was enhanced by positively charged residues on the N-terminal side (intermembrane side) of the hydrophobic sequence. As these studies were conducted in an in vitro system, it remains to be tested whether an absolute requirement for the N-terminal positive charges might be observed in vivo, as we have found for VAMP-1B targeting. The mechanism inserting signal-anchored proteins to mitochondria overlaps to a degree with that of matrix-directed proteins, but differs in important aspects, particularly in the initial step of recognition (Millar and Shore, 1996). The mitochondrial membrane and/or cytosolic component(s) mediating this recognition have yet to be identified, and the discrete sequence requirements identified here could be useful in that identification.
VAMP-1 joins a growing list of proteins with multiple isoforms, one of which preferentially localizes to mitochondria. These include the endopeptidase 24.16 that localizes either to cytosol or to mitochondria, depending on the splice isoform expressed (Kato et al., 1997), and P4501A1, normally microsomal, in which a cryptic mitochondrial sorting signal is exposed by a proteolytic clip (Addya et al., 1997). Cytochrome b5 is a carboxy-terminal–anchored protein like VAMP-1 and is expressed in two isoforms encoded by different genes. Analogous to the VAMP-1 localization reported here, one of these isoforms is localized to the endoplasmic reticulum, and the other to the outer membrane of mitochondria. The carboxy-terminal region of the mitochondrial isoform is responsible for mitochondrial targeting, as we have demonstrated for VAMP-1B, although the precise elements required for that targeting have not been delineated (De Silvestris et al., 1995). Although functionally diverse, these protein pairs demonstrate how the cell has capitalized on the specificity of the mitochondrial targeting machinery to generate differential localization with a minimal perturbation of protein structure.
The localization of a VAMP protein to mitochondria is unexpected and provocative since, from a protein-trafficking perspective, mitochondria are generally regarded to be a modified prokaryotic symbiont that is independent of the vesicular trafficking pathways of the cell. What would be the function of VAMP-1 in mitochondria? A number of possibilities can be envisioned, such as a vesicular pathway originating in mitochondria to deliver mitochondrial products of lipid metabolism to other organelles. Another potential function would be to mediate a transient fusion between the endoplasmic reticulum and mitochondria in the transport of phosphatidylserine for mitochondrial phosphatidylethanolamine synthesis (Voelker, 1990). However, we feel a likely function of VAMP-1B is to mediate homotypic fusion events between mitochondria. Fusion of individual mitochondria to form mitochondrial reticula or large mitochondrial structures is a widespread phenomenon (Bereiter-Hahn, 1990). Cytoplast fusion studies have shown that a mitochondrial network involves virtually every mitochondrion in Hela cells (Hayashi, 1994; however, see also Yoneda et al., 1994). However, the molecular underpinning of this fusion is almost completely unknown. The regulation of mitochondrial fusion, and the extent to which it occurs, varies widely between tissues and organisms (reviewed by Warren and Wickner, 1996; and Hales and Fuller, 1997). Fusion of mitochondria apparently occurs developmentally in diaphragm muscle in the rat (Bakeeva et al., 1981), after mitosis in Euglena (Calvayrac et al. 1974), before mitosis in scorpion (reviewed in Warren and Wicker, 1996), and after mating in Saccharomyces cerevisiae (Nunnari et al., 1997). The fusion of mitochondria in mammalian cells has been inferred from the extended filamentous morphology observed in a number of cell types (Johnson et al. 1980) and tissues (Brandt et al., 1974; Kirkwood et al., 1986). A particularly well characterized mitochondrial fusion occurs in the developing Drosophila spermatid. In these cells mitochondria fuse postmeiotically to form large structures termed Nebenkern. The only known molecular participant in mitochondrial fusion has been identified by a genetic analysis of mutants in this fusion event (Hales and Fuller, 1997). This protein, termed Fzo, is a predicted membrane-bound guanosine triphosphatase (GTPase) without significant homology (other than in the GTPase domain) to any characterized protein. Fzo associates with mitochondria in the developing spermatid only when they are in the process of fusion. The Fzo GTPase activity is apparently required for its fusion-related activity, since mutation of key conserved GTP-binding residues inactivates the protein. Whether Fzo has a proofreading function, analogous to the rab GTP-binding proteins (reviewed by Aridor and Balch, 1996), or a more direct mechanical role in fusion, analogous to function of dynamin in membrane fission (reviewed by Urrutia et al., 1997), remains to be seen. In yeast a number of genes have been identified that affect mitochondrial inheritance and morphology and could conceivably be involved in mitochondrial fusion (Berger et al., 1997; Fisk and Yaffe, 1997). However, a direct role for fusion of these proteins has not been demonstrated.
Given the wide range of vesicle fusion events that utilize the N-ethylmaleimide-sensitive factor (NSF)/SNARE mechanism (reviewed by Hay and Scheller, 1997), it would not be surprising if mitochondria had appropriated this apparatus to accomplish membrane fusion. One prediction of this concept is that a target SNARE capable of binding to VAMP-1, such as syntaxins 1 or 4 and/or SNAP-25 or SNAP-23, should also be found on mitochondrial membranes. No such target SNARE has been reported in mitochondria, but previously there was no impetus to look for one. Not all evidence supports a role for SNAREs in mitochondrial fusion, however. First, analysis of the complete yeast genome has identified only eight syntaxin homologues (Weimbs et al., 1997; Holthius et al., 1998), none of which are localized to mitochondria. Second, a prediction for SNARE-mediated fusion of mitochondria is that the NSF would also be required in the process to rearrange the SNARE complex before fusion. However, when temperature-sensitive mutants of NSF and NSF homologues in yeast were examined for fusion of mitochondria after mating, no effect of the mutations was detected (Nunnari et al., 1997). This may be due to differences between mitochondrial fusion in yeast and mammalian cells, but another interesting possibility is that the mitochondrial VAMP serves a role in docking that is not connected to a fusion event. The levels of VAMP-1B protein are low and could potentially be below a threshold level required for fusion. In this way VAMP-1B could be involved in tethering mitochondria to target-SNARE–containing sites, e.g., the plasma membrane, but not in fusion at those sites. Ultimately, functional tests of VAMP-1B in mitochondrial will define its role there. We are currently exploring the effects of ablating VAMP-1B on mitochondrial morphology and function.
ACKNOWLEDGMENTS
The advice of Paul Moretti on the PCR experiments is gratefully acknowledged. We thank the staff in the delivery wards of Women’s and Children’s Hospital (North Adelaide, South Australia) and Burnside War Memorial Hospital (Adelaide, South Austalia) for collection of umbilical cords. We gratefully acknowledge Mr. James Roberts for the preparation of the HUVEC cDNA library. We also thank participants of the 1998 Hanson Satellite meeting for helpful suggestions. This work was supported by grants from the National Health and Medical Research Council and by a Royal Adelaide Hospital Florey Fellowship (to B.W.). This work is dedicated to the memory of Dr. David F. Silbert, a membrane biochemist of the first rank.
Footnotes
The abbreviations used are: HUVEC, human umbilical vein endothelial cells; NSF, N-ethylmaleimide sensitive factor; RT-PCR, reverse transcription-PCR; SNARE, SNAP receptors; VAMP, vesicle-associated membrane protein.
REFERENCES
- Aalto MK, Ronne H, Kerαanen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addya S, Anandatheerthavarada HK, Biswas G, Bhagwat SV, Mullick J, Vadhani NG. Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J Cell Biol. 1997;139:589–599. doi: 10.1083/jcb.139.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer BR, III, Özçelik T, Jahn R, Francke U, Südhof TC. Structures and chromosomal localizations of two human genes encoding synaptobrevins 1 and 2. J Biol Chem. 1990;265:17267–17273. [PubMed] [Google Scholar]
- Aridor M, Balch WE. Timing is everything. Nature. 1996;383:220–221. doi: 10.1038/383220a0. [DOI] [PubMed] [Google Scholar]
- Bakeeva E, Chentsov YS, Skulachev VP. Ontogenesis of mitochondrial reticulum in rat diaphragm muscle. Eur J Cell Biol. 1981;25:175–181. [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Rabouille C, Warrren G, Pelham HRB. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt JT, Martin AP, Lucas FV, Vorbeck ML. The structure of rat liver mitochondria: a reevaluation. Biochem Biophys Res Commun. 1974;59:1097–1103. doi: 10.1016/s0006-291x(74)80091-4. [DOI] [PubMed] [Google Scholar]
- Broadie K, Prokop A, Bellen HJ, O’Kane CJ, Schulze KL, Sweeney ST. Syntaxin and synaptobrevin function downstream of vesicle docking in Drosophila. Neuron. 1995;15:663–673. doi: 10.1016/0896-6273(95)90154-x. [DOI] [PubMed] [Google Scholar]
- Calakos N, Bennett MK, Peterson KE, Scheller RH. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- Calvayrac R, Bertaux O, Lefort-Tran M, Valencia R. Generalization of the mitochondrial cycle in synchronous Euglena gracilis Z. during heterotrophic and phototrophic growth. Protoplasma. 1974;80:355–370. doi: 10.1007/BF01276351. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- De Silvestris M, D’Arrigo A, Borgese N. The targeting information of the mitochondrial outer membrane isoform of cytochrome b5 is contained within the carboxyl-terminal region. FEBS Lett. 1995;370:69–74. doi: 10.1016/0014-5793(95)00797-d. [DOI] [PubMed] [Google Scholar]
- Elferink LA, Trimble WS, Scheller RH. Two vesicle-associated membrane protein genes are differentially expressed in the rat central nervous system. J Biol Chem. 1989;264:11061–11064. [PubMed] [Google Scholar]
- Fisk HA, Yaffe MP. Mutational analysis of Mdm1; function in nuclear and mitochondrial inheritance. J Cell Biol. 1997;138:485–494. doi: 10.1083/jcb.138.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Hao JC, Bennett MK, Kelly RB. A targeting signal in VAMP regulating transport to synaptic vesicles. Cell. 1995;81:518–589. doi: 10.1016/0092-8674(95)90079-9. [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hanson B, Nuttal S, Hoogenraad N. A receptor for the import of proteins into human mitochondria. Eur J Biochem. 1996;235:750–753. doi: 10.1111/j.1432-1033.1996.t01-1-00750.x. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Hirling H, Scheller RH. Mammalian vesicle trafficking proteins of the endoplasmic reticulum and Golgi apparatus. J Biol Chem. 1996;271:5671–5679. doi: 10.1074/jbc.271.10.5671. [DOI] [PubMed] [Google Scholar]
- Hay JC, Scheller RH. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Hayashi J-I, Takemitsu M, Goto Y-I, Nonaka I. Human mitochondria and mitochondrial genome function as a single dynamic cellular unit. J Cell Biol. 1994;125:43–50. doi: 10.1083/jcb.125.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthius JCM, Nichols BJ, Dhruvakumar S, Pelham HRB. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt JM, Bommert K, Charlton MP, Kister A, Habermann E, Augustine GJ, Betz H. A post-docking role for synaptobrevin in synaptic vesicle fusion. Neuron. 1994;12:1269–1279. doi: 10.1016/0896-6273(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Belyavsky AV. Identification of differentially expressed genes by restriction endonuclease-based gene expression fingerprinting. Nucleic Acids Res. 1995;23:2954–2958. doi: 10.1093/nar/23.15.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Sugiura N, Hosoiri T, Hasue H, Hirose S. Targeting of endopeptidase 24.16 to different subcellular compartments by alternative promoter usage. J Biol Chem. 1997;272:15313–15322. doi: 10.1074/jbc.272.24.15313. [DOI] [PubMed] [Google Scholar]
- Kirkwood SP, Munn EA, Brooks GA. Mitochondrial reticulum in limb skeletal muscle. Am J Physiol. 1986;251:C395–C402. doi: 10.1152/ajpcell.1986.251.3.C395. [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–213. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–74. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Mandic R, Trimble WS, Lowe AW. Tissue-specific alternative RNA splicing of rat vesicle-associated membrane protein-1 (VAMP-1) Gene. 1997;199:173–179. doi: 10.1016/s0378-1119(97)00244-8. [DOI] [PubMed] [Google Scholar]
- McBride HM, Millar DG, Li J-M, Shore GC. A signal-anchor sequence selective for the mitochondrial outer membrane. J Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar DG, Shore GC. Signal anchor sequence insertion into the outer mitochondrial membrane. Comparison with porin and the matrix protein targeting pathway. J Biol Chem. 1996;271:25823–25829. doi: 10.1074/jbc.271.42.25823. [DOI] [PubMed] [Google Scholar]
- Nagahama M, Orci L, Ravazzola M, Amherdt M, Lacomis L, Tempst P, Rothman JE, Söllner TH. A v-SNARE implicated in intra-Golgi transport. J Cell Biol. 1996;133:507–516. doi: 10.1083/jcb.133.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOX1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H, Blasi J, Reinhard J. Clostridial neurotoxins: new tools for dissecting exocytosis. Trends Cell Biol. 1994;4:179–184. doi: 10.1016/0962-8924(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Nishina Y, Nakagoshi H, Imamoto F, Gonda TJ, Ishii S. Trans-activation by the c-myb proto-oncogene. Nucleic Acids Res. 1989;17:107–117. doi: 10.1093/nar/17.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Transport vesicle docking: SNAREs and associates. Annu Rev Cell Dev Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Pelham HRB. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regazzi R, Sadoul K, Meda P, Kelly RB, Halban PA, Wollheim CB. Mutational analysis of VAMP domains implicated in Ca2+-induced insulin exocytosis. EMBO J. 1996;15:6951–6959. [PMC free article] [PubMed] [Google Scholar]
- Rossetto O, Gorza L, Schiavo G, Schiavo N, Scheller RH, Montecucco C. Vamp/synaptobrevin isoforms 1 and 2 are widely and differentially expressed in nonneuronal tissues. J Cell Biol. 1996;132:167–179. doi: 10.1083/jcb.132.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–62. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases. Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker DR. Characterization of phosphatidylserine synthesis and translocation in permeabilized animal cells. J Biol Chem. 1990;265:14340–14346. [PubMed] [Google Scholar]
- Wall RT, Harker LA, Quadracci LJ, Striker GE. Factors influencing endothelial cell proliferation in vitro. J Cell Physiol. 1978;96:203–213. doi: 10.1002/jcp.1040960209. [DOI] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Ellenberg J, Bonifacino JS, Weissman AM. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]
- Yoneda M, Miyatake T, Attardi G. Complementation of mutant and wild-type human mitochondrial DNAs coexisting since the mutation event and lack of complementation of DNAs introduced separately into a cell within distinct organelles. Mol Cell Biol. 1994;14:2699–2712. doi: 10.1128/mcb.14.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]