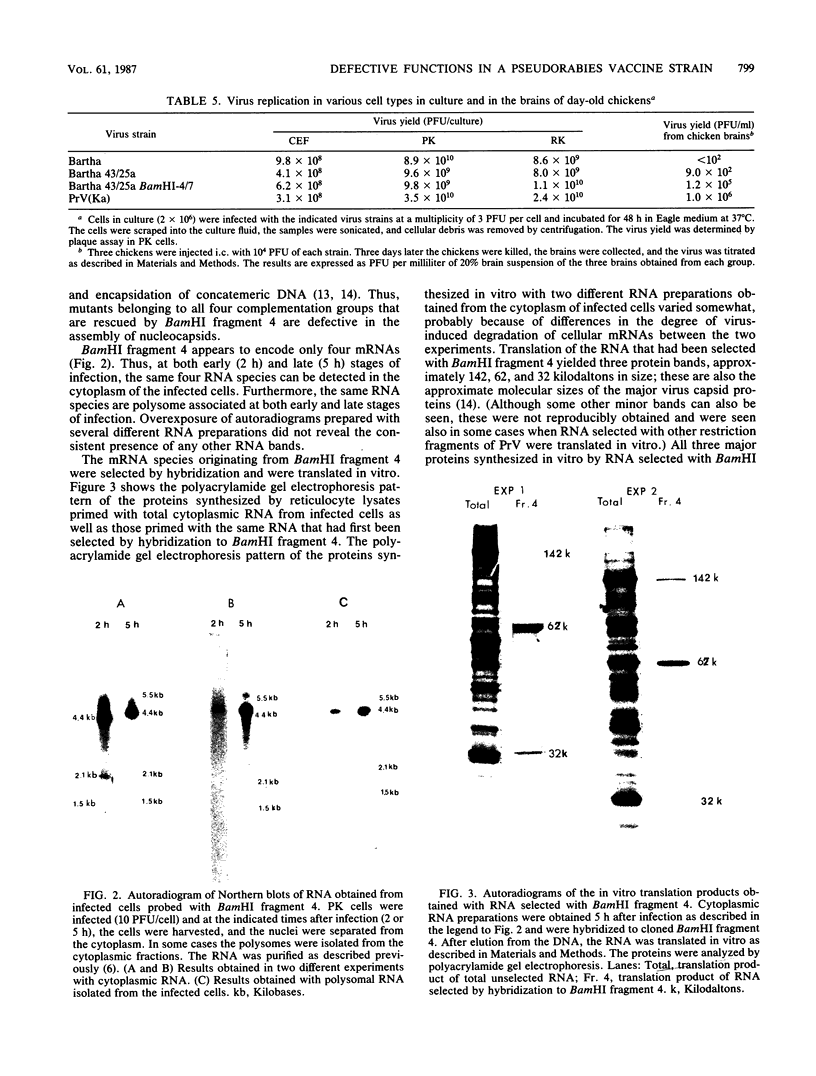
Abstract
We have shown previously (Lomniczi et al., J. Virol. 52:198-205, 1984) that the Bartha vaccine strain of pseudorabies virus has a deletion in the short unique (Us) region of its genome--a deletion that is related to the absence of virus virulence. This strain is, however, also defective in other genes involved in virulence. We show here that virulence can be restored by marker rescue of the Bartha strain to which an intact Us has been restored (but not to the parental Bartha strain) by sequences derived from approximate map units 0.460 and 0.505 of the wild-type virus genome. No difference in the ability to grow in cell culture was observed between parental Bartha, Bartha 43/25a (Bartha to which an intact Us has been restored), or the doubly rescued Bartha strains. However, only the doubly rescued Bartha strain was virulent for both chickens and pigs and replicated to high titers when inoculated directly into the brains of chickens. The sequences that could restore virulence to the Bartha 43/25a strain encode four genes, all of which are involved in processes leading to the assembly of nucleocapsids. Since these sequences rescue virulence, it appears that a function that plays a role in nucleocapsid assembly is defective in the Bartha strain and that this defect contributes to the lack of virulence of this virus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., DeMarchi J., Pendrys J., Veach R. A., Kaplan A. S. Proteins specified by the short unique region of the genome of pseudorabies virus play a role in the release of virions from certain cells. J Virol. 1986 Jan;57(1):191–196. doi: 10.1128/jvi.57.1.191-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Hoffmann P., Brown L., Feldman L., Blankenship M. L. Partial characterization of temperature-sensitive mutants of pseudorabies virus. Virology. 1982 Oct 30;122(2):251–267. doi: 10.1016/0042-6822(82)90225-2. [DOI] [PubMed] [Google Scholar]
- Berns A., van den Ouweland A., Quint W., van Oirschot J., Gielkens A. Presence of markers for virulence in the unique short region or repeat region or both of pseudorabies hybrid viruses. J Virol. 1985 Jan;53(1):89–93. doi: 10.1128/jvi.53.1.89-93.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibor-Hardy V., Pouchelet M., St-Pierre E., Herzberg M., Simard R. The nuclear matrix is involved in herpes simplex virogenesis. Virology. 1982 Sep;121(2):296–306. doi: 10.1016/0042-6822(82)90169-6. [DOI] [PubMed] [Google Scholar]
- Deatly A. M., Ben-Porat T. Relation between the levels of mRNA abundance and kinetics of protein synthesis in pseudorabies virus-infected cells. Virology. 1985 Jun;143(2):558–568. doi: 10.1016/0042-6822(85)90394-0. [DOI] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Hampl H., Ben-Porat T., Ehrlicher L., Habermehl K. O., Kaplan A. S. Characterization of the envelope proteins of pseudorabies virus. J Virol. 1984 Nov;52(2):583–590. doi: 10.1128/jvi.52.2.583-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S., Ladin B. F., Ben-Porat T. Comparison of the physical and genetic maps of pseudorabies virus shows that the genetic map is circular. Virology. 1982 Oct 30;122(2):268–278. doi: 10.1016/0042-6822(82)90226-4. [DOI] [PubMed] [Google Scholar]
- Isle H. B., Venkatesan S., Moss B. Cell-free translation of early and late mRNAs selected by hybridization to cloned DNA fragments derived from the left 14 million to 72 million daltons of the vaccinia virus genome. Virology. 1981 Jul 15;112(1):306–317. doi: 10.1016/0042-6822(81)90636-x. [DOI] [PubMed] [Google Scholar]
- KAPLAN A. S., VATTER A. E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959 Apr;7(4):394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Ladin B. F., Blankenship M. L., Ben-Porat T. Replication of herpesvirus DNA. V. Maturation of concatemeric DNA of pseudorabies virus to genome length is related to capsid formation. J Virol. 1980 Mar;33(3):1151–1164. doi: 10.1128/jvi.33.3.1151-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladin B. F., Ihara S., Hampl H., Ben-Porat T. Pathway of assembly of herpesvirus capsids: an analysis using DNA+ temperature-sensitive mutants of pseudorabies virus. Virology. 1982 Jan 30;116(2):544–561. doi: 10.1016/0042-6822(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Lomniczi B., Blankenship M. L., Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J Virol. 1984 Mar;49(3):970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomniczi B., Watanabe S., Ben-Porat T., Kaplan A. S. Genetic basis of the neurovirulence of pseudorabies virus. J Virol. 1984 Oct;52(1):198–205. doi: 10.1128/jvi.52.1.198-205.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]