Abstract
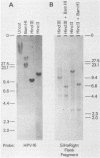
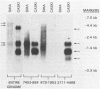
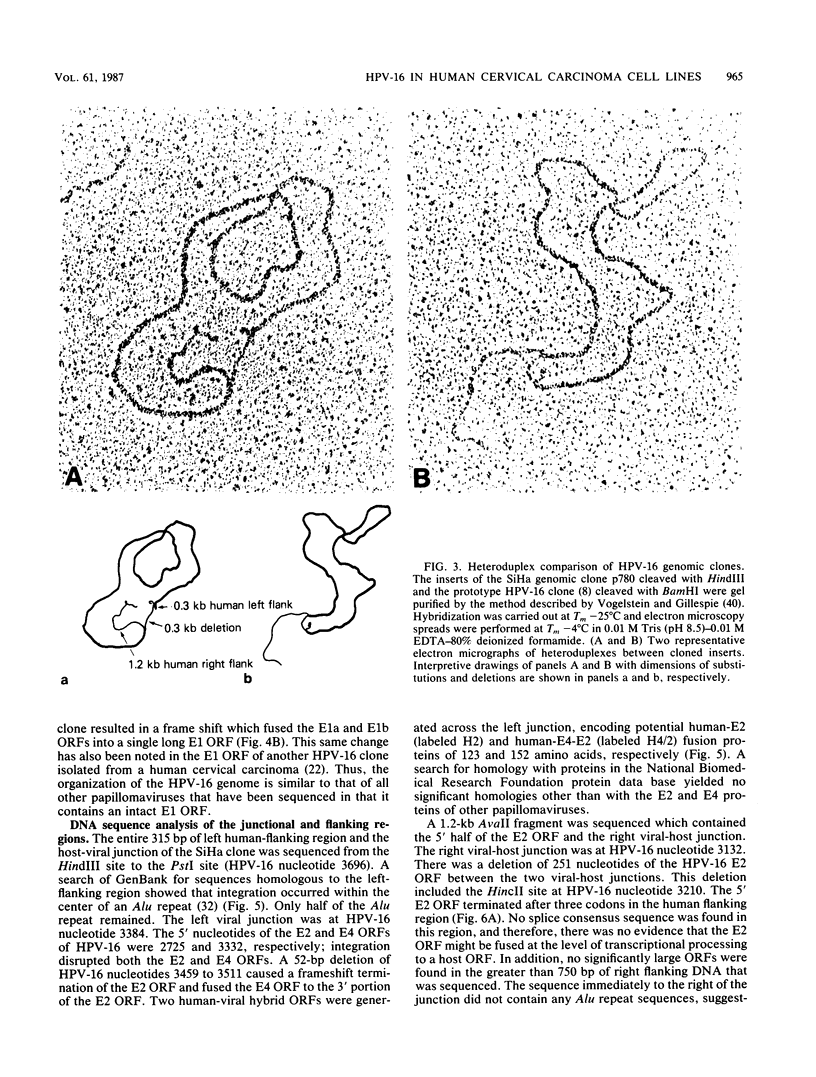
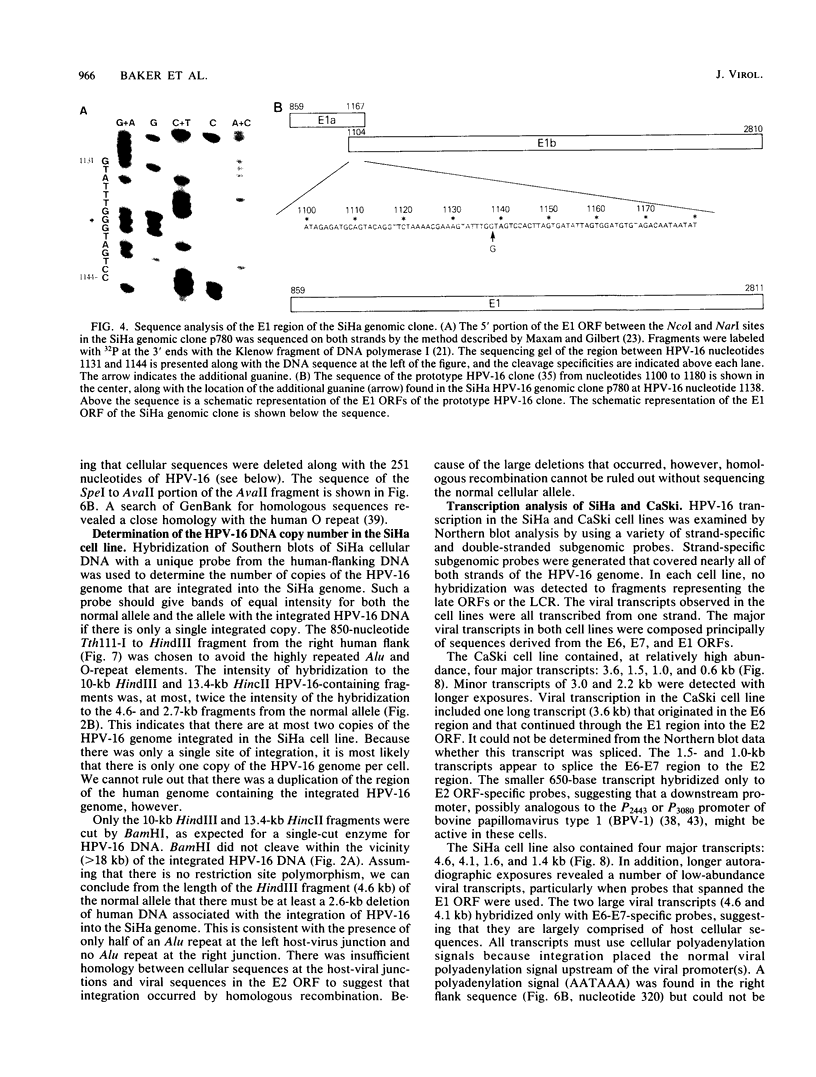
We cloned and analyzed the integrated human papillomavirus type 16 (HPV-16) genomes that are present in the human cervical carcinoma cell lines SiHa and CaSki. The single HPV-16 genome in the SiHa line was cloned as a 10-kilobase (kb) HindIII fragment. Integration of the HPV-16 genome occurred at bases 3132 and 3384 with disruption of the E2 and E4 open reading frames (ORFs). An additional 52-base-pair deletion of HPV-16 sequences fused the E2 and E4 ORFs. the 5' portion of the disrupted E2 ORF terminated immediately in the contiguous human right-flanking sequences. Heteroduplex analysis of this cloned integrated viral genome with the prototype HPV-16 DNA revealed no other deletions, insertions, or rearrangements. DNA sequence analysis of the E1 ORF, however, revealed the presence of an additional guanine at nucleotide 1138, resulting in the fusion of the E1a and E1b ORFs into a single E1 ORF. Sequence analysis of the human flanking sequences revealed one-half of an Alu sequence at the left junction and a sequence highly homologous to the human O repeat in the right-flanking region. Analysis of the three most abundant BamHI clones from the CaSki line showed that these consisted of full-length, 7.9-kb HPV-16 DNA; a 6.5-kb genome resulting from a 1.4-kb deletion of the long control region; and a 10.5-kb clone generated by a 2.6-kb tandem repeat of the 3' early region. These HPV-16 genomes were arranged in the host chromosomes as head-to-tail, tandemly repeated arrays. Transcription analysis revealed expression of the HPV-16 genome in each of these two cervical carcinoma cell lines, albeit at significantly different levels. Preliminary mapping of the viral RNA with subgenomic strand-specific probes indicated that viral transcription appeared to be derived primarily from the E6 and E7 ORFs.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M., Gissmann L., Ikenberg H., Kleinheinz A., Scheurlen W., zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984 May;3(5):1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum C. P., Ikenberg H., Richart R. M., Gissman L. Human papillomavirus type 16 and early cervical neoplasia. N Engl J Med. 1984 Apr 5;310(14):880–883. doi: 10.1056/NEJM198404053101403. [DOI] [PubMed] [Google Scholar]
- Crum C. P., Mitao M., Levine R. U., Silverstein S. Cervical papillomaviruses segregate within morphologically distinct precancerous lesions. J Virol. 1985 Jun;54(3):675–681. doi: 10.1128/jvi.54.3.675-681.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürst M., Gissmann L., Ikenberg H., zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gissmann L., Schwarz E. Persistence and expression of human papillomavirus DNA in genital cancer. Ciba Found Symp. 1986;120:190–207. doi: 10.1002/9780470513309.ch13. [DOI] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Jarrett W. F., McNeil P. E., Grimshaw W. T., Selman I. E., McIntyre W. I. High incidence area of cattle cancer with a possible interaction between an environmental carcinogen and a papilloma virus. Nature. 1978 Jul 20;274(5668):215–217. doi: 10.1038/274215a0. [DOI] [PubMed] [Google Scholar]
- Jarrett W. F., Murphy J., O'Neil B. W., Laird H. M. Virus-induced papillomas of the alimentary tract of cattle. Int J Cancer. 1978 Sep 15;22(3):323–328. doi: 10.1002/ijc.2910220316. [DOI] [PubMed] [Google Scholar]
- Law M. F., Lowy D. R., Dvoretzky I., Howley P. M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci U S A. 1981 May;78(5):2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lusky M., Botchan M. R. Genetic analysis of bovine papillomavirus type 1 trans-acting replication factors. J Virol. 1985 Mar;53(3):955–965. doi: 10.1128/jvi.53.3.955-965.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura T., Kanda T., Furuno A., Yoshikawa H., Kawana T., Yoshiike K. Cloning of monomeric human papillomavirus type 16 DNA integrated within cell DNA from a cervical carcinoma. J Virol. 1986 Jun;58(3):979–982. doi: 10.1128/jvi.58.3.979-982.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCance D. J., Campion M. J., Clarkson P. K., Chesters P. M., Jenkins D., Singer A. Prevalence of human papillomavirus type 16 DNA sequences in cervical intraepithelial neoplasia and invasive carcinoma of the cervix. Br J Obstet Gynaecol. 1985 Nov;92(11):1101–1105. doi: 10.1111/j.1471-0528.1985.tb03019.x. [DOI] [PubMed] [Google Scholar]
- McCance D. J., Clarkson P. K., Dyson J. L., Walker P. G., Singer A. Human papillomavirus types 6 and 16 in multifocal intraepithelial neoplasias of the female lower genital tract. Br J Obstet Gynaecol. 1985 Nov;92(11):1093–1100. doi: 10.1111/j.1471-0528.1985.tb03018.x. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Pater M. M., Pater A. Human papillomavirus types 16 and 18 sequences in carcinoma cell lines of the cervix. Virology. 1985 Sep;145(2):313–318. doi: 10.1016/0042-6822(85)90164-3. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rous P., Kidd J. G. THE CARCINOGENIC EFFECT OF A VIRUS UPON TARRED SKIN. Science. 1936 May 15;83(2159):468–469. doi: 10.1126/science.83.2159.468. [DOI] [PubMed] [Google Scholar]
- Sarver N., Rabson M. S., Yang Y. C., Byrne J. C., Howley P. M. Localization and analysis of bovine papillomavirus type 1 transforming functions. J Virol. 1984 Nov;52(2):377–388. doi: 10.1128/jvi.52.2.377-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J. T., Vass W. C., Lowy D. R. Identification of a second transforming region in bovine papillomavirus DNA. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7880–7884. doi: 10.1073/pnas.81.24.7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. W., Jelinek W. R. The Alu family of dispersed repetitive sequences. Science. 1982 Jun 4;216(4550):1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- Schneider-Gädicke A., Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 1986 Sep;5(9):2285–2292. doi: 10.1002/j.1460-2075.1986.tb04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E., Freese U. K., Gissmann L., Mayer W., Roggenbuck B., Stremlau A., zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985 Mar 7;314(6006):111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- Seedorf K., Krämmer G., Dürst M., Suhai S., Röwekamp W. G. Human papillomavirus type 16 DNA sequence. Virology. 1985 Aug;145(1):181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- Smotkin D., Wettstein F. O. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalholz B. A., Yang Y. C., Howley P. M. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell. 1985 Aug;42(1):183–191. doi: 10.1016/s0092-8674(85)80114-8. [DOI] [PubMed] [Google Scholar]
- Stenlund A., Zabielski J., Ahola H., Moreno-Lopez J., Pettersson U. Messenger RNAs from the transforming region of bovine papilloma virus type I. J Mol Biol. 1985 Apr 20;182(4):541–554. doi: 10.1016/0022-2836(85)90240-2. [DOI] [PubMed] [Google Scholar]
- Sun L., Paulson K. E., Schmid C. W., Kadyk L., Leinwand L. Non-Alu family interspersed repeats in human DNA and their transcriptional activity. Nucleic Acids Res. 1984 Mar 26;12(6):2669–2690. doi: 10.1093/nar/12.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Ikenberg H., Boehm N., Gissmann L. Identification of human papillomavirus in cervical swabs by deoxyribonucleic acid in situ hybridization. Obstet Gynecol. 1984 Dec;64(6):767–772. [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. C., Okayama H., Howley P. M. Bovine papillomavirus contains multiple transforming genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1030–1034. doi: 10.1073/pnas.82.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee C., Krishnan-Hewlett I., Baker C. C., Schlegel R., Howley P. M. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985 Jun;119(3):361–366. [PMC free article] [PubMed] [Google Scholar]