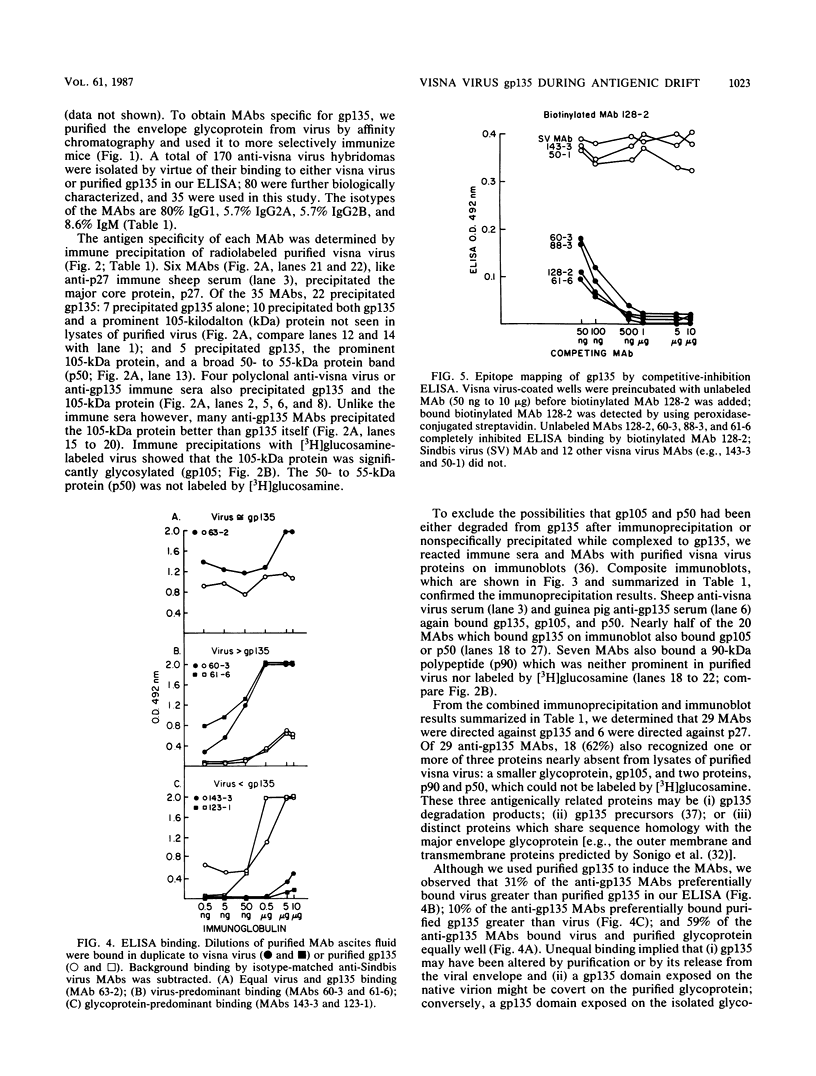
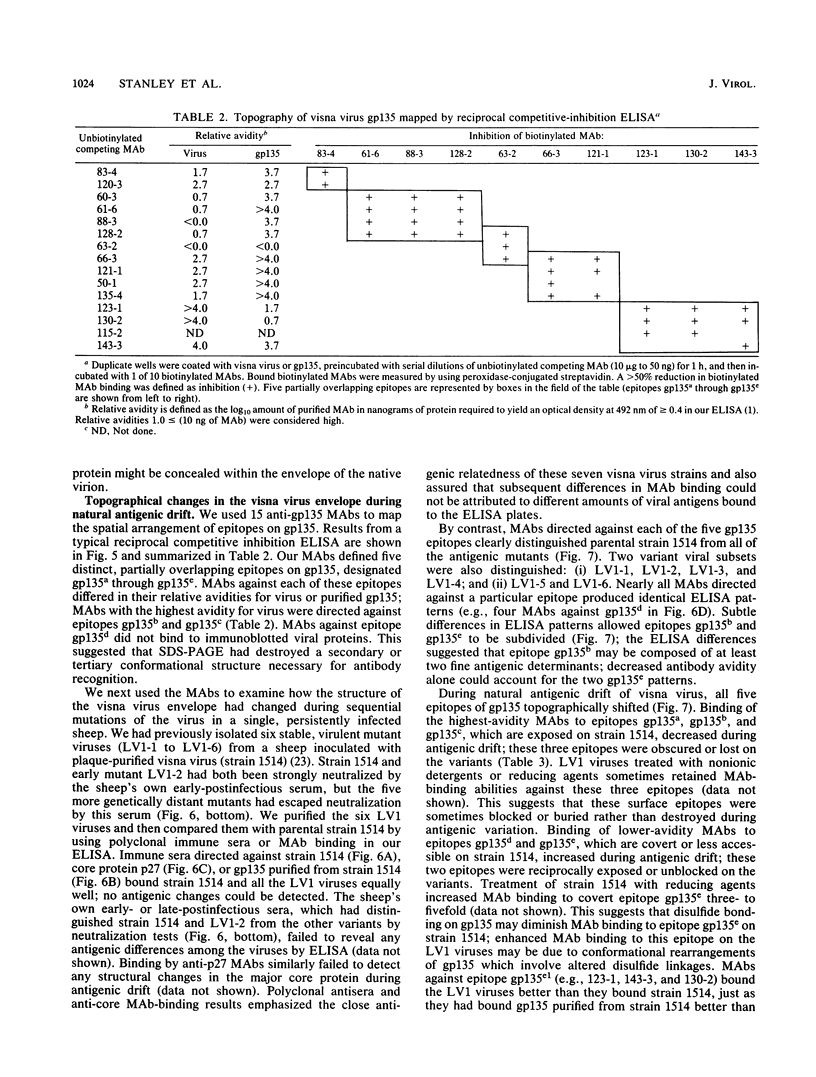
Abstract
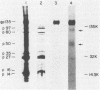
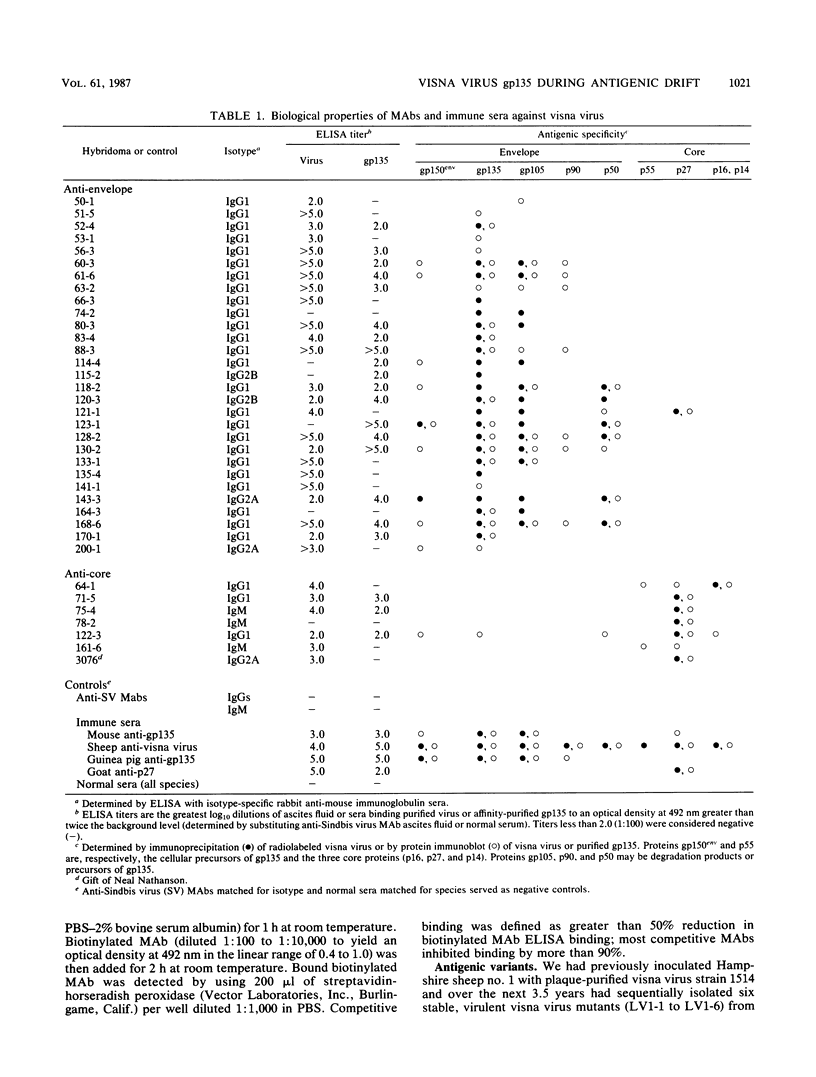
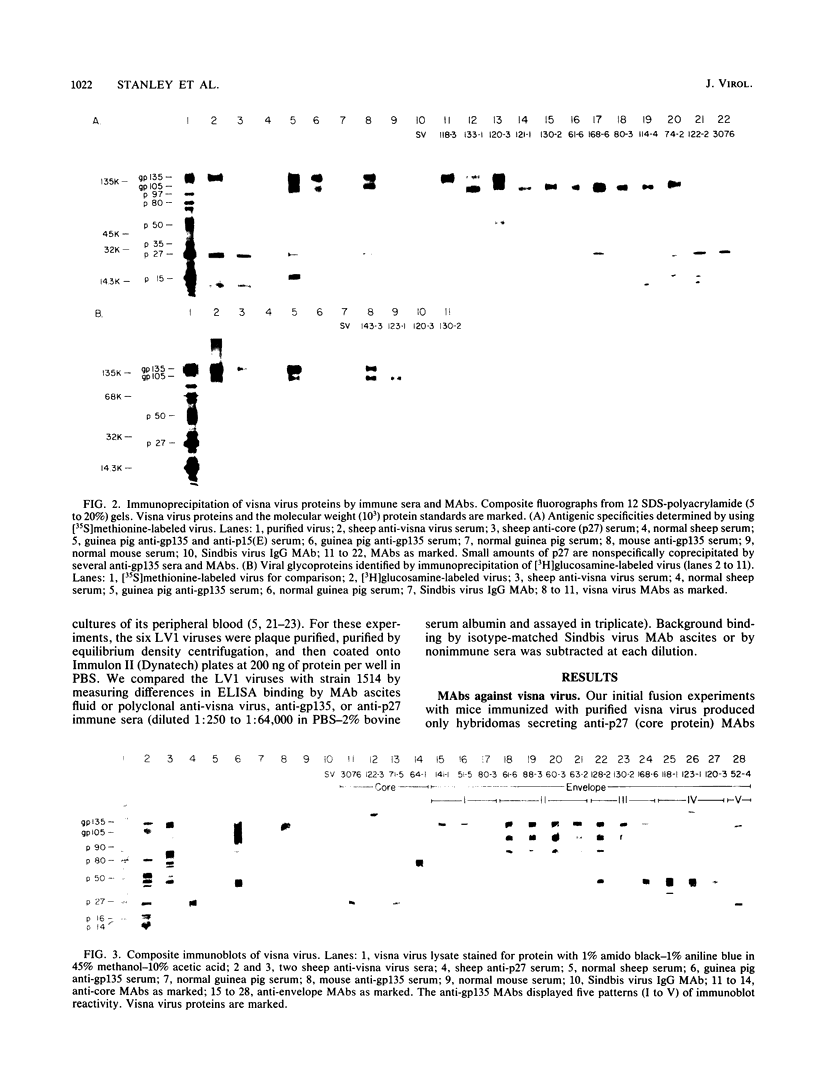
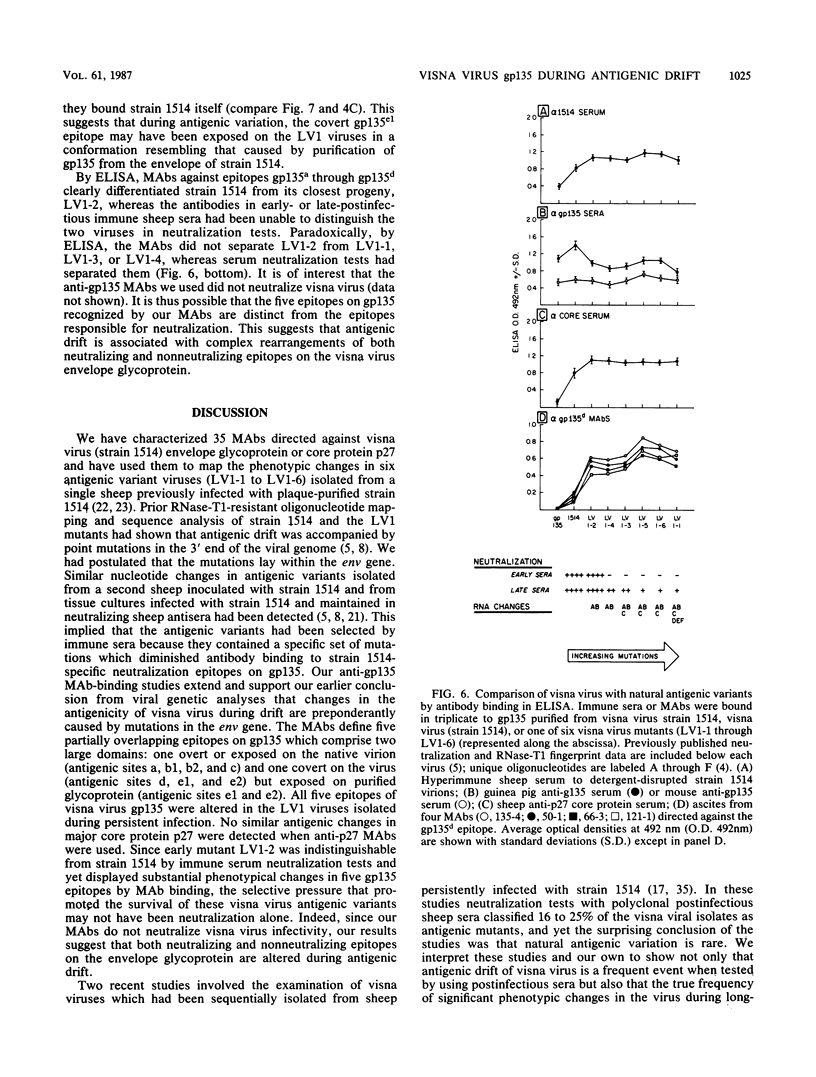
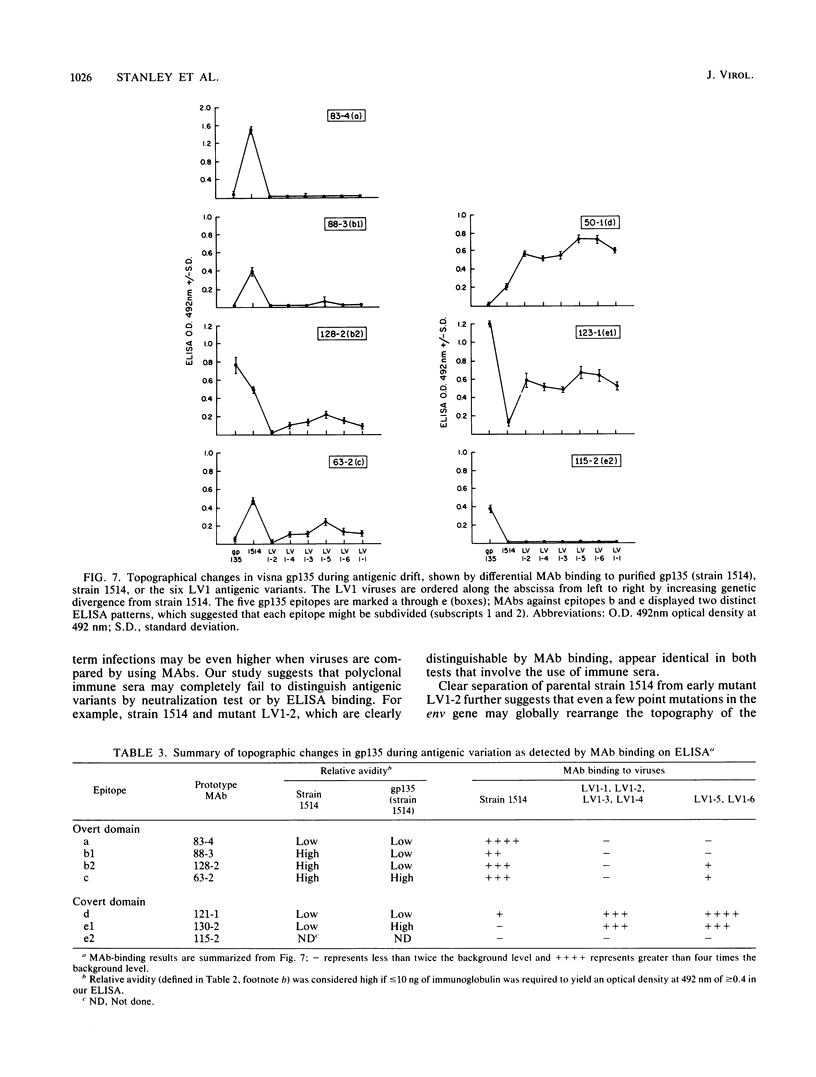
Visna virus undergoes antigenic drift during persistent infection in sheep and thus eludes neutralizing antibodies directed against its major envelope glycoprotein, gp135. Antigenic variants contain point mutations in the 3' end of the genome, presumably within the envelope glycoprotein gene. To localize the changes in the viral proteins of antigenic mutants, we isolated 35 monoclonal antibodies (MAbs) against the envelope glycoprotein gp135 or the major core protein p27 of visna virus. The MAbs defined five partially overlapping epitopes on gp135. We used the MAbs and polyclonal immune sera directed against visna virus, gp135, or p27 in enzyme-linked immunosorbent assays to compare visna virus (strain 1514) with antigenic mutants (LV1-1 to LV1-6) previously isolated from a single sheep persistently infected with plaque-purified strain 1514. Polyclonal immune sera and anti-core p27 MAbs failed to distinguish antigenic differences among the viruses. By contrast, the anti-gp135 MAbs detected changes in all five epitopes of the envelope glycoprotein. Three gp135 epitopes, prominently exposed on strain 1514, were lost or obscured on the mutants; two covert gp135 epitopes, poorly exposed on strain 1514, were reciprocally revealed on the mutants. Even virus LV1-2, which is indistinguishable from parental strain 1514 by serum neutralization tests and which differs from it by only two unique oligonucleotides on RNase-T1 fingerprinting, displayed global changes in gp135. Our data suggest that visna virus variants may emerge more frequently during persistent infection than can be detected by serological tests involving the use of polyclonal immune sera, and the extent of phenotypic changes in their envelope glycoproteins may be greater than predicted by the small number of genetic changes previously observed. We suggest that topographical rearrangements in the three-dimensional structure of gp135 may magnify the primary amino acid sequence changes caused by point mutations in the env gene. This may complicate strategies to construct lentiviral vaccines by using the envelope glycoprotein.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boere W. A., Harmsen T., Vinjé J., Benaissa-Trouw B. J., Kraaijeveld C. A., Snippe H. Identification of distinct antigenic determinants on Semliki Forest virus by using monoclonal antibodies with different antiviral activities. J Virol. 1984 Nov;52(2):575–582. doi: 10.1128/jvi.52.2.575-582.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brahic M., Stowring L., Ventura P., Haase A. T. Gene expression in visna virus infection in sheep. Nature. 1981 Jul 16;292(5820):240–242. doi: 10.1038/292240a0. [DOI] [PubMed] [Google Scholar]
- Clements J. E., D'Antonio N., Narayan O. Genomic changes associated with antigenic variation of visna virus. II. Common nucleotide sequence changes detected in variants from independent isolations. J Mol Biol. 1982 Jul 5;158(3):415–434. doi: 10.1016/0022-2836(82)90207-8. [DOI] [PubMed] [Google Scholar]
- Clements J. E. Hypothesis on the molecular basis of nononcogenic retroviral diseases. Rev Infect Dis. 1985 Jan-Feb;7(1):68–74. doi: 10.1093/clinids/7.1.68. [DOI] [PubMed] [Google Scholar]
- Clements J. E., Narayan O., Griffin D. E., Johnson R. T. The synthesis and structure of visna virus DNA. Virology. 1979 Mar;93(2):377–386. doi: 10.1016/0042-6822(79)90242-3. [DOI] [PubMed] [Google Scholar]
- Clements J. E., Pedersen F. S., Narayan O., Haseltine W. A. Genomic changes associated with antigenic variation of visna virus durig persistent infection. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4454–4458. doi: 10.1073/pnas.77.8.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galfrè G., Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 1981;73(Pt B):3–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- Gendelman H. E., Narayan O., Kennedy-Stoskopf S., Kennedy P. G., Ghotbi Z., Clements J. E., Stanley J., Pezeshkpour G. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J Virol. 1986 Apr;58(1):67–74. doi: 10.1128/jvi.58.1.67-74.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman H. E., Narayan O., Molineaux S., Clements J. E., Ghotbi Z. Slow, persistent replication of lentiviruses: role of tissue macrophages and macrophage precursors in bone marrow. Proc Natl Acad Sci U S A. 1985 Oct;82(20):7086–7090. doi: 10.1073/pnas.82.20.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda M. A., Wong-Staal F., Gallo R. C., Clements J. E., Narayan O., Gilden R. V. Sequence homology and morphologic similarity of HTLV-III and visna virus, a pathogenic lentivirus. Science. 1985 Jan 11;227(4683):173–177. doi: 10.1126/science.2981428. [DOI] [PubMed] [Google Scholar]
- Gudnadóttir M. Visna-maedi in sheep. Prog Med Virol. 1974;18(0):336–349. [PubMed] [Google Scholar]
- Haase A. T., Baringer J. R. The structural polypeptides of RNA slow viruses. Virology. 1974 Jan;57(1):238–250. doi: 10.1016/0042-6822(74)90124-x. [DOI] [PubMed] [Google Scholar]
- Haase A. T., Varmus H. E. Demonstration of a DNA provirus in the lytic growth of visna virus. Nat New Biol. 1973 Oct 24;245(147):237–239. doi: 10.1038/newbio245237a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lutley R., Pétursson G., Pálsson P. A., Georgsson G., Klein J., Nathanson N. Antigenic drift in visna: virus variation during long-term infection of Icelandic sheep. J Gen Virol. 1983 Jul;64(Pt 7):1433–1440. doi: 10.1099/0022-1317-64-7-1433. [DOI] [PubMed] [Google Scholar]
- McDougal J. S., Kennedy M. S., Sligh J. M., Cort S. P., Mawle A., Nicholson J. K. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986 Jan 24;231(4736):382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- Montelaro R. C., Parekh B., Orrego A., Issel C. J. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J Biol Chem. 1984 Aug 25;259(16):10539–10544. [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Cabradilla C. D., Benton C. V., Lasky L. A., Capon D. J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985 Feb 7;313(6002):450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- Narayan O., Clements J. E., Griffin D. E., Wolinsky J. S. Neutralizing antibody spectrum determines the antigenic profiles of emerging mutants of visna virus. Infect Immun. 1981 Jun;32(3):1045–1050. doi: 10.1128/iai.32.3.1045-1050.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan O., Griffin D. E., Chase J. Antigenic shift of visna virus in persistently infected sheep. Science. 1977 Jul 22;197(4301):376–378. doi: 10.1126/science.195339. [DOI] [PubMed] [Google Scholar]
- Narayan O., Griffin D. E., Clements J. E. Virus mutation during 'slow infection': temporal development and characterization of mutants of visna virus recovered from sheep. J Gen Virol. 1978 Nov;41(2):343–352. doi: 10.1099/0022-1317-41-2-343. [DOI] [PubMed] [Google Scholar]
- Narayan O., Wolinsky J. S., Clements J. E., Strandberg J. D., Griffin D. E., Cork L. C. Slow virus replication: the role of macrophages in the persistence and expression of visna viruses of sheep and goats. J Gen Virol. 1982 Apr;59(Pt 2):345–356. doi: 10.1099/0022-1317-59-2-345. [DOI] [PubMed] [Google Scholar]
- Peluso R., Haase A., Stowring L., Edwards M., Ventura P. A Trojan Horse mechanism for the spread of visna virus in monocytes. Virology. 1985 Nov;147(1):231–236. doi: 10.1016/0042-6822(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Pyper J. M., Clements J. E., Gonda M. A., Narayan O. Sequence homology between cloned caprine arthritis encephalitis virus and visna virus, two neurotropic lentiviruses. J Virol. 1986 May;58(2):665–670. doi: 10.1128/jvi.58.2.665-670.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyper J. M., Clements J. E., Molineaux S. M., Narayan O. Genetic variation among lentiviruses: homology between visna virus and caprine arthritis-encephalitis virus is confined to the 5' gag-pol region and a small portion of the env gene. J Virol. 1984 Sep;51(3):713–721. doi: 10.1128/jvi.51.3.713-721.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson A. B., Martin M. A. Molecular organization of the AIDS retrovirus. Cell. 1985 Mar;40(3):477–480. doi: 10.1016/0092-8674(85)90189-8. [DOI] [PubMed] [Google Scholar]
- SIGURDSSON B., PALSSON P., GRIMSSON H. Visna, a demyelinating transmissible disease of sheep. J Neuropathol Exp Neurol. 1957 Jul;16(3):389–403. doi: 10.1097/00005072-195707000-00010. [DOI] [PubMed] [Google Scholar]
- Scott J. V., Stowring L., Haase A. T., Narayan O., Vigne R. Antigenic variation in visna virus. Cell. 1979 Oct;18(2):321–327. doi: 10.1016/0092-8674(79)90051-5. [DOI] [PubMed] [Google Scholar]
- Shaw G. M., Hahn B. H., Arya S. K., Groopman J. E., Gallo R. C., Wong-Staal F. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science. 1984 Dec 7;226(4679):1165–1171. doi: 10.1126/science.6095449. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985 Aug;42(1):369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- Stanley J., Cooper S. J., Griffin D. E. Alphavirus neurovirulence: monoclonal antibodies discriminating wild-type from neuroadapted Sindbis virus. J Virol. 1985 Oct;56(1):110–119. doi: 10.1128/jvi.56.1.110-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R. M., Casey J. W., Rice N. R. Equine infectious anemia virus gag and pol genes: relatedness to visna and AIDS virus. Science. 1986 Feb 7;231(4738):589–594. doi: 10.1126/science.3003905. [DOI] [PubMed] [Google Scholar]
- Thormar H., Barshatzky M. R., Arnesen K., Kozlowski P. B. The emergence of antigenic variants is a rare event in long-term visna virus infection in vivo. J Gen Virol. 1983 Jul;64(Pt 7):1427–1432. doi: 10.1099/0022-1317-64-7-1427. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne R., Filippi P., Quérat G., Sauze N., Vitu C., Russo P., Delori P. Precursor polypeptides to structural proteins of visna virus. J Virol. 1982 Jun;42(3):1046–1056. doi: 10.1128/jvi.42.3.1046-1056.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxham M. N., Wolinsky J. S. Detailed immunologic analysis of the structural polypeptides of rubella virus using monoclonal antibodies. Virology. 1985 May;143(1):153–165. doi: 10.1016/0042-6822(85)90104-7. [DOI] [PubMed] [Google Scholar]
- Yaniv A., Dahlberg J. E., Tronick S. R., Chiu I. M., Aaronson S. A. Molecular cloning of integrated caprine arthritis-encephalitis virus. Virology. 1985 Sep;145(2):340–345. doi: 10.1016/0042-6822(85)90169-2. [DOI] [PubMed] [Google Scholar]