Abstract
The spindle pole body (SPB) is the major microtubule-organizing center of budding yeast and is the functional equivalent of the centrosome in higher eukaryotic cells. We used fast-frozen, freeze-substituted cells in conjunction with high-voltage electron tomography to study the fine structure of the SPB and the events of early spindle formation. Individual structures were imaged at 5–10 nm resolution in three dimensions, significantly better than can be achieved by serial section electron microscopy. The SPB is organized in distinct but coupled layers, two of which show ordered two-dimensional packing. The SPB central plaque is anchored in the nuclear envelope with hook-like structures. The minus ends of nuclear microtubules (MTs) are capped and are tethered to the SPB inner plaque, whereas the majority of MT plus ends show a distinct flaring. Unbudded cells containing a single SPB retain 16 MTs, enough to attach to each of the expected 16 chromosomes. Their median length is ∼150 nm. MTs growing from duplicated but not separated SPBs have a median length of ∼130 nm and interdigitate over the bridge that connects the SPBs. As a bipolar spindle is formed, the median MT length increases to ∼300 nm and then decreases to ∼30 nm in late anaphase. Three-dimensional models confirm that there is no conventional metaphase and that anaphase A occurs. These studies complement and extend what is known about the three-dimensional structure of the yeast mitotic spindle and further our understanding of the organization of the SPB in intact cells.
INTRODUCTION
Microtubule-organizing centers (MTOCs) nucleate and anchor microtubules (MTs) during cell differentiation and division (for review, see Brinkley, 1985; Rose et al., 1993; Kellogg et al., 1994; Pereira and Schiebel, 1997). Although MTOCs vary widely in structure, their functions are largely conserved. In metazoa, the principal MTOC is the centrosome, which organizes both the interphase MTs and those of the mitotic spindle. In the yeast Saccharomyces cerevisiae, this role is served by the spindle pole body (SPB), a complex and dynamic organelle that undergoes significant structural changes during the yeast cell cycle (for review, see Winey and Byers, 1993; Kilmartin, 1994; Snyder, 1994). Nuclear MTs, which form the meiotic or mitotic spindles, attach to an inner plaque of the SPB, whereas cytoplasmic MTs, important for nuclear movements and position, are attached to an outer plaque (Moens and Rapport, 1971; Byers and Goetsch, 1974, 1975; Rabinow and Marak, 1966). Recent studies of isolated SPBs by cryomicroscopy and electron tomography have shown that SPBs are organized from six major layers, including a central crystalline core that contains the SPC42 gene product (Bullitt et al., 1997).
Duplication of the SPB during the cell cycle begins with the formation of a “satellite” on the cytoplasmic face of the half-bridge, a specialized region of the nuclear envelope that lies immediately adjacent to the existing SPB. By processes not yet described in detail, the satellite develops into an SPB, which is then inserted into the nuclear envelope, again by a poorly defined mechanism. There it initiates new nuclear MTs and contributes to the process of spindle formation (Byers and Goetsch, 1974; Byers, 1981).
A combination of experimental approaches has improved our understanding of SPB components, and more recently localization studies using immunoelectron microscopy have mapped a number of these proteins to the SPB (Wigge et al., 1998). To date 27 proteins have been identified, and 18 have been localized to subregions of the spindle pole (Rout and Kilmartin, 1990; Spang et al., 1996; Sundberg et al., 1996; Wigge et al., 1998). A high percentage of these proteins have predicted coiled-coil domains, including Spc42p, which forms a central crystalline core (Donaldson and Kilmartin, 1996; Bullitt et al., 1997), and Spc110p, which acts as a molecular spacer between the central and inner plaques (Kilmartin et al., 1993; Kilmartin and Goh, 1996). The amino terminus of Spc110p in turn interacts with Spc98p (Geissler et al., 1996; Sundberg and Davis, 1997; Nguyen et al., 1998), which along with Spc97p (Knopp and Schiebel, 1997) and Tub4p (Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996) make up the γ-tubulin complex at the MT ends at both the inner and outer plaques. Spc72p and Cnm67p localize to the outer plaque of the SPB (Wigge et al., 1998). Although biochemical and genetic analyses of these SPB components are revealing how components of the SPB interact with one another, a detailed understanding of SPB fine structure and how its components are organized is still in its infancy.
Some of the requisite structural analyses have been carried out on isolated SPBs. As a result, however, interactions of the SPB with components in the intact cell have been disrupted, and assigning the SPB under study to a particular stage in the cell cycle has not been possible. It has also been difficult to understand the relationship between the SPB and the three-dimensional (3-D) geometry of forming mitotic spindles. Indeed, the forming yeast spindle has presented a formidable challenge, even to serial section microscopy of well-fixed cells, because MTs are growing from side-by-side SPBs in virtually all directions. These difficulties have motivated a structural study of SPBs in situ using high-voltage electron tomography.
In this paper we have used dual tilt-axis tomography of semithick sections to study the fine structure of the SPB and the events of early spindle formation. The increased resolution achieved by applying these methods to well-fixed material has allowed us to elucidate the 3-D fine structure of both SPBs and forming mitotic spindles. Our results provide insight into how kinetochore MTs are organized as the mitotic spindle is forming. They should also provide a benchmark for comparisons between wild-type and mutant strains.
MATERIALS AND METHODS
Yeast Strains Used
The yeast strains used were the haploid S288c (MATα). The diploid strain was DDY759 (MATa/MATα; cry1/cry1, ade2-1/ade2-1, his3-11,15/his3-11, 15, leu2-3, 112/leu2-3112, ura3-1/ura3-1, trp1-1,trp1-1, can1-100/can1-100). The cells were grown in YEPD at 30°C in a shaking water bath and harvested in midlog phase at a density of 5 × 106 cells/ml.
Preparation of Cells, Freezing, and Freeze-Substitution
The yeast were frozen using the method described previously (Winey et al., 1995). Briefly, aliquots from log-phase cultures were collected by vacuum filtration, the cell paste was transferred to sample holders, and the samples were frozen in a Balzers (BAL-TEC, Liechtenstein) HPM010 high-pressure freezer. The frozen cells were freeze-substituted in 1 or 3% glutaraldehyde and 0.1% uranyl acetate in acetone at −90°C for 3 d then low-temperature embedded in Lowicryl HM20 (Electron Microscopy Sciences, Fort Washington, PA). Cells embedded in Lowicryl HM20 contained SPBs and MTs that were well contrasted; thus the use of this resin greatly facilitated finding cells that contained single SPBs and developing spindles (McDonald, personal communication). Serial semithick (250- to 400-nm) sections were cut using a Reichart (Leica) Ultracut-E microtome and collected onto Formvar-coated copper slot grids. The sections were poststained with 2% uranyl acetate in 70% methanol and Reynold’s lead citrate. Fifteen-nanometer colloidal gold particles were affixed to both surfaces of the section to serve as fiducial markers for subsequent image alignment.
Microscopy and Tomographic Reconstruction
Sections were first imaged in a Philips CM10 TEM (Philips Electronic Instruments, Mahwah, NJ) operating at 100 kV to identify cells in various stages of SPB duplication and early spindle formation. Low-magnification overview maps were taken to facilitate locating these same cells in the high-voltage electron microscope (HVEM), where contrast is greatly reduced. For tomography, the grids were placed in a tilt–rotate specimen holder (model 650; Gatan, Pleasanton, CA) and imaged in a JEM-1000 HVEM (JEOL USA) operating at 1 MeV. Images were taken every 1.5° over a ±60° range on Agfa (Mortsel, Belgium) Scientia film. The grids were then rotated 90°, and a second tilt series was imaged. The film was developed in D19 (Eastman Kodak, Rochester, NY) diluted 1:1 in water for 8 min. The negatives were digitized using a cooled charge-coupled device camera (STAR-1; Photometrix, Tuscon, AZ) into a Silicon Graphics (Mountain View, CA) INDY computer at a pixel size of 2.3 nm. The tilted views were aligned using the positions of the gold particles, and tomograms were computed for each tilt axis using the R-weighted back-projection algorithm (Gilbert, 1972). The two tomograms were aligned to each other and combined using the methods described by Mastronarde (1997).
Image Analysis
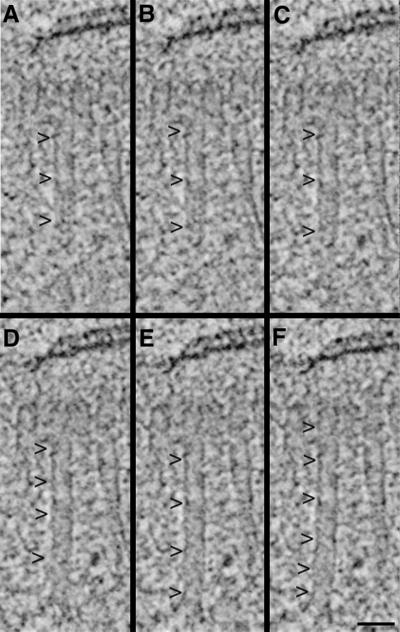
The tomograms were displayed and analyzed using the IMOD software package developed in our laboratory (Kremer et al., 1996). This program allows an operator to step through serial slices extracted from the tomogram and to track or model objects of interest in three dimensions. An “image slicer” window in IMOD was used to display a slice extracted from the 3-D image data in any position or orientation. This feature of the program was particularly useful for tracking oblique MTs. Figure 1A shows a 2.3-nm tomographic slice that contains an oblique MT (arrowheads). The orientation at which the slice was extracted was then rotated about an axis perpendicular to the MT until the entire fiber could be seen in a single view (Figure 1F, arrowheads; a total of 4° in this case; Figure 1, B–F). This method allowed unambiguous tracking of MTs in three dimensions to characterize the MTs of the forming mitotic spindle and enabled a clear view of MT end morphology.
Figure 1.
Rotating the orientation at which a slice of image data is extracted to view a MT along its length. A tomographic slice is first extracted perpendicular to the electron beam axis; here it contains an oblique MT (A, arrowheads). The orientation of the slice was then rotated a total of 4° about a horizontal axis to bring the image of the entire MT into one view (B–F, arrowheads). Note that the SPB, represented by dark, horizontal lines at the top of each panel, changes little during this process. Bar, 50 nm.
The slicer window was also used for analyzing the fine structure of the SPB itself. The orientation of a slice could be rotated to view a particular region of the SPB en face. Because the IMOD program displays the orientation of each slice by a line drawn in its main image window, it was easy to make a direct correlation between a tilted slice and a particular region of the SPB in its vertical orientation. Once features in the tomogram were modeled, a projection of the 3-D model was displayed and rotated to study its 3-D geometry. Quantitative results, such as the lengths of MTs or of individual SPB components, were extracted from the model data using a companion program, IMOD INFO. The angular packing of components of the SPB layers was examined using a neighbor density analysis, as described in McDonald et al., (1992).
A limitation of electron tomography with plastic sections is the collapse of the section that occurs in the electron beam, largely during the first seconds of irradiation before data collection has begun (Luther et al., 1988). Sections with a nominal thickness of 250, 300, or 400 nm were reconstructed. The resulting tomograms were significantly thinner than was indicated by either the microtome setting or the interference color of the section. We have found that comparable amounts of thinning occur with both Lowicryl HM20 and Epon-embedded samples (Mastronarde, 1997). Because extensive previous work has shown that this thinning is a collapse of the section’s thickness, not a loss of material from its surface (Luther et al., 1988; Braunfeld et al., 1994), we used the ratio of the microtome setting to the section’s measured thickness to calculate a thinning factor. This factor was used to correct the tomogram’s dimension along the beam axis (z-axis) to obtain accurate proportions for displaying models and for measuring length distributions.
RESULTS
Electron Tomography Reveals the Fine Structure of SPBs In Situ
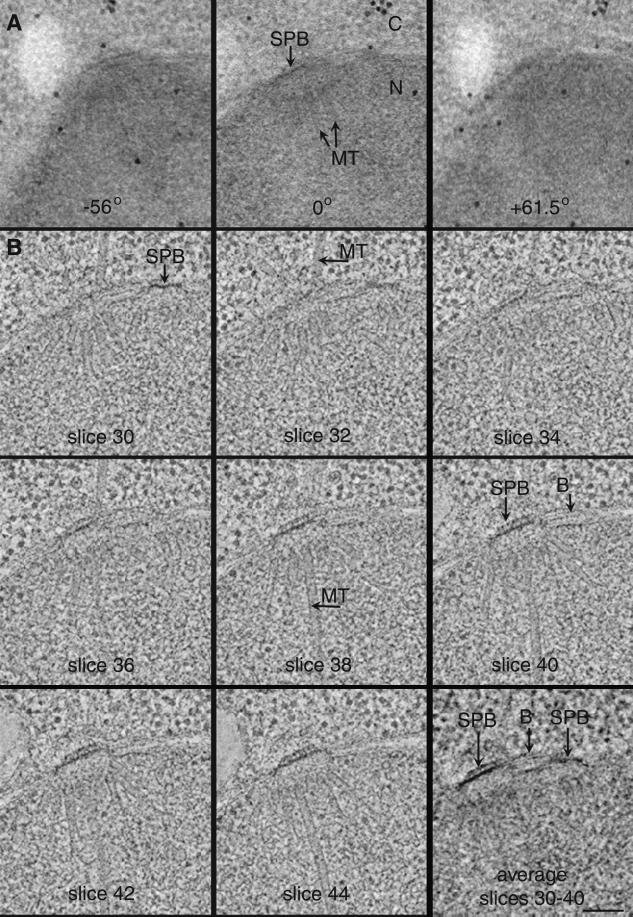
We have used dual tilt-axis HVEM tomography to obtain computed 3-D image data describing 3 diploid and 15 haploid SPBs. In this method, a thick section (250–400 nm) containing part of a fast-frozen, freeze-substituted yeast cell is imaged in the HVEM, and serial tilted views of appropriate areas are used as raw data from which to calculate a tomographic reconstruction. Figure 2A and Video Sequence 1 show images of a 250-nm-thick section with their tilt angles indicated. Little detail is apparent in the thick section, because so much cellular material is superimposed within the volume of the section at any given view. The nucleoplasm (N) is very dense and uniform, and the cytoplasm (C) is filled with ribosomes. The dark circles are gold particles on the section surfaces used for alignment of the tilted views. In some views (Figure 2A, 0°), MTs can be detected in the nucleoplasm. The central plaque of an SPB can be seen embedded in the nuclear envelope (Figure 2A, 0°), but its fine structure is not resolved. At higher tilts (Figure 2A, −56° and +61.5°) the apparent thickness of the section nearly doubles along the electron beam axis, making for even greater difficulty in detecting detail within the section. Video Sequence 1 shows a movie of an aligned tilt series containing 77 serially tilted views separated by 1.5°. The thickness of the section is evident when viewing the positions of gold particles, on the top and bottom surfaces of the section, which appear to move in opposite directions relative to each other. Although an MT array can be detected in the tilt series, a clear understanding of its 3-D fine structure is not possible.
Figure 2.
Tomographic reconstructions are calculated by back-projection of a series of tilted views of a single thick section. (A) 250-nm-thick section of a diploid cell. Shown here are views at 0° and at high tilt (−56° and +61.5°). Gold particles on the section’s surfaces were used to align the tilted views. N, nucleoplasm; C, cytoplasm. (B) 2.3-nm slices computed from the tomogram at different levels along the beam axis, as indicated. This cell contains duplicated SPBs connected by a bridge. Individual MTs, bridge (B), and SPBs are clearly resolved. Slices 30–40 were computationally averaged to produce an image that corresponds better to a standard 40-nm-thick section. Bar, 100 nm. Video Sequence 1 shows a movie of the complete, aligned tilt series displaying serial tilts collected every 1.5° from −56 to +61.5°. When viewing video sequences, select “play every frame” in your movie software. Video Sequence 2 shows a movie of 64 2.3-nm serial tomographic slices. Gold particles are present at the top and bottom surfaces of the section, and MTs are seen radiating in diverse orientations from the SPBs. For Video Sequence 3, modeling software was used to place model points along each nuclear MT originating from the SPBs (green and pink); cytoplasmic MTs too are marked (light blue).
A tomographic reconstruction of this same volume is shown in Figure 2B and Video Sequences 2 and 3. The relative positions of each 2.3-nm slice extracted from the volume of the tomographic reconstruction is indicated by the slice number. Now a tremendous amount of structural detail can be seen. In this diploid cell, two SPBs are found (slices 30 and 40); they are connected to each other by a bridge (B). Individual MTs can be clearly seen in the nucleus (slice 38) and cytoplasm (slice 32). A clear view of both SPBs in any one 2.3-nm tomographic slice was not obtained. It is interesting to note, however, that an ultrathin section (∼40 nm), cut using a microtome for standard electron microscopy, would contain all of the information in slices 30–40 in Figure 2B. This sort of image can be approximated by averaging slices 30–40 (Figure 2B), which shows two side-by-side SPBs connected by a bridge (Figure 2B; average slices 30–40). Video Sequence 2 is a movie of 64 computed serial 2.3-nm tomographic slices from the top surface of the original thick section, through the cell, to the bottom surface of the section. The surfaces of the section contain 15-nm gold particles, which appear as electron-dense spheres.
When one is stepping through the serial slices of the reconstructed volume, the nuclear MT array is evident. The trajectories of both nuclear and cytoplasmic MTs were modeled using software developed in our laboratory (Kremer et al., 1996). Video Sequence 3 is a movie of the 64 tomographic slices of Sequence 2 with model points superimposed to mark the nuclear MTs (green and pink) as well as cytoplasmic MTs (light blue). The high density of MTs present in this nucleus could not have been modeled with accuracy by standard serial section (∼50 nm) reconstruction methods (Winey et al., 1995), because the MTs emanate from the SPB in virtually all directions, making it impossible to track them all. Thus, tomographic reconstruction is a useful method with which to study the 3-D geometry of complex biological structures such as the forming spindles of yeast.
The Yeast SPB Is Organized from Distinct Layers
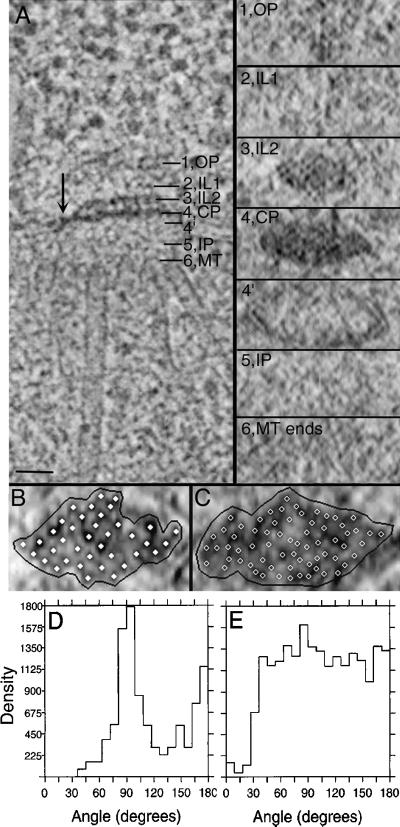
Fast-frozen and freeze-substituted cells show an excellent preservation of morphology and offer a unique opportunity to study the fine structure of SPBs in situ. The “vertical” architecture of the S. cerevisiae SPB (order along the spindle axis) is shown in Figure 3A, left. Similar to what has been reported for isolated, frozen-hydrated SPBs (Bullitt et al., 1997), the SPB in situ is organized from six major layers. Following the nomenclature of Bullitt et al. (1997), these layers are defined as follows: an outer plaque (OP; Figure 3A, 1), which connects to the cytoplasmic MTs; a first intermediate layer (IL1; Figure 3A, 2) and an electron-dense second intermediate layer (IL2; Figure 3A, 3); an electron-dense central plaque (CP; Figure 3A, 4), which is at the level of the nuclear envelope (Figure 3A, 4′) and is connected to it by hook-like structures (arrow); an ill-defined inner plaque (IP; Figure 3A, 5); and a layer of the inner plaque that contains capped nuclear MT ends (MT; Figure 3A, 6).
Figure 3.
The SPB is organized from six major layers. (A). 2.3-nm tomographic slice showing the vertical architecture of a diploid yeast SPB (left). These major layers include an outer plaque (OP), a first intermediate layer (IL1), a second intermediate layer (IL2), a central plaque (CP), an inner plaque (IP), and the nuclear MT ends (MT). The slicer tool in the IMOD software package was used to extract slices from the volume of image data at an orientation perpendicular to that shown on the left, sampling specific regions of the SPB at positions 1–6, as marked in the left panel. The corresponding slices through layers 1–6 are shown en face in the right panel. The outer plaque (1) connects to cytoplasmic MTs but does not appear electron dense in these preparations. Layer 3 may correspond to the Spc-42p–containing central crystal reported by Bullitt et al., (1997). The central plaque (4) is built from an array of electron-dense subunits and is anchored into the nuclear envelope with “hook-like” structures (4′ and A, arrow). Vertical striations can be seen spanning layers 3 and 4. The inner plaque (5) connects the nuclear MTs (6), but neither layer is structurally well defined. Bar, 50 nm. (B and C) IL2 (B) and CP (C) displayed with model points placed on subunits within these layers (solid and open circles). The boundary used for neighbor density analysis is shown in black. (D and E) Neighbor density analyses showing the distributions of angles between vectors that connect adjacent subunits for the IL2 layer (D) and CP (E). The IL2 layer shows a peak in its angular distribution of 90° (D). Although subunits are closely packed in the CP, no clear peak in the angular distribution was detected (E).
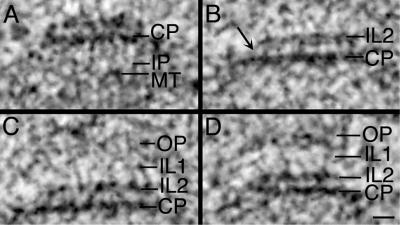
Figure 4.
SPB layers are physically coupled. (A) In this haploid SPB, vertical strands can be detected in the IP that connect the CP to the capped MT ends. (B) In this diploid SPB, connections between the CP and IL2 are seen (arrow); (C) IL1 often appears as a beaded layer connected to IL2. (D) Vertical connections between IL2 and IL1, as well as between IL1 and the OP are shown in another haploid SPB. Bar, 25 nm.
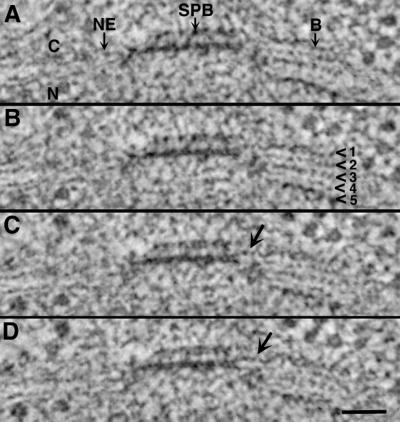
Figure 5.
Fine structure of the bridge from a diploid cell. Serial 2.3-nm tomographic slices resolve the double membrane of the nuclear envelope (NE). The bridge (B) consists of at least five layers (arrowheads), which appear particularly electron dense at the cytoplasmic (C) and nuclear (N) faces. Layers 2–5 appear to derive from the membranes of the NE, and layer 1 is added to the cytoplasmic surface of the NE; it connects directly to the SPB (C and D, arrows). Bar, 50 nm.
Figure 6.
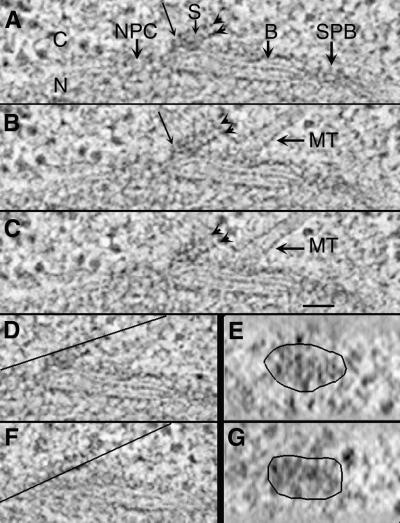
Serial 2.3-nm tomographic slices (A–C) through a satellite (S). An SPB, a bridge (B), an MT, and a nuclear pore complex (NPC) are indicated. Slices through each layer of the satellite (D and F, lines) were rotated and outlined to display en face views (E and G). The fine structure of the satellite is similar to that of the central region of the SPB (see Figure 2) with ordered layers and hooks (A, thin arrow). N, nucleus; C, cytoplasm. Bar, 50 nm.
Using the IMOD modeling software, one can slice a tomogram of the SPB perpendicular to the spindle axis (and perpendicular to the plane shown in Figure 3A, left) and then view it at any chosen level (Figure 3A, right). When seen in this en face view, both the CP and IL2 layers show an organized packing of subunits. The IL2 layer has been reported to contain the SPC42 gene product, organized as a hexagonally packed crystal (Bullitt et al., 1997).
The packing of IL2 subunits in our images is closer to square than hexagonal (Figure 3A, right), suggesting that section thinning before tomography has distorted the organization of this layer. We analyzed the packing of subunits within the IL2 layer in this en face view, using algorithms described elsewhere (McDonald et al., 1992). Model points were placed on the individual densities within this layer (Figure 3B) and on IL2 layers from six additional cells. Their angular distributions were computed and pooled in the graph shown in Figure 3D. The strong peak at 90° confirms the square packing in this region. The oval shape of the SPB en face view is also consistent with a section thinning of ∼40%. Studies are currently under way to examine profiles of SPBs sectioned en face to test this interpretation.
The CP layer of SPBs reconstructed by tomography in situ also exhibits a close packing of subunits, but its arrangement is distinct from that of IL2 (Figure 3C). Both the size and the shape of the unit cells are different. The neighbor density analysis of this region shows that the subunits are more closely spaced and that this region is more electron dense compared with the IL2 layer (Figure 3C). Although this layer shows a close packing of subunits, a strong peak in its angular distribution was not found (Figure 3E). Accurate description of the CP layer must await the discovery of specimens that are not distorted by section thinning.
A slice taken directly above the CP and rotated to view en face shows that the SPB is bounded at its periphery by the membrane of the nuclear envelope Figure 3A, right). The other layers (MT ends, IP, IL1, and OP) do not show ordered packing in these tomograms (Figure 3A, right).
The vertical architecture of the yeast SPB appears to be maintained through physical connections between its six major layers. Examples of vertical connections between SPB layers are shown in Figure 4 from two haploid cells (Figure 4, A and D) and a diploid cell (Figure 4, B and C). Often seen in the inner plaque are vertical strands bridging the CP to the capped ends of the nuclear MTs (Figure 4A, IP). Figure 4B shows vertical striations 10.8 ± 1.45 nm long that connect the CP with IL2. Although IL2 is slightly smaller than the CP, these two layers are connected diagonally at their ends (Figure 4B, arrow). Connections between the two intermediate layers, IL1 and IL2, can also be detected (Figure 4, C and D). In these preparations, the OP is not densely stained but is seen adjacent to IL1; in some views, vertical connections 10.12 ± 1.65 nm long could be detected between them (Figure 4D).
The Bridge Is Structurally Distinct from the Nuclear Envelope
Situated beside each SPB is a “half-bridge” or bridge depending on the cell’s stage in its division cycle. These structures are built as specializations of the nuclear envelope. The fine structure of a bridge is revealed in the serial 2.3-nm tomographic slices shown in Figure 5. In contrast to the double membrane of the nuclear envelope itself, seen where the continuous bilayers turn at the SPB edge (Figure 5A, NE), the bridge is made up of at least five separate layers (Figure 5B, arrowheads). Like the membrane bilayers that form the nuclear envelope, layer 2 is continuous with 5, and layer 3 is continuous with 4 (Figure 5B, arrowheads). Layer 1, which lies at the cytoplasmic face of the bridge, does not appear continuous with any of the other membrane bilayers. It is associated with a region between the central plaque and IL2 at the edge of the SPB (Figure 5, C and D, arrows). Bridge layers 1 and 5 are especially electron dense at the cytoplasmic and nuclear faces, respectively. Cytoplasmic MTs (two or three in number) were detected at bridge layer 1 in satellite-bearing cells and in cells with duplicated but not separated SPBs.
A Satellite Consists of Two Distinct Layers and Is Assembled on the Cytoplasmic Face of the Bridge
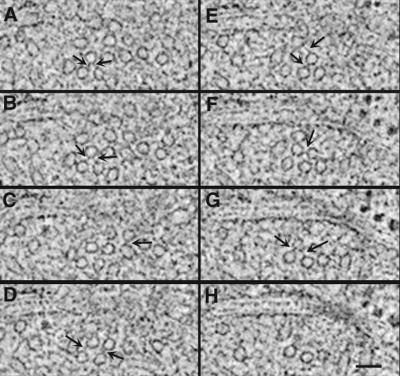
A satellite forms during G1 at the SPB-distal end of the bridge. This structure is thought to be the precursor to the daughter SPB; it is often appears as an electron-dense sphere in chemically fixed preparations (Byers and Goetsch, 1974; Byers, 1981) and in some cells prepared by our methods. In our preparations, we have also seen a layered structure at the distal end of the bridge. This geometry has been detected twice in thin section and once in thick section for tomography; it may represent a transient intermediate in SPB formation. Figure 6, A–C, shows serial 2.3-nm tomographic slices through the SPB and bridge (B) of a diploid cell containing such a structure. The satellite (S) is situated near one end of the cytoplasmic face of the bridge. In this view, the bridge is positioned between an SPB and a nuclear pore complex (NPC). Cytoplasmic MTs run approximately parallel to the satellite and appear to interact with an electron-dense layer of the bridge (Figure 6, B and C). This satellite consists of two distinct layers (arrowheads). The fine structures of these layers resemble those of the central plaque and of IL2 in the mature SPB shown in Figure 3. A close association of the satellite with the bridge is evident (Figure 6, A and B, arrows), suggesting a physical link between these structures. Slices through each of the two satellite layers (Figures 6, D and F, lines) were rotated to view their structural organization en face (Figure 6, E and G). Some evidence of ordered subunit packing is apparent. These images suggest that the satellite has a structural polarity and that components of the central plaque and IL2 are assembled in the cytoplasm before their insertion into the nuclear envelope.
Nuclear MT Ends Have Specialized Structures
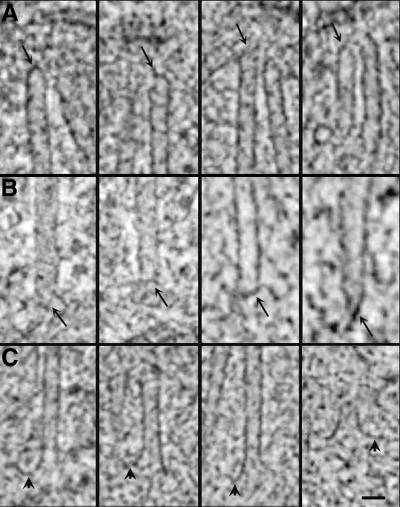
The morphology of MT ends was studied using the slicer window of IMOD to sample the data in planes that contained each MT axis (see Figure 1 and MATERIALS AND METHODS). MT minus ends, i.e., those proximal to the SPB, were capped (Figure 7, A and B, arrows). Closed ends on nuclear MTs have been described by others using isolated SPBs (Byers et al., 1978; Rout and Kilmartin, 1990; Bullitt et al., 1997), but here we can see that these capped ends are often tapered and have electron-dense material associated with them (Figure 7, A and B, arrows). In general, a pair of MTs was detected at the outer plaque, and these minus ends were also capped. In fact, the morphologies of MT minus ends at the inner and outer plaques were remarkably similar (Figure 7, A and B, respectively), given that there are ∼10 times more nuclear MTs than cytoplasmic MTs in haploid cells.
Figure 7.
MT end morphology. (A and B) Nuclear (A) and cytoplasmic (B) MT minus ends are capped and often appear pointed or tapered. Electron-dense material can be seen associated with MT ends and the inner plaque (arrows). (C) The majority of MTs (>70%) have a distinct flaring at their plus ends. These flared ends are present in cells from every cell cycle stage examined. Bar, 25 nm.
Nuclear MT plus ends, i.e., those distal to the SPB, were distinctly flared (Figure 7C). Nuclear MT ends whose plus ends could be clearly identified were extracted from two unbudded cells containing a single SPB (n = 35 MTs), two cells with duplicated but not separated SPBs (n = 43 MTs), three cells with bipolar spindles (n = 38 MTs), and a cell in late anaphase (12 MTs). The ends were then scored as flared, not flared, or ambiguous. In all stages examined, >70% of the MTs had a flared morphology. These flared plus ends were characteristically curved, showing an electron-dense extension off one side of the MT end (Figure 7C, arrowheads). Flared MT ends were also present in late anaphase cells, where the MTs are only slightly longer than they are wide (Figure 7C, right). The plus ends of cytoplasmic MTs were not examined, because these MTs tend to be quite long (Yeh et al., 1995) and were not present in the volume of the original thick section.
The 3-D Geometry of Early Mitotic Spindle Formation in S. cerevisiae
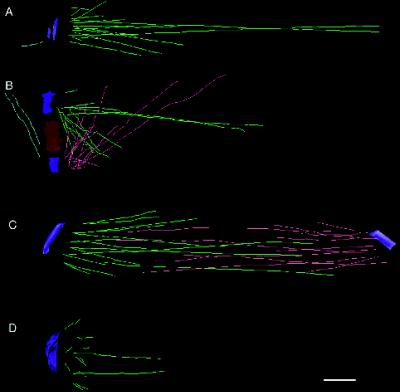
Figure 8 shows representative 3-D models from four haploid cells in various stages of spindle formation. Nuclear MTs are present in all stages of the cell cycle (Kilmartin and Adams, 1984). In unbudded cells containing a single SPB, 16 MTs can be seen radiating in many directions from the nuclear surface of the SPB (Figure 8A). After SPB duplication, while the SPBs are still connected by a bridge, the MTs from each SPB interdigitate at sharp angles (Figure 8B). After SPB separation, a bipolar spindle is formed (Figure 8C) containing MTs that form a core bundle, as well as numerous shorter MTs. A half-spindle from a cell in late anaphase (Figure 8D) contains numerous short MTs situated at the face of the SPB. The two longer MTs shown in Figure 8D are only fragments, because their plus ends were not in in the volume of the original thick section; probably they represent the MTs that formed the core bundle of the anaphase interzone (Winey et al., 1995). A common feature of the 3-D models is the presence of several MTs (two to four per SPB) that are long relative to a group of shorter MTs. The 3-D models also show no evidence for a conventional metaphase plate.
Figure 8.
Representative 3-D models from haploid cells. (A) Model from an unbudded cell containing a single SPB (blue) and 16 nuclear MTs (green). (B) Early spindle from a cell with duplicated SPBs (blue) connected by a bridge (red). Nuclear MTs (green and pink) interdigitate over the bridge. (C) Bipolar spindle that is 1 μm in length. (D) Half-spindle from a cell in late anaphase. In each stage, the spindles contain several long MTs and a group of shorter MTs of similar length. Bar, 0.1 μm.
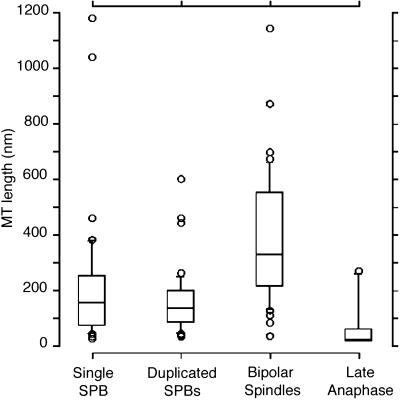
The lengths of MTs whose plus and minus ends could be identified within the volume of the section were measured, and their length distributions were pooled in the Tukey plots shown in Figure 9. Tukey plots are useful for the display of distributions in which there are several outliers relative to a group of similar lengths, like the MTs shown in Figure 8. In these plots, the outside borders of the box represent the 25th and 75th percentiles, and the middle line is the 50th percentile or median length. The top and bottom horizontal lines outside the box show the 90th and 10th percentiles, and the circles outside the box represent outliers to the distribution. MTs from unbudded cells containing a single SPB have a median length of ∼150 nm (n = 35). In cells with duplicated but unseparated SPBs, the median MT length is similar to that of cells containing single SPBs; median length is ∼130 nm (n = 43). As a bipolar spindle is formed, the median MT length increases to ∼300 nm (n = 38). The median MT length in cells during late anaphase decreases sharply to only ∼30 nm (n = 12) or just slightly longer than an MT’s width (also see Figure 7C, right). These very short MTs lie at the face of the SPB; they are invisible in standard thin sections (∼50–70 nm) and thus were not detected in our previous study of yeast spindle structure (Winey et al., 1995). Very short MTs lying at the SPB face confirm that anaphase A occurs in this organism.
Figure 9.
Tukey plots displaying MT length distributions from cells in various stages. The top and bottom of each rectangle represent the 75th and 25th percentiles, respectively; the center line of the rectangle marks the 50th percentile or median length. The top and bottom horizontal marks show the 90th and 10th percentiles, respectively. Circles represent outlying data points. MTs from unbudded cells with a single SPB present have a median length of ∼150 nm (n = 44). MTs from cells with duplicated but unseparated SPBs have a median length of ∼120 nm (n = 57). As a bipolar spindle is formed, the median MT length increases to ∼300 nm (n = 68). The median MT length in cells during late anaphase decreases dramatically to only ∼30 nm (n = 13).
MTs Growing from Duplicated but Unseparated SPBs Interdigitate over the Bridge
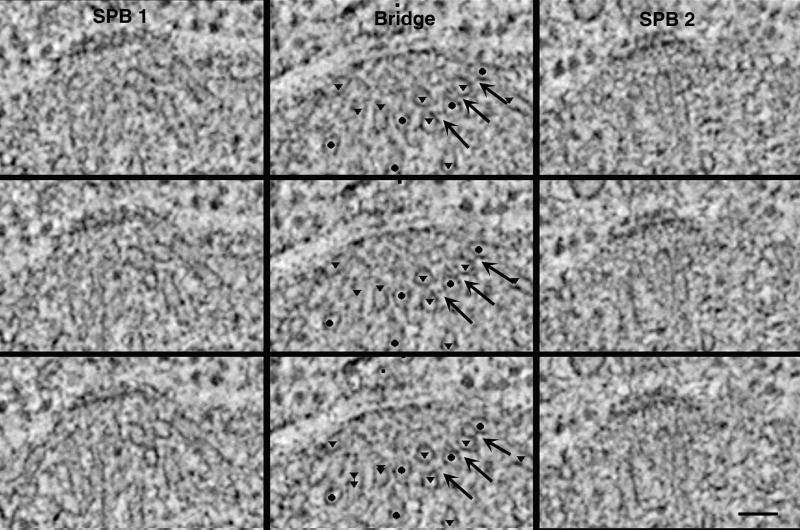
Serial 2.3-nm tomographic slices from the cell in Figure 8B show that MTs from opposite poles interdigitate over the bridge (Figure 10, Video Sequences 4 and 5). Three serial 2.3-nm cross-sections through SPB1, the bridge, and then SPB2 (Figure 10, top to bottom) show that MTs emanate from the poles at oblique angles. The positions of the SPBs are represented in the video by blue lines, and the position of the bridge is represented by red lines. The MTs originating from the two SPBs are marked by triangles (green in the video) and circles (pink in the video). MT arrays can be seen originating from either SPB at oblique angles and then crossing and interdigitating over the bridge. The complete 3-D model from the cell in Figure 10 and displayed in Figure 8B is rotated 360° about the y-axis in Video Sequence 5, illustrating the 3-D geometry of the MT arrays emanting from either SPB. MTs whose plus ends could be identified within the volume of the original thick section are marked with yellow circles; in the rotating model it is apparent that MT ends from opposite poles do not pair. Cross-bridges between MTs from opposite poles can be detected over the bridge (Figure 10, arrows). Connections between MTs from opposite poles are 11.18 ± 3.20 (n = 38) in length. Shown in Video Sequence 4 is a movie of 64 computed 2.3-nm tomographic slices through SPB1, the bridge, and SPB2. Cross-bridges between MTs from the same pole can be detected in the region over the bridge but close to the SPB (Figure 11, arrows). They have an average length of 11.28 ± 2.52 (n = 44). Such images suggest that the 3-D geometry of the early spindles may be partly due to the presence of physical linkages between MTs.
Figure 10.
Three adjacent tomographic slices through one SPB, the bridge, and the second SPB from the cell in Figure 8B. MT cross-sections are prevalent over the bridge; circles mark MTs that originated from SPB1, and triangles mark MTs from SPB2. Cross-bridges between MTs from opposite SPBs are seen (arrows). Bar, 50 nm. Video Sequence 4 is a movie of 64 computed 2.3-nm tomographic slices through a cell containing duplicated but not separated SPBs (blue lines) connected by a bridge (red lines) from the cell shown in Figures 8B and 10. The MTs originating from each SPB are marked by green triangles and pink circles, respectively. Video Sequence 5 is a rotating 3-D model from the cell shown in Video Sequence 4. MT arrays from each SPB, represented by green and pink lines, interdigitate over the bridge at sharp angles. MTs whose plus ends could be identified within the volume of the section are marked with yellow circles. In this forming mitotic spindle, MTs from opposite poles do not pair.
Figure 11.
Adjacent serial tomographic slices through a haploid cell show cross-bridges (arrows) between MTs originating from the same SPB. Bar, 50 nm.
DISCUSSION
Tomography is a useful method for looking at complex 3-D structures such as the centrosome (Moritz et al., 1995), the Golgi apparatus (Ladinsky et al., 1994, 1999), kinetochores (McEwen et al., 1993), and mitochondria (Mannella, 1997; Perkins et al., 1997). The technique enables one to examine 3-D image data at a resolution better than that obtained by serial sections and to slice through one organelle in many orientations, permitting a better understanding of the geometry of a cellular subsystem. Our study has used tomography to examine the 3-D architecture of both the SPB and the forming mitotic spindle of budding yeast.
We confirm a layered architecture for the SPB, as described in the literature based both on sectioned material and on tomography of isolated, frozen-hydrated SPBs. Our methods, which allow resampling of the same material with slices cut in several different orientations, reveal order in two layers of the SPB. Connections are seen between all SPB layers; these probably help define rigorous order in the SPB along the spindle axis. We also detect hook-like connections between the SPB and the nuclear envelope that surrounds its perimeter. Our work identifies the fine structure of the “bridge” that forms beside the SPB as a specialization of the nuclear envelope. The satellite and the new SPB that form at the distal end of this bridge also contain ordered layers that resemble the order at the SPB core. Tomographic slices of SPBs at different stages of the cell cycle have allowed us to characterize the nuclear MT arrays that grow from first the old SPB and then the new one, as it enters the nuclear envelope. MTs in these preparations have a structural polarity; their SPB-proximal ends are capped, whereas the majority of MT ends projecting into the nucleus (presumably the plus ends) are distinctly flared. We see no pairing between the distal ends of nuclear MTs growing from side-by-side SPBs, suggesting that sister kinetochores are not both attached to MTs at this early stage. The 3-D geometries of early spindles are described for the first time, and the changes in spindle MT structure that accompany mitosis are defined, including strong evidence for an anaphase shortening of kinetochore-associated MTs.
Structural Organization of the SPB In Situ
Our study complements and extends data from isolated SPBs in several important ways and has provided a context for the structural organization of the SPB in situ. Tomographic reconstruction has been used previously to study the fine structure of isolated, heparin-stripped SPBs, a preparation that leads to the loss of the inner plaque, the outer plaque, and the first intermediate layer (Bullitt et al., 1997). Both the data presented in this study and those by Bullitt et al. (1997) support the assignment that IL2 is made largely of Spc42p. However, order in the CP, which is evident in our tomograms but was lost in heparin-stripped samples, suggests that heparin removed a component that is essential for ordered packing in this layer. Our study shows that the CP order is different from that seen in IL2, so proteins in addition to Spc42p are to be expected. Indeed, biochemical and genetic studies have shown that the carboxyl terminus of Spc110p along with calmodulin is present at the central plaque (Sundberg et al., 1996) in a complex with Spc42p and a 35-kDa protein (Knopp and Schiebel, 1997). The amino terminus of Spc110p links the central plaque to the inner plaque through an interaction with Spc98p, a component of the yeast γ-tubulin complex (Geissler et al., 1996; Sundberg and Davis, 1997; Nguyen et al., 1998). Because of the physical interactions of Spc110p with both the central and inner plaques, the absence of the inner plaque in the heparin-stripped cores could have led to a disruption of the organization of proteins in this region. Alternatively, the hook-like structures present in our images and lost during isolation of SPB cores may also be important in stabilizing order in this region.
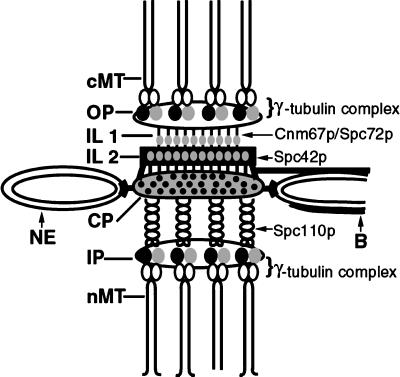
The structural data presented in this study, together with recent genetic and biochemical analyses, permit a tentative identification of several SPB substructures (Figure 12). Our images provide evidence for vertical connections between the different layers of the SPB. The central coiled-coil domain of Spc110p appears to act as a vertical linker between the central and inner plaques, and studies have shown that truncations in this region cause a shortening of the distance between the central and inner plaques (Kilmartin et al., 1993). The vertical strands observed in our tomograms between the central plaque and the nuclear MT ends at the SPB inner plaque are likely the central coiled-coil domain of Spc110p that bridges these two layers (Figure 12).
Figure 12.
Summary model of the putative locations of components that may serve a structural role in maintaining the vertical architecture of the yeast SPB (also see Figure 3).
The morphology of MT minus ends at the inner and outer plaques was remarkably similar, given that the density of MTs at the inner plaque is 10 times higher, suggesting that the closed, tapered morphology is not due to a close packing of MTs. Biochemical analyses and immunoelectron microscopy have shown that the γ-tubulin–like protein Tub4p forms a complex with Spc98p and Spc97p, and the resulting γ-tubulin complex localizes to the inner and outer SPB plaques (Rout and Kilmartin, 1990; Spang et al., 1996). It is likely that the electron-dense material forming the caps on the minus ends of MTs at the inner and outer plaques comprises, in part, this γ-tubulin complex (Figure 12).
Recent studies have shown that mutations in CNM67 and SPC72 genes cause structural defects in or even a complete absence of the outer plaque, suggesting that these gene products may serve a role in maintaining connections between the outer plaque and the rest of the SPB (Brachat et al., 1998; Chen et al., 1998; Knopp and Schiebel, 1998). Antibodies to both Spc72p and Cnm67p localize to the region of the SPB, where we have detected vertical strands connecting the outer plaque to the IL1 and the IL1 to IL2 (Figure 12; Wigge et al., 1998). It seems likely that the connections observed in our tomograms are, in part, made of these proteins.
Our structural data support the hypothesis proposed by Bullitt et al. (1997) that the central crystalline layer (IL2) may provide a scaffold to which other proteins are added, providing a mechanism to allow changes in the horizontal dimension that occur between the haploid and diploid states. The differences in the order of IL2 and the central plaque described in this study may provide a vernier that helps to specify the area of the SPB in the plane of the nuclear envelope. Alternatively, the hook-like structures that appear to anchor the central plaque to the nuclear envelope may not only tether the SPB to its cellular location but also provide a means of capping the growth of the central crystalline core.
How does the central crystal form? Our data show that the satellite, or perhaps a very early stage in the formation of a daughter SPB from the satellite, consists of distinct layers that are similar in structure to the central plaque and IL2 of mature SPBs. The cytoplasmic-facing layer of the satellite and the IL2 of the mature SPB have similar organizations of subunits. It is therefore possible that some components of the central crystalline core of the central plaque and IL2 are assembled in the cytoplasm before insertion into the nuclear envelope. However, the central plaque of the mature SPB appears more electron dense than the two-layered satellite structure, so other components of the central plaque may be added after its insertion into the nuclear envelope.
Structural Organization of Forming Mitotic Spindles
Our previous work on spindle structure in budding yeast used reconstructions from serial thin sections to document the 3-D geometry of bipolar spindles, but we were not able to track the MTs growing from side-by-side SPBs (Winey et al., 1995). The tomographic reconstruction method used here, however, allows one to section the same volume in multiple orientations, which has let us characterize the 3-D geometry of nuclear MT arrays in interphase cells and in forming mitotic spindles, including MT arrays from putative old and new SPBs.
Nuclear MTs are present in S. cerevisiae throughout the cell cycle. Our models show that cells with one SPB contain enough MTs to connect to each of the 16 chromosomes. Late anaphase cells, too, contain about this number of short or very short MTs. It is therefore possible that nuclear MTs from a mother SPB remain attached to kinetochores throughout the cell cycle.
A feature common to all of these models is the presence of a few long MTs and a group of short MTs of similar length. The long MTs may make up the “core bundle” of MTs in bipolar spindles, which probably participates in spindle elongation during anaphase B (Winey et al., 1995). Alternatively, the longer MTs may hold certain kinetochores at an uncommonly great distance from the SPB throughout the cell cycle.
In cells containing duplicated but not separated SPBs, equal numbers of MTs arise from the two SPBs and interdigitate over the bridge. Cross-bridges were seen between MTs arising from the same SPB and between MTs from opposite SPBs. Although the dimensions of these structures were similar, their positions relative to the SPB were different. Cross-bridges between parallel MTs were prevalent close to the SPB, whereas cross-bridges between MTs from opposite poles were more common in the middle of the spindle, over the bridge itself. The presence of these structures suggests that they may have a structural role in maintaining the 3-D geometry of the spindle MTs as they interdigitate at sharp angles over the bridge. Alternatively, the bridging molecules detected between MTs from opposite poles may serve a role during SPB separation. For example, the mitotic kinesins Cin8p and Kip1p contribute to SPB separation and spindle elongation (Hoyt et al., 1992; Roof et al., 1992). Although their location at the ultrastructural level is not yet known, they may correspond to the cross-bridges we have observed. Future localization studies on well-fixed material will be needed to examine this possibility.
The flared morphology of MT plus ends detected in the tomograms from all cells examined suggests that the majority of MTs may be in a state of dynamic activity. Indeed, open-ended MTs have been detected in yeast using other preparative techniques (Byers et al., 1978; Bullitt et al., 1997). Flared MT ends are indicative of depolymerizing MTs (Mandelkow et al., 1991; Chretien et al., 1995) in which protofilaments appear to “peel off,” forming a flared morphology at their ends. Dynamic activity of spindle MTs has been documented at the light microscopic level using GFP-tubulin (Straight et al., 1997), in which centromeres appeared to oscillate along spindles ∼2.5 μm in length. Many of the flared MTs shown in this study were from MTs <200 nm in length and thus below the limit of resolution of the light microscope.
The GFP studies of living yeast cells suggest that kinetochores must remain attached to MTs during this dynamic activity. Kinesin-related proteins have been implicated in tethering kinetochores to disassembling MTs in other systems. For example, the centromere-associated protein CENP-E has been localized to the kinetochore corona (Yen et al., 1992; Yao et al., 1997); it is important in maintaining connection of kinetochores to dynamic MTs in Xenopus extracts (Wood et al., 1997) and in more purified systems (Lombillo et al., 1995). A detachment of kinetochores from the MT would activate the spindle checkpoint (Rudner and Murray, 1996; Straight, 1997), and although it is likely that kinetochores remain attached to depolymerizing MTs via linker proteins, such a model in yeast cells is yet to be established.
It was not possible to identify kinetochores in our images, because there was no discernible structure at the plus ends of MTs in the tomographic slices. However, because there are 16 chromosomes in a haploid S. cerevisiae, spindles probably contain a single MT connected to each of the chromosomes plus a few extra nonkinetochore MTs to form the core bundle (Peterson and Ris, 1976; Winey et al., 1995). Thus, the majority of spindle MTs modeled are likely kinetochore MTs.
A conventional metaphase plate has not been detected either in our models or in studies using living cells (Straight et al., 1997) or using fluorescence in situ hybridization (Guacci et al. 1997). In addition, our models did not show any pairing of MTs originating from opposite poles, as would be expected if sister kinetochores were attached at this stage. The 3-D geometry of the spindle at this early stage and the cross-bridges between MTs may be important for pole separation but not necessarily for centromere separation. Indeed it has been shown that some centromeres do not separate until a bipolar spindle of 2.5 μm in length is formed (Straight et al., 1997).
Although S. cerevisiae spindle behavior does not follow classic eukaryote models, a similar mitotic process has been described for the oomycete fungus Saprolegnia ferax (Heath and Rethoret, 1981). In S. ferax, kinetochores remain attached to nuclear MTs throughout the cell cycle. Cytoplasmic centrioles with associated centrosomal material duplicate at the end of G1, and the kinetochore MTs become distributed equally between the side-by-side centrosomes. The kinetochores replicate during S phase, during which time the MT number does not increase. It is only when a bipolor spindle is formed that MT number increases and attachments to sister kinetochores are made, although a conventional metaphase plate never forms. Anaphase then proceeds with separation of chromosomes and elongation of the spindle. As in S. ferax, nuclear MTs are present in all stages of the S. cerevisiae cell cycle, and anaphase separation of chromosomes proceeds in the absence of a conventional metaphase plate. However, because yeast chromosomes do not condense to form a morphologically discernible structure, it is not yet known whether the MTs present in interphase nuclei have kinetochores attached.
In conclusion, tomographic reconstruction has provided new structural insights into the MTOC of budding yeast and novel information on how early mitotic spindles are arranged. We plan to apply this method to strains containing mutations in genes that encode key SPB components to identify structure–function relationships in this organism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kent McDonald for the preparation of the diploid strain and for advice on freeze–substitution, Dave Drubin for providing the diploid strain, Mary Morphew for preparing the haploid strain, David Mastronarde for training and help with computer analysis, and Andrew Staehelin for the use of the high-pressure freezer. This work was supported by National Institutes of Health grant RR-00592 (to J.R.M.) and National Science Foundation grant MBC-09357033 (to M.W.).
Abbreviations used:
- CP
central plaque
- HVEM
high-voltage electron microscope
- IL
intermediate layer
- IP
inner plaque
- MT
microtubule
- MTOC
microtubule-organizing center
- OP
outer plaque
- SPB
spindle pole body
- 3-D
three-dimensional
Footnotes
REFERENCES
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunfeld MB, Koster AJ, Sedat JW, Agard DA. Cryo automated tomography: toward high-resolution reconstructions of plastic-embedded structures. J Microsc. 1994;174:75–84. doi: 10.1111/j.1365-2818.1994.tb03451.x. [DOI] [PubMed] [Google Scholar]
- Brinkley BR. Microtubule organizing centers. Annu Rev Cell Biol. 1985;1:145–172. doi: 10.1146/annurev.cb.01.110185.001045. [DOI] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Byers B. In: Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In: Molecular Genetics in Yeast. Alfred Benson Symposium. von Wettstein D, Friis D, J, Kielland-Brandt M, Stenderup A, editors. Copenhagen: Munksgaard; 1981. pp. 119–131. [Google Scholar]
- Byers B, Goetsch L. Duplication of the spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of the spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC. The yeast spindle pole component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component SPC98p interacts with the γ tubulin Tub4p of Saccharomyces cerevisiae at sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gilbert PFC. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. II. Direct methods. Proc R Soc Lond B Biol Sci. 1972;182:89–102. doi: 10.1098/rspb.1972.0068. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;6:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath IB, Rethoret K. Nuclear cycle of Saprolegnia ferax. J Cell Sci. 1981;49:353–367. doi: 10.1242/jcs.49.1.353. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular-organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. Genetic and biochemical approaches to spindle function and chromosome segregation in eukaryotic microogansisms. Curr Opin Cell Biol. 1994;6:50–54. doi: 10.1016/0955-0674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiae spindle pole body whose transcript is cell cycle-regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Goh PY. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knopp M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopp M, Schiebel E. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh JR, Howell KE. HVEM tomography of the trans-golgi network: structural insights and identification of a lace-like vesicle coat. J Cell Biol. 1994;127:29–38. doi: 10.1083/jcb.127.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK, Lawrence MC, Crowther RA. A method for monitoring the collapse of plastic sections as a function of electron dose. Ultramicroscopy. 1988;24:7–18. doi: 10.1016/0304-3991(88)90322-1. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella CA. Reconsidering mitochondrial structure: new views of an old organelle. Trends Biochem Sci. 1997;22:37–38. doi: 10.1016/s0968-0004(96)30050-9. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in PtK cells. J Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Arena JT, Frank J, Rieder CL. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J Cell Biol. 1993;120:301–312. doi: 10.1083/jcb.120.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae. J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Vinh DBN, Crawford DK, Davis TN. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Centrosome-microtubule nucleation. J Cell Sci. 1997;110:295–300. doi: 10.1242/jcs.110.3.295. [DOI] [PubMed] [Google Scholar]
- Perkins G, Renken C, Martone ME, Young SJ, Ellisman M, Frey T. Electron tomography of neuronal mitochondria: three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Rabinow CF, Marak J. A fiber apparatus in the nucleus of the yeast cell. J Cell Biol. 1966;29:129–151. doi: 10.1083/jcb.29.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose M. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Biggins S, Satterwhite LL. Unravelling the tangled web at the microtubule-organizing center. Curr Opin Cell Biol. 1993;5:105–115. doi: 10.1016/s0955-0674(05)80015-8. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8:773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- Snyder M. The spindle pole body of yeast. Chromosoma. 1994;103:369–380. doi: 10.1007/BF00362281. [DOI] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:429–441. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle function. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF. Cell cycle: checkpoint proteins and kinetochores. Curr Biol. 1997;7:13–16. doi: 10.1016/s0960-9822(06)00315-0. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sundberg HA, Davis TN. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2575–2590. doi: 10.1091/mbc.8.12.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle body components. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DM, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LSB, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.