Abstract
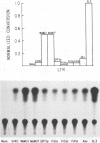
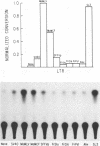
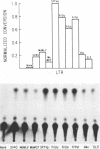
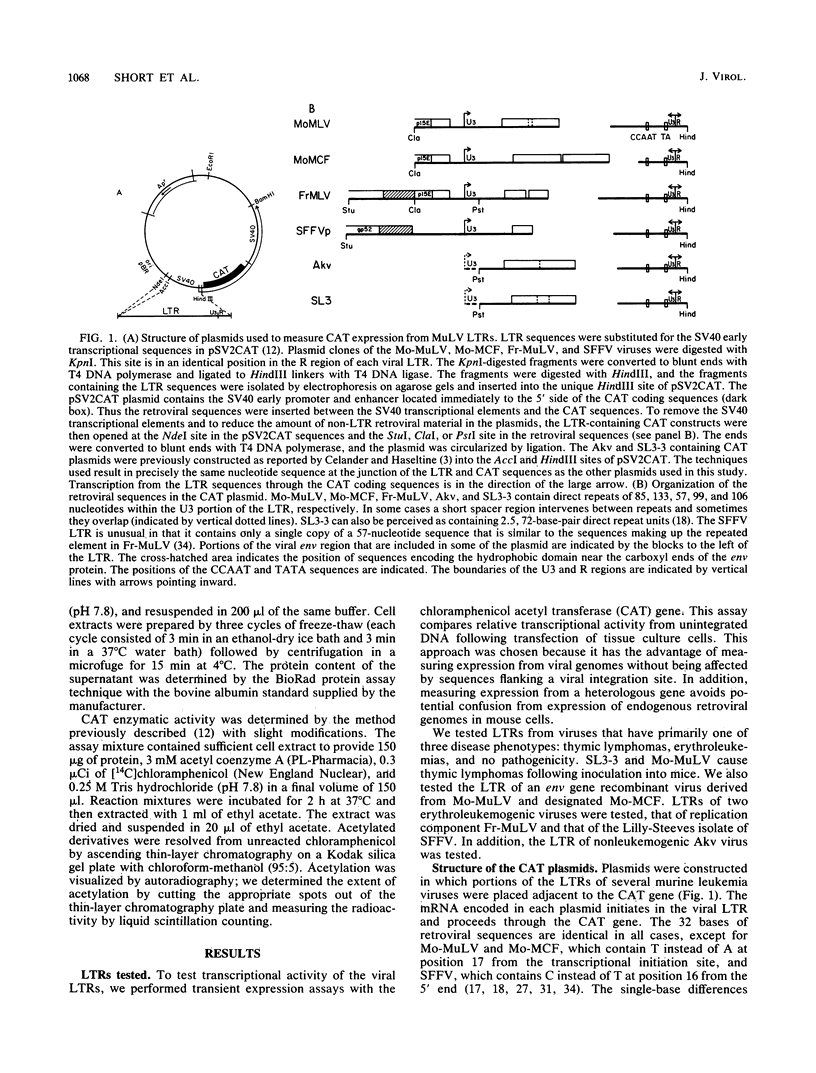
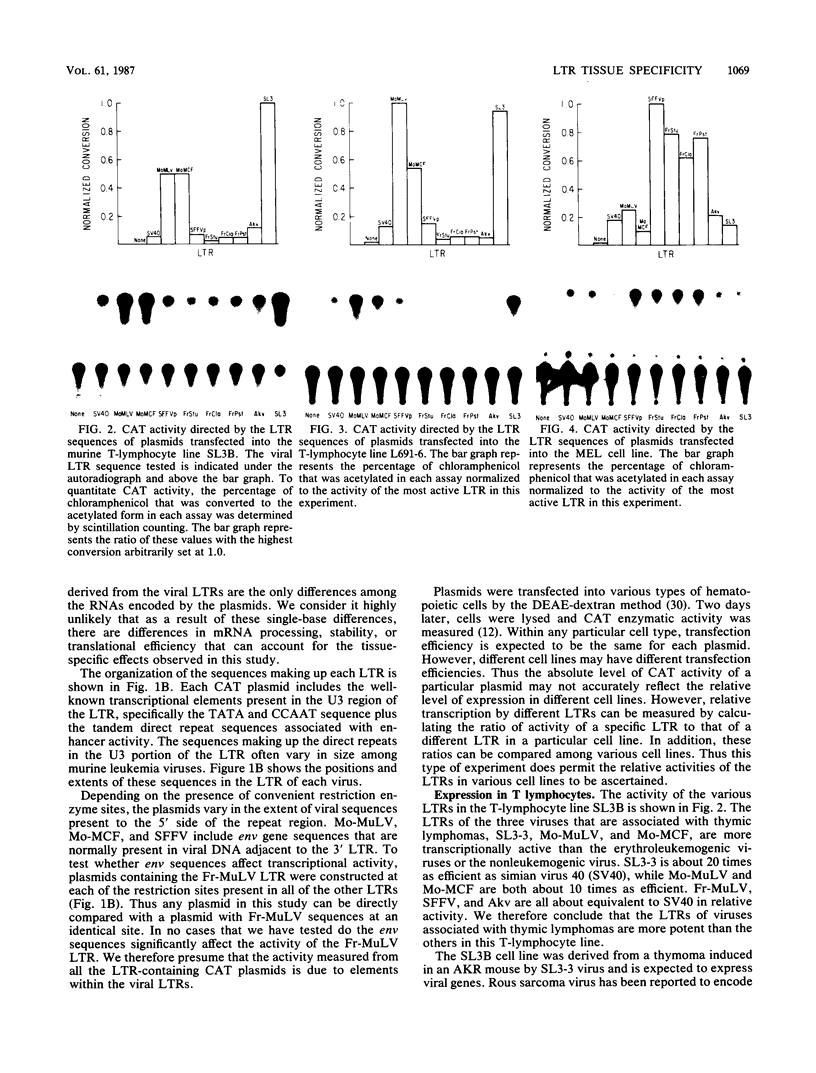
Recombination studies have established that retroviral long terminal repeats (LTRs) are important genetic determinants of the viral capacity to induce hematopoietic tumors and to specify the type of cell making up the tumor. Plasmids containing LTRs of several murine leukemia viruses linked to the chloramphenicol acetyltransferase gene were tested in transient assays to measure relative rates of transcriptional activity in different types of hematopoietic cells. LTRs of the thymomagenic viruses SL3-3, Moloney leukemia virus, and a Moloney mink cell focus-forming virus all expressed to higher levels than other LTRs in T-lymphocyte cell lines. Conversely, the LTRs of Friend leukemia virus and a polycythemic spleen focus-forming virus expressed to higher levels than other LTRs in erythroleukemia cells. The LTR of nonleukemogenic Akv virus induced a relatively low level of activity compared with the others in all cells tested. Thus the relative level of LTR-driven expression in various types of cells corresponds to the type of tumor caused by the intact virus in vivo. These results provide direct evidence that the tissue specificity of the transcriptional activity of LTRs plays a critical role in determining the target cell for retroviral oncogenesis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broome S., Gilbert W. Rous sarcoma virus encodes a transcriptional activator. Cell. 1985 Mar;40(3):537–546. doi: 10.1016/0092-8674(85)90202-8. [DOI] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Dandolo L., Blangy D., Kamen R. Regulation of polyoma virus transcription in murine embryonal carcinoma cells. J Virol. 1983 Jul;47(1):55–64. doi: 10.1128/jvi.47.1.55-64.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Villemur R., Jolicoeur P. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3' end of env. J Virol. 1983 Jul;47(1):24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays E. F., Margaretten N., Swanson S. K. Spontaneous leukemia viruses: lymphomagenic ecotropic viruses of AKR mice. J Natl Cancer Inst. 1982 Nov;69(5):1077–1082. [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985 Feb;141(1):30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Janowski M., Merregaert J., Boniver J., Maisin J. R. Proviral genome of radiation leukemia virus: molecular cloning of biologically active proviral DNA and nucleotide sequence of its long terminal repeat. J Virol. 1985 Jul;55(1):251–255. doi: 10.1128/jvi.55.1.251-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney S., Natarajan V., Strike D., Khoury G., Salzman N. P. JC virus enhancer-promoter active in human brain cells. Science. 1984 Dec 14;226(4680):1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Linemeyer D. L., Ruscetti S. K., Menke J. G., Scolnick E. M. Recovery of biologically active spleen focus-forming virus from molecularly cloned spleen focus-forming virus-pBR322 circular DNA by cotransfection with infectious type C retroviral DNA. J Virol. 1980 Sep;35(3):710–721. doi: 10.1128/jvi.35.3.710-721.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E., Davis B., Overhauser J., Chao E., Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. 1984 Mar 29-Apr 4Nature. 308(5958):470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- McGrath M. S., Pillemer E., Kooistra D., Weissman I. L. The role of MuLV receptors on T-lymphoma cells in lymphoma cell proliferation. Contemp Top Immunobiol. 1980;11:157–184. doi: 10.1007/978-1-4684-3701-0_5. [DOI] [PubMed] [Google Scholar]
- Oliff A. I., Hager G. L., Chang E. H., Scolnick E. M., Chan H. W., Lowy D. R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980 Jan;33(1):475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985 Jul;4(7):1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. E., Fredrickson T. N. Susceptibility to Friend helper virus leukemias in CXB recombinant inbred mice. J Exp Med. 1983 Nov 1;158(5):1693–1702. doi: 10.1084/jem.158.5.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford J., Queen C. Cell-type specific expression of a transfected immunoglobulin gene. Nature. 1983 Nov 3;306(5938):77–79. doi: 10.1038/306077a0. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R., Rassart E., DesGroseillers L., Jolicoeur P. Molecular cloning of viral DNA from leukemogenic Gross passage A murine leukemia virus and nucleotide sequence of its long terminal repeat. J Virol. 1983 Feb;45(2):539–546. doi: 10.1128/jvi.45.2.539-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Swift S., Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985 Jul;55(1):184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff L., Kaminchik J., Hankins W. D., Ruscetti S. K. Sequence comparisons of the anemia- and polycythemia-inducing strains of Friend spleen focus-forming virus. J Virol. 1985 Feb;53(2):570–578. doi: 10.1128/jvi.53.2.570-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]